Abstract
The protein factor U2 snRNP Auxiliary Factor (U2AF) 65 is an essential component required for splicing and involved in the coupling of splicing and 3′ end processing of vertebrate pre-mRNAs. Here we have addressed the mechanisms by which U2AF 65 stimulates pre-mRNA 3′ end processing. We identify an arginine/serine-rich region of U2AF 65 that mediates an interaction with an RS-like alternating charge domain of the 59 kDa subunit of the human cleavage factor I (CF Im), an essential 3′ processing factor that functions at an early step in the recognition of the 3′ end processing signal. Tethered functional analysis shows that the U2AF 65/CF Im 59 interaction stimulates in vitro 3′ end cleavage and polyadenylation. These results therefore uncover a direct role of the U2AF 65/CF Im 59 interaction in the functional coordination of splicing and 3′ end processing.
Introduction
Prior to translation to protein, most eukaryotic mRNAs follow a complex program that consists of various co- and post-transcriptional processing steps. This includes the formation of the 5′ cap structure, the splicing of introns, processing of the 3′ end and transport to the cytoplasm. Several studies have shown that these different steps of gene expression do not proceed independently of each other. Several functional coupling and molecular links exist between these steps (Maniatis and Reed, 2002). Specifically, functional interactions have been described between 3′ end processing and splicing of the terminal intron of pre-mRNAs (Niwa et al, 1990; Niwa and Berget, 1991; Nesic et al, 1993, 1995; Wassarman and Steitz, 1993; Nesic and Maquat, 1994; Berget, 1995; Cooke and Alwine, 1996; Gunderson et al, 1997; Antoniou et al, 1998; Bauren et al, 1998; Cooke et al, 1999; Vagner et al, 2000). However, the mechanistic details of this coupling are not fully understood yet.
The 3′ end processing of pre-mRNAs consists of endonucleolytic cleavage of the primary transcript followed by the addition of a poly(A) tail to the 5′ cleavage product. It allows the correct positioning of the 3′ end of the pre-mRNA, the release of the newly transcribed RNA from the site of transcription and the protection of the mRNA against 3′–5′ exonucleolytic activities (see Colgan and Manley, 1997; Custodio et al, 1999; Minvielle-Sebastia and Keller, 1999; Wahle and Rüegsegger, 1999). Pre-mRNA 3′ end processing is accomplished through recognition of several cis-acting signals by protein complexes. The highly conserved AAUAAA or AUUAAA hexanucleotide, which lies 10–30 nucleotides upstream of the cleavage and polyadenylation site (poly(A) site), is recognised by the cleavage and polyadenylation specificity factor (CPSF). A more degenerate GU- or U-rich sequence located downstream of the poly(A) site is recognised by the cleavage stimulatory factor (CstF). Finally, a set of UGUAA elements flanking and overlapping the AAUAAA sequence are recognised by the mammalian cleavage factor I (CF Im) (Brown and Gilmartin, 2003; Venkataraman et al, 2005). To stabilise the binding of these three sequence-specific RNA-binding factors to RNA, cooperative interactions between these factors are formed. For instance, the interaction between CPSF and CstF is involved in the concerted recognition of the RNA (Takagaki and Manley, 2000), whereas CF Im interacts with CPSF and stabilises the binding of CPSF to the RNA (Rüegsegger et al, 1996, 1998; Venkataraman et al, 2005). After the initial cotranscriptional assembly of the CPSF, CstF and CF Im complexes to the RNA substrate, the cleavage and polyadenylation reactions require the additional recruitment of the mammalian cleavage factor II (CF IIm) and poly(A) polymerase (PAP).
Several reports have demonstrated that 3′ end processing is coupled to the splicing of the terminal intron of a gene. Specifically, mutations in the intronic polypyrimidine tract reduce 3′ end processing efficiency in vitro and in vivo (Niwa et al, 1990; Cooke et al, 1999; Cooke and Alwine, 2002; Millevoi et al, 2002). The polypyrimidine tract is located between the branch point and the 3′ splice site and is essential for splicing. It is recognised by the 65 kDa subunit of the U2 snRNP Auxiliary Factor (U2AF) complex at an early step in the assembly of the spliceosome (Reed, 2000). In addition to its crucial role in splicing, we have demonstrated that U2AF 65 bound to the polypyrimidine tract of the second and last intron of β-globin pre-mRNA promotes 3′ end processing (Millevoi et al, 2002). We have also previously demonstrated that U2AF 65 interacts with PAP (Vagner et al, 2000). Through this interaction, PAP is able to recruit U2AF 65 to an adjacent upstream 3′ splice site, thereby stimulating splicing (Vagner et al, 2000). The direct interaction between U2AF 65 and PAP may serve as a primary molecular link to support the stimulatory effect of the intronic polypyrimidine tract on 3′ end processing. However, PAP is one of the last factors that assemble on the 3′ end processing machinery and it is therefore possible that other protein components of the cleavage/polyadenylation machinery may also constitute good candidates in this process. Particularly, CF Im has been identified as a component of purified human spliceosomes (Rappsilber et al, 2002; Zhou et al, 2002) and also constitutes one of the earliest complexes to assemble to pre-mRNA 3′ end processing signals (Rüegsegger et al, 1998; Venkataraman et al, 2005). There are three polypeptides of 25, 59 and 68 kDa and a minor polypeptide of 72 kDa that copurify with CF Im activity (Rüegsegger et al, 1996). Reconstitution of CF Im activity with recombinant proteins suggests that CF Im is a heterodimer composed of the 25 kDa subunit and a large subunit of 59, 68 or 72 kDa (Rüegsegger et al, 1998). Interestingly, both the 59 and 68 kDa subunits contain a C-terminal RS-like alternating charged domain (Rüegsegger et al, 1998). RS domains are found in the SR family of proteins that function in constitutive and alternative splicing but also in some other splicing factors such as U2AF 65 (Graveley, 2000). The SR domains are thought to mediate protein–protein interactions and the RS-like domain of the 68 kDa subunit of CF Im has been found to interact with the SRp20, 9G8 and hTra2β members of the SR family of splicing factors (Dettwiler et al, 2004).
In this report, we have addressed the mechanism by which U2AF 65 promotes 3′ end cleavage and polyadenylation. Our experiments reveal that a 17-amino-acid-long region composed of five RS dipeptides of U2AF 65 interacts with the RS-like alternating charged domain of the 59 kDa subunit of CF Im and thereby assists in the recruitment of the CF Im 59/25 heterodimer to cleavage/polyadenylation signals. Our results are consistent with a mechanism whereby CF Im physically links the splicing and 3′ end processing machineries by interacting simultaneously with U2AF 65 (our data) and both hFip1 and PAP (Kim and Lee, 2001; Kaufmann et al, 2004).
Results
A 17-amino-acid-long region of U2AF 65, when tethered to cleavage/polyadenylation substrates, is necessary and sufficient to promote 3′ end cleavage in vitro
In order to delineate the region of U2AF 65 required for its stimulatory effect on 3′ end cleavage of pre-mRNAs, we adopted an in vitro tethered approach that previously helped us to establish the role of U2AF 65 in 3′ end processing (Millevoi et al, 2002). Tethering assays to investigate the function of U2AF 65 have also been used successfully to demonstrate the role of the RS-rich domain of U2AF 65 in promoting splicing (Philipps et al, 2003) or in inhibiting polyadenylation (Ko and Gunderson, 2002). We expressed in Escherichia coli and purified under native conditions a series of hybrid proteins in which the R17/MS2 RNA-binding domain was fused to various regions of U2AF 65 (Figure 1A). The activities of the fusion proteins in promoting 3′ end processing were analysed with two pre-mRNA substrates in which the R17 binding site was inserted in place of the polypyrimidine tract 3′ splice site and upstream of the adenovirus L3 or the human β-globin AAUAAA cleavage/polyadenylation signals (Millevoi et al, 2002).
Figure 1.
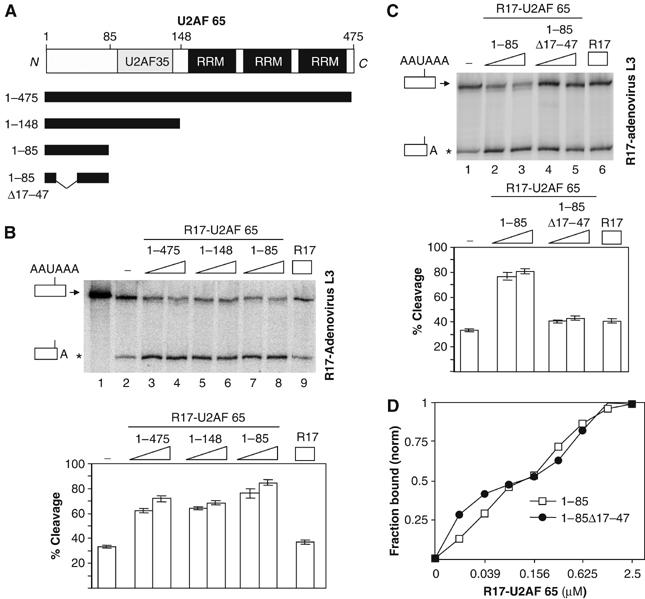
The region spanning residues 17–47 of U2AF 65 is necessary to promote in vitro 3′ end cleavage in HeLa cell nuclear extracts. (A) Illustration of the domain organisation of the U2AF 65 protein showing a central U2AF 35-interacting domain (U2AF 35) and three carboxyl-terminal RRMs. The extent of the full-length and various deletion mutants of U2AF 65 used in this investigation are shown. (B) In vitro cleavage reactions using the R17-adenovirus L3 RNA cleavage substrate. 32P-labelled RNA substrate was incubated with HeLa nuclear extract (lanes 2–9) in the presence of an increasing amount (7 or 14 pmol) of the various R17-U2AF 65 fusion proteins (lanes 3–8) or the R17 protein (14 pmol) (lane 9). Lane 1 corresponds to input RNA. Identities of the uncleaved and cleaved products are shown on the left. (C) In vitro cleavage reactions. The R17-adenovirus L3 32P-labelled RNA substrate was incubated with HeLa nuclear extract in the presence of an increasing amount (7 or 14 pmol) of the various R17-U2AF 65 fusion proteins (lanes 2–5) or the R17 protein (14 pmol) (lane 6). Identities of the uncleaved and cleaved products are shown on the left. These representative experiments were repeated at least six times. Increasing the amount of all the added fusion proteins up to 30 pmol did not further activate the efficiency of 3′ end cleavage showing that the maximum stimulatory effects were reached with 14 pmol of added proteins. The percentage of cleavage as determined by the ratio of the cleaved to uncleaved product are represented on the histograms in panels B and C. (D) Filter binding assays; the normalized fraction of filter-bound RNA is plotted as function of the protein concentration. Curves were fitted to average data points of three independent experiments. The asterisk shows the position of migration of the cleaved product.
These pre-mRNAs were labelled and then incubated in HeLa cell nuclear extracts either alone or in the presence of the various R17-U2AF 65 proteins. A cleaved RNA product appeared upon incubation of the R17-adenovirus L3 RNA substrate in HeLa nuclear extract (Figure 1B, lanes 1 and 2). No cleavage product was observed with an RNA substrate containing an AAUAAA to AAGAAA mutation in the adenovirus L3 cleavage/polyadenylation site (data not shown; Millevoi et al, 2002). Consistent with our previous study (Millevoi et al, 2002), addition of increasing amounts of R17-U2AF 65, but not R17, to HeLa cell nuclear extracts increased cleavage efficiency (Figure 1B, lanes 3, 4 and 9). This stimulation is dependent on the binding of the R17-U2AF 65 protein to the R17-containing RNA cleavage substrate since the stimulatory effect was not observed with an RNA cleavage substrate devoid of the R17 binding site (Millevoi et al, 2002).
Several deletion mutants of the U2AF 65 protein were then used in such cleavage assays. A U2AF 65 deletion mutant containing the first 148 amino acids but lacking the carboxyl-terminal RNA recognition motifs (RRMs) of U2AF 65 was still able to stimulate 3′ end processing (Figure 1B, lanes 5 and 6). Since U2AF 65 was shown to form a stable heterodimeric complex with U2AF 35 (Zamore and Green, 1989) through residues located at positions 85–112 (Rudner et al, 1998; Kielkopf et al, 2001), we wanted to analyse the requirement of the U2AF 35 interaction in the ability of U2AF 65 to stimulate 3′ end processing. A deletion mutant containing the first 85 amino acids of U2AF 65 but lacking the U2AF 35 interaction domain was still able to stimulate 3′ end cleavage to the same degree (Figure 1B, lanes 7 and 8). This is in agreement with our previous results showing that U2AF 35 did not stimulate cleavage (Millevoi et al, 2002). Since we and others reported that a region spanning amino acids 17–47 of U2AF 65 was potentially implicated in creating a molecular link between the splicing and cleavage/polyadenylation machineries (Vagner et al, 2000; Ko and Gunderson, 2002), we analysed the effect of deleting this region on the ability of U2AF 65 to stimulate 3′ end processing. Our results show that a deletion mutant containing residues 1–85 but lacking amino acids 17–47 had lost its stimulatory effect on 3′ end cleavage (Figure 1C, compare lanes 2 and 3 with lanes 4 and 5). Since this protein was not affected in its ability to bind to the RNA cleavage substrate, as judged by filter binding assay (Figure 1D), we concluded that amino acids located between residues 17–47 are crucial to stimulate 3′ end cleavage. In conclusion, these data demonstrate that a region spanning residues 1–85 is able to stimulate 3′ end cleavage to the same extent as the full-length protein.
We next tested whether the region located between residues 17–47 alone was sufficient to stimulate 3′ end processing. An R17 fusion protein containing amino acids 17–47 of U2AF 65 was able to stimulate 3′ end cleavage efficiency (Figure 2B, lanes 4 and 5), showing that this region was sufficient to promote this process. A striking feature of this region is that it contains a sequence with five arginine/serine dipeptides between positions 30 and 47 (Figure 2A). We therefore assessed whether the ability of this domain to stimulate 3′ end processing was due to its RS-rich subdomain. A fusion protein containing amino acids 30–47 was still able to increase the efficiency in 3′ end cleavage, while a fusion protein containing only amino acids 17–30 was unable to do so (Figure 2B, lanes 6–9), although its ability to bind RNA was not impaired (data not shown).
Figure 2.
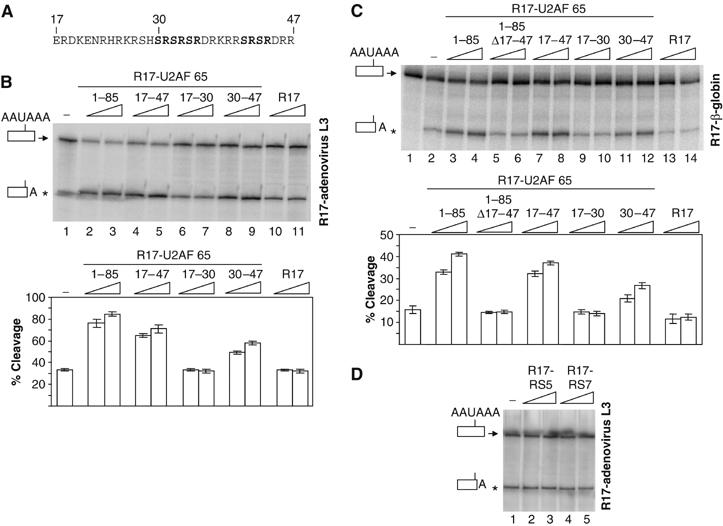
The RS-rich region located between residues 30 and 47 of U2AF is sufficient to promote in vitro cleavage in HeLa cell nuclear extracts, whereas peptides containing five or seven RS repetitions do not stimulate cleavage. (A) The amino-acid sequence of the region located between residues 17 and 47 of U2AF 65. (B–D) In vitro cleavage reactions using the R17-adenovirus L3 (B, D) or R17-β-globin (C) RNA cleavage substrates. 32P-labelled RNA substrates were incubated with HeLa nuclear extract in the absence (lane 1 of panels B and D; lane 2 of panel C) or in the presence of increasing amounts (7 or 14 pmol) of the R17 protein (lanes 10–11 of panel B and lanes 13–14 of panel C), the various R17-U2AF 65 fusion proteins as indicated (lanes 2–9 of panel B and lanes 3–12 of panel C) or R17 fusion proteins containing five or seven RS dipeptides (lanes 2–5 of panel D). Identities of the uncleaved and cleaved products are shown on the left of the autoradiogram. The percentage of cleavage as determined by the ratio of the cleaved to uncleaved product are represented on the histograms in panels B and C. These representative experiments were repeated at least five times. Increasing the amount of all the added fusion proteins up to 30 pmol did not further activate the efficiency of 3′ end cleavage showing that the maximum stimulatory effects were reached with 14 pmol of added proteins. The asterisk shows the position of migration of the cleaved product.
In order to extend these results and to show that the stimulatory effect of the RS-rich region of U2AF 65 is not substrate-specific, the same experiments were performed with a human β-globin cleavage/polyadenylation substrate. Here again, addition of the R17-U2AF fusion proteins containing the 1–85, the 17–47 or the 30–47 domains stimulated 3′ end cleavage (Figure 2C, lanes 3 and 4, 7 and 8 and 11 and 12), whereas the R17 fusion proteins lacking the 17–47 domains (1–85 Δ17–47; Figure 2C, lanes 5 and 6) or that containing the amino acids 17–30 (Figure 2C, lanes 9 and 10) failed to stimulate cleavage above basal levels.
We therefore concluded that a U2AF 65 domain rich in RS dipeptides (17–47) is necessary and sufficient to stimulate 3′ end cleavage. It is also evident that the R17-U2AF fusion protein containing the 30–47 domains alone stimulated 3′ end cleavage to a weaker extent when compared to the fusion protein containing the 1–85 domains. This could be explained by the partial inactivity of the domain when fused to R17. Alternatively, this could indicate that the stimulatory activity does not rely only on the RS content.
To further examine the relationship between RS content and 3′ end processing stimulation, we tested the ability of R17 hybrid proteins containing five or seven RS dipeptides to increase cleavage efficiency. As shown in Figure 2D, none of these proteins stimulated 3′ end processing, indicating that the richness in RS dipeptides is not the sole determinant of 3′ end processing stimulation.
Tethering U2AF 65 to the adenovirus L3 pre-mRNA cleavage substrate reduces the lag phase of the cleavage reaction
The previous experiments (Figures 1 and 2) demonstrated that a tethered U2AF 65 moiety stimulated the efficiency of 3′ pre-mRNA cleavage. However, they did not address a possible effect of U2AF 65 on the kinetics of the cleavage reaction. To investigate whether tethering U2AF 65 could lead to a faster assembly of the 3′ end processing complex and to an enhancement of the rate of the cleavage reaction, we performed a time-course experiment of an in vitro cleavage reaction in either the presence or absence of the R17-U2AF 65 (1–85) fusion protein, which is the minimal sequence that retained maximal stimulation of cleavage efficiency (Figure 1). We observed that the presence of R17-U2AF 65 (1–85) not only enhanced the rate at every time point but also reduced the lag phase of the cleavage reaction from 5 to less than 1 min (Figure 3). These data indicate that U2AF 65 might promote a very early step in the assembly of the cleavage/polyadenylation machinery.
Figure 3.
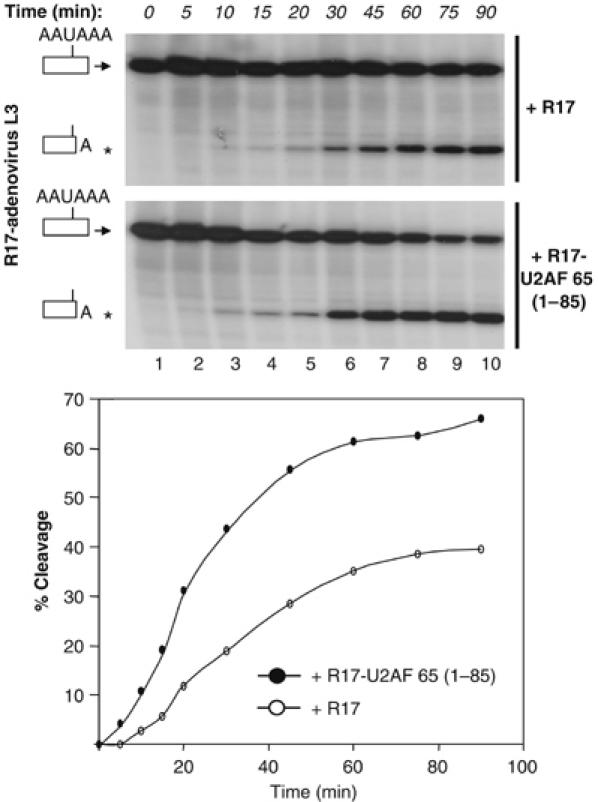
U2AF 65 enhances the rate of 3′ end cleavage and reduces the lag phase of the cleavage reaction. A time course of in vitro cleavage reactions was performed for the R17-adenovirus L3 RNA cleavage substrate using HeLa cell nuclear extracts in the presence of 14 pmol of either the R17 protein or the R17-U2AF 65 (1–85) fusion proteins. The percentages of cleavage as determined by the ratio of the cleaved to uncleaved product were plotted on the graph for each time point. These representative experiments were repeated at least three times. The asterisk shows the position of migration of the cleaved product.
U2AF 65 interacts with the 59 kDa subunit of CF Im
It has previously been shown that one of the earliest steps in the assembly of the 3′ end processing machinery is the interaction of CF Im with pre-mRNA that in turn facilitates the recruitment of other processing factors. Indeed, pre-incubation of CF Im with RNA results in a reduction of the lag phase of the cleavage reaction (Rüegsegger et al, 1998). Furthermore, two of the CF Im subunits, namely CF Im 59 and CF Im 68, contain RS domains that may interact with the RS-rich region of U2AF 65 involved in the stimulation of 3′ end processing.
The ability of the amino-terminal RS-containing domain (1–85) of U2AF 65 to interact with the RS domains of CF Im 59 and CF Im 68 was examined in a glutathione S-transferase (GST) pull-down experiment. The E. coli-produced GST-R17-U2AF 65 (1–85) proteins but not the GST-R17 control protein was able to form a complex with the in vitro translated 35S-methionine-labelled RS domain of CF Im 59 kDa subunit, but not with the RS domain of CF Im 68 (Figure 4A, lanes 1–6).
Figure 4.
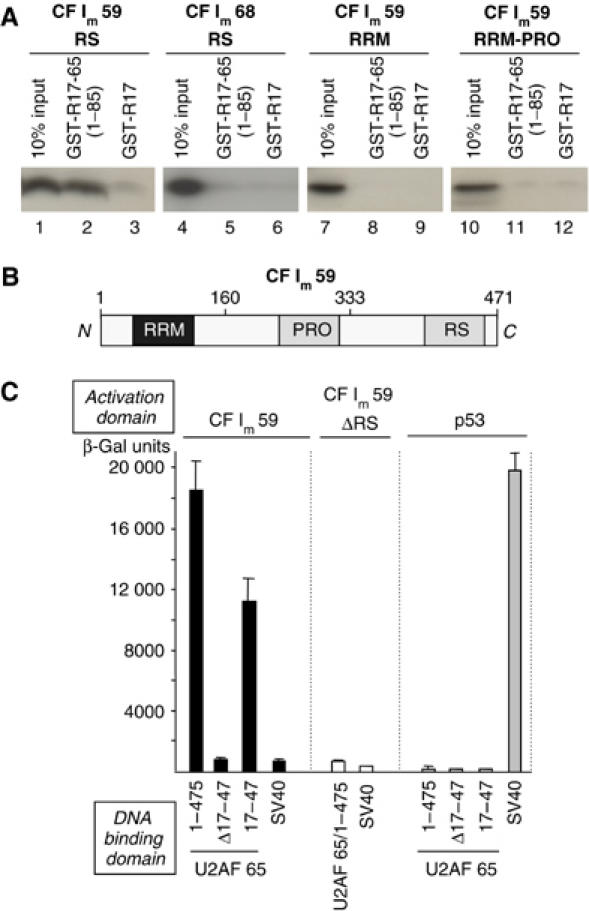
U2AF 65 interacts with the 59 kDa subunit of CF Im. (A) In vitro protein–protein interaction assays using GST pull down. The indicated GST fusion proteins (GST-R17, GST-R17-U2AF 65 and GST-R17-U2AF 65 (1–85)) were bound to glutathione-agarose beads and used in binding assays with 35S-methionine-labelled domains of the 59 and 68 kDa subunits of CF Im as indicated. Ten percent of the input was loaded. (B) Illustration of the domain organisation of the 59 kDa subunit of CF Im containing an amino-terminal RRM, a central proline-rich domain (PRO) and the carboxyl- terminal RS-like alternating charge domain (RS). All the GST pull-down experiments were performed at least three times to ensure reproducibility. (C) In vivo protein–protein interaction assay using yeast two-hybrid experiments. Yeast were transformed with combinations of the plasmids encoding the indicated proteins and grown on Leu−Trp− media prior to assessment of β-galactosidase activity.
The 59 kDa subunit of CF Im is composed of an amino-terminal RRM, a central proline-rich region (PRO) and a carboxyl-terminal RS-like domain (Rüegsegger et al, 1998; Figure 4B). In order to determine the contribution of each of the other domains of CF Im 59 to interact with the RS-containing domain of U2AF 65, we performed GST pull-down experiments with in vitro translated domains of CF Im 59 corresponding to the RRM alone or the RRM-PRO. As shown in Figure 4A, we found that both of these domains were unable to interact with the R17-U2AF 65 (1–85) protein (Figure 4A, lanes 7–12). We concluded that the RS domain of CF Im 59 was necessary and sufficient to interact specifically with the amino-terminal region (1–85) of U2AF 65.
We next confirmed the interaction between U2AF 65 and CF Im 59 by performing yeast two-hybrid experiments with the full-length proteins. As judged by a liquid β-gal assay presented in Figure 4C, we found that an interaction occurred between U2AF 65 and CF Im 59 and that the regions 17–47 of U2AF 65 was sufficient for interaction with CF Im 59. However, no interaction was observed when either the regions 17–47 of U2AF 65 or the RS domain of CF Im 59 were deleted (Figure 4C).
Taken together, these data provide in vitro and in vivo evidence for an interaction between U2AF 65 and CF Im 59 and implicate the RS domains present in both proteins as being crucial in this process.
U2AF 65 recruits CF Im 59 to the cleavage/polyadenylation site
Our results described above reveal that U2AF 65 enhances the rate of cleavage and interacts with CF Im 59. U2AF 65 may therefore help in the recruitment of CF Im 59 to RNA. To investigate this possibility, we performed UV crosslinking experiments with the two RNA cleavage substrates and a recombinant CF Im 59–CF Im 25 complex (CF Im59/25). The 59 and 25 kDa subunits of CF Im were coexpressed in Sf9 cells following baculovirus infection. The 25 kDa subunit was expressed with an N-terminal hexahistidine tag for efficient purification of the heterodimeric complex. We obtained a pure fraction of CF Im59/25 complex as judged by Coomassie staining (Figure 5A (i)) and Western blot (Figure 5A (ii)). Two crosslinked bands corresponding to the CF Im 59 and 25 kDa subunits were observed with the CF Im59/25 complex (Figure 5B, lane 1). Crosslinking of CF Im 59 to the R17-adenovirus L3 substrate was more efficient in the presence of an R17 fusion protein containing a domain able to interact with CF Im 59 (Figure 5B, lanes 2 and 4) as compared with crosslinking carried out in the presence of the R17 fusion proteins unable to interact with CF Im 59 (Figure 5B, lanes 3 and 5). The same effect was also observed with the R17-β-globin RNA substrate (Figure 5C) showing that the effect is not substrate-specific. It is noteworthy that the stimulatory effect on CF Im 25 crosslinking was weaker as compared to the effect observed with CF Im 59. This might be due to the excess of CF Im 25 in this preparation (see Figure 5A (i)).
Figure 5.
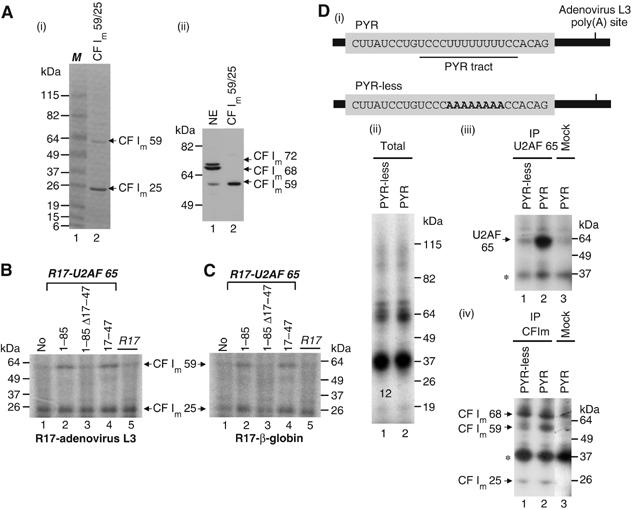
U2AF 65 recruits CF Im 59 to the adenovirus L3 cleavage/polyadenylation site. (A) (i) Coomassie blue staining of the recombinant CF Im59/25 complex. M: protein markers. (ii) Western blot using the antibody raised against CF Im 68. NE: nuclear extract. (B, C) UV crosslinking experiments were performed with a uniformly 32P-labelled R17-adenovirus L3 RNA substrate (B) or a uniformly 32P-labelled R17-β-globin RNA substrate (C). The labelled RNA was incubated with CF Im59/25 (6 pmol) in the absence (B, C, lane 1) or the presence (B, C, lanes 2–5) of 8 pmol of the various R17-U2AF 65 fusion proteins. Following incubation, crosslinking was performed and the crosslinked polypeptides were resolved by SDS–PAGE. This experiment was performed at least four times. The poor quality of the migration of the crosslinking reaction is due to the presence of polyvinylalcohol in the reaction. (D) (i) A 32P-labelled RNA (PYR) containing the 3′ splice site of the adenovirus I major late intron upstream of the adenovirus L3 polyadenylation site (described in Vagner et al, 2000) and a 32P-labelled RNA (PYR-less) in which eight uridines were mutated to adenines were incubated in HeLa nuclear extract and crosslinked by UV irradiation. Crosslinked polypeptides were either fractionated directly by SDS–PAGE (ii) or immunoprecipitated before fractionation with the MC3 monoclonal antibody directed against U2AF 65 (Gama-Carvalho et al, 1997) (iii, lanes 1 and 2) or a polyclonal antibody against CF Im (iv, lanes 1 and 2). Mock columns for immunoprecipitation are shown in panels (iii) and (iv) lanes 3 and show a nonspecific immunoprecipitation of a protein migrating at 37 kDa (*).
The recruitment of CF Im 59 by U2AF 65 was also tested using a pre-mRNA containing the natural pyrimidine tract (the authentic U2AF 65 binding site) whose functional implication in 3′ end processing has been well established by us and others in vitro and in vivo (see Introduction). To perform this experiment, we used two RNAs containing the adenovirus L3 cleavage/polyadenylation site and upstream 3′ splice sites, which differ in their pyrimidine content and therefore in their ability to bind U2AF 65 (Figure 5D (i)). These RNAs were incubated in HeLa nuclear extract. We analysed the binding of CF Im subunits by UV crosslinking followed by immunoprecipitation with an antibody directed against CF Im 68. This antibody is also able to recognise CF Im 59 (Rüegsegger et al, 1996). The complex crosslinking pattern (Figure 5D (ii)) observed with these two RNAs was resolved by immunoprecipitation. Immunoprecipitation with a monoclonal antibody against U2AF 65 (Gama-Carvalho et al, 1997) indeed showed that this protein was bound to the PYR RNA containing the pyrimidine tract, but not to the PYR-less RNA in which eight pyrimidines were mutated to purines (Figure 5D (iii)). Immunoprecipitation with the antibody against CF Im 68 gave three bands corresponding to the three CF Im subunits and an additional nonspecific band that also appeared in the ‘mock' immunoprecipitation (Figure 5D (iv)). According to our expectation, the crosslinking of CF Im 59 but not CF Im 68 was stronger with the PYR RNA as compared to the PYR-less RNA (Figure 5D (iv), lanes 1 and 2). A slight effect could also be observed on CF Im 25 crosslinking. This slight effect can be explained by the fact that CF Im 25 is in a complex with both CF Im 68 and CF Im 59. Thus, the effect is masked by CF Im 25 in a complex with CF Im 68.
Altogether, these results demonstrate that U2AF 65 is able to recruit CF Im 59 to an RNA cleavage substrate.
Tethered U2AF 65 promotes the extent and efficiency of polyadenylation
Since CF Im has been shown to play a role in the polyadenylation step of 3′ end processing (Brown and Gilmartin, 2003; Venkataraman et al, 2005) and to interact with several polyadenylation components (PAP and hFip1) (Kim and Lee, 2001; Kaufmann et al, 2004), we examined the impact of tethered U2AF 65 on poly(A) addition in HeLa cell nuclear extracts. Both the extent and efficiency of poly(A) addition to the precleaved R17-adenovirus L3 substrates were enhanced upon tethering of the amino-terminal (1–85) regions of U2AF 65 (Figure 6A, compare lanes 3 and 4 with lane 2). This effect was strictly dependent on the presence of the region of interaction with PAP and CF Im 59 since addition of the R17-U2AF 65 (1–85 Δ17–47) proteins did not lead to an activation of polyadenylation (data not shown).
Figure 6.
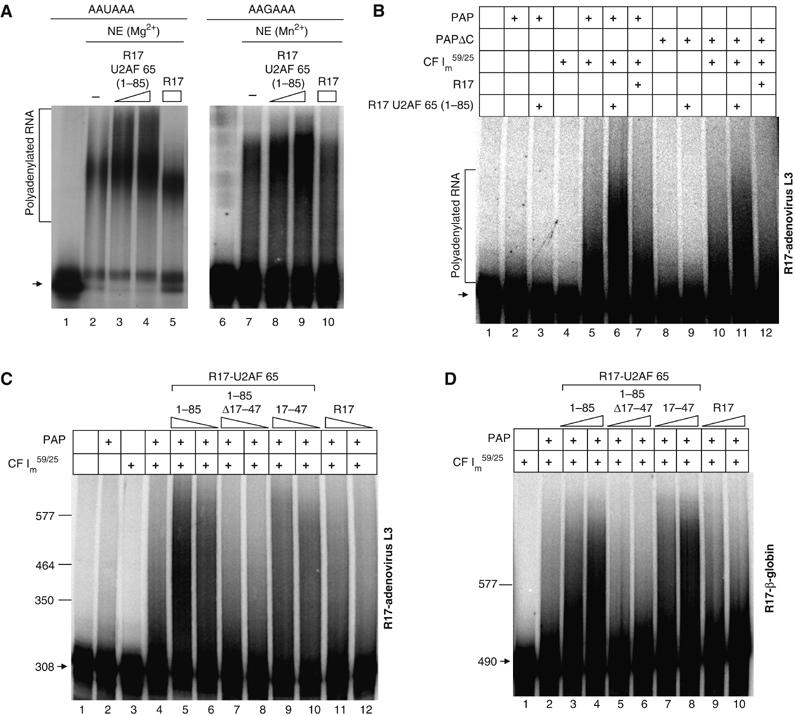
The interaction between U2AF 65 and CF Im59/25 promotes polyadenylation. (A) Polyadenylation assays using HeLa cell nuclear extracts (NE). 32P-labelled precleaved R17-adenovirus L3 RNA substrates containing either a AAUAAA (lanes 1–5) or a AAGAAA (lanes 6–10) sequence were incubated in HeLa nuclear extracts without (lanes 2 and 7) or with the addition of R17 (lanes 5 and 10) or R17-U2AF 65 (1–85) fusion proteins (lanes 3, 4 and 8, 9) as indicated above each lane. Lanes 1 and 2 are the input RNAs. The polyadenylation reaction was performed in the presence of Mg2+ (lanes 1–5) or Mn2+ (lanes 6–10). RNAs were analysed on 6% (19:1) denaturing PAGE gels. (B–D) Reconstituted polyadenylation assays using recombinant PAP, PAPΔC and the CF Im59/25 complex and the adenovirus L3 precleaved substrate. In all experiments, RNAs were analysed on 6% (19:1) denaturing PAGE gels. The positions of migration of RNA markers are indicated on the left. Experiments were repeated at least three times. (B) Polyadenylation reactions were performed using recombinant PAP (lanes 2, 3 and 5–7; 0.1 pmol) or PAPΔC (lanes 8–12; 0.1 pmol) in the presence of the CF Im59/25 complex (lanes 4–7 and 10–12; 2 pmol) and 1 pmol of the R17 (lanes 7 and 12) or the R17-U2AF 65 (1–85) fusion proteins (lanes 3, 6, 9 and 11). (C) Reconstituted polyadenylation assays carried out with the adenovirus L3 precleaved RNA substrate and using recombinant PAP (lanes 2 and 4–12; 0.1 pmol) in the presence of CF Im59/25 (lanes 3–12; 2 pmol) and decreasing amounts (1 and 0.5 pmol) of the indicated R17-U2AF 65 fusion proteins. Arrow: input RNA. (D) Reconstituted polyadenylation assays carried out with the β-globin pre-cleaved RNA substrate and using recombinant PAP (lanes 2–10; 0.1 pmol) in the presence of CF Im59/25 (lanes 2–10; 2 pmol) and increasing amounts (0.5 and 1 pmol) of the indicated R17-U2AF 65 fusion proteins. Arrow: input RNA.
Similar results were obtained in CPSF-independent conditions assays, using an RNA substrate containing a mutated AAGAAA hexamer. Stimulation of polyadenylation was still observed upon tethering of the amino-terminal (1–85) regions of U2AF 65 to the RNA substrate (Figure 6A, lanes 8 and 9) in the presence of Mn2+, showing that the stimulatory effect of U2AF 65 on polyadenylation is CPSF-independent.
Since we previously reported an interaction between U2AF 65 and PAP (Vagner et al, 2000), we investigated whether this interaction might be responsible for the stimulatory effect of U2AF 65 on polyadenylation. We performed experiments employing a reconstituted polyadenylation assay with a wild-type PAP and a carboxy-terminal deletion mutant of PAP unable to interact with U2AF 65 (PAP ΔC). These two PAPs had the same activity in basal polyadenylation (Supplementary Figure 1). The reactions were carried out with the R17-adenovirus L3 precleaved substrate in conditions of a 10-fold excess of RNA over PAP to minimise the reassociation of PAP with the same RNA (Figure 6B, lane 2). Addition of R17-U2AF 65 (1–85) did not stimulate the polyadenylation activity of PAP or PAPΔC (Figure 6B, compare lanes 2 and 3 or 8 and 9). We could even observe a slight decrease in the polyadenylation activity of PAP upon addition of the R17-U2AF 65 (1–85) proteins that would reflect the inhibitory effect of this interaction as published by Ko and Gunderson (2002). This led us to conclude that the U2AF 65–PAP interaction is not involved in the stimulatory effect of U2AF 65 on polyadenylation and led us to hypothetise that the U2AF 65–CF Im 59 interaction might be involved.
We therefore performed experiments employing a reconstituted polyadenylation assay with recombinant PAP and CF Im59/25. As expected, the CF Im59/25 complex alone had no polyadenylation activity (Figure 6B, lane 4). Addition of CF Im59/25 to PAP led to a stimulation of polyadenylation (Figure 6B, lane 5). This may reflect the role of the known interaction between the 25 kDa subunit of CF Im and PAP (Kim and Lee, 2001) that would help in the recruitment of PAP to the RNA substrate. Addition of the R17-U2AF 65 (1–85) fusion proteins containing a domain able to interact with CF Im 59 but not the control R17 protein increased polyadenylation efficiency (Figure 6B, lanes 6 and 7). The stimulatory effect of U2AF 65 was also observed in a reconstituted polyadenylation assay containing a recombinant PAP deleted of its carboxyl-terminal region (PAP ΔC; Figure 6B, lane 11). Since PAP ΔC is not able to interact with U2AF 65 (Vagner et al, 2000), this shows again that the stimulatory effect of U2AF 65 on polyadenylation does not involve the U2AF 65–PAP interaction.
The stimulatory effect of U2AF 65 was not observed when the experiment was performed in a reconstituted polyadenylation assay containing PAP and the 25 kDa subunit of CF Im (Supplementary Figure 2) showing that the presence of CF Im 59 in the assay was essential to observe the stimulatory effect.
Furthermore, this enhancing effect of U2AF 65 was not observed when the reconstituted polyadenylation assay was performed with an R17 fusion protein unable to interact with CF Im 59 (R17-U2AF 65 1–85 Δ17–47; Figure 6C, lanes 7 and 8) as compared to the effects observed with the R17-U2AF 65 1–85 or 17–47 proteins (Figure 6C, lanes 5–6 and 9–10).
To demonstrate that the CF Im 59-dependent stimulatory effect of U2AF 65 on polyadenylation was not RNA substrate-specific, we performed the same experiment with the R17-β-globin precleaved RNA substrate (Figure 6D). Again, addition of an R17 fusion protein containing a domain able to interact with CF Im 59 increased polyadenylation efficiency (Figure 6D, lanes 3, 4 and 7, 8), whereas addition of an R17 fusion protein unable to interact with CF Im 59 had no effect (Figure 6D, lanes 5, 6 and 9, 10).
Taken together, these data demonstrate that the interaction between U2AF 65 and CF Im is required to support the stimulatory effect of U2AF 65 on polyadenylation.
Discussion
It has been a long-standing proposal that splicing and 3′ end processing are tightly coupled reactions, but little is known about the mechanistic details of the stimulatory effect of splicing signals and factors on the cleavage/polyadenylation process. In this report, we have performed a series of in vitro experiments to reconstitute and understand the stimulatory effect of the U2AF 65 splicing factor on 3′ end processing. We have found that a tethered U2AF 65 protein stimulates both cleavage and polyadenylation of the adenovirus L3 and β-globin 3′ end processing signals. Our observation that the region 17–47 of U2AF 65 physically associates with CF Im 59 provides some insights into the coupling of splicing and 3′ end processing. Importantly, we have demonstrated that the ability of U2AF 65 and CF Im to interact with each other is required to observe the stimulatory effect of U2AF 65 on 3′ end processing. This supports a model in which U2AF 65 bound to a 3′ splice site can interact with CF Im 59 to help tether the CF Im59/25 heterodimer to the cleavage/polyadenylation signal, thereby stimulating 3′ end processing (Figure 7A).
Figure 7.
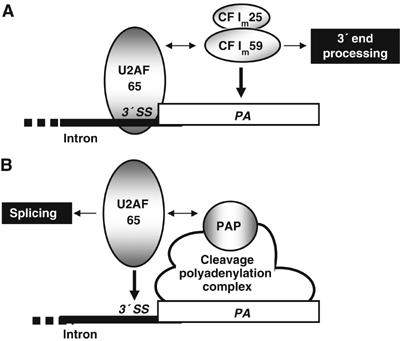
Illustration suggesting a model for the participation of U2AF 65 in the coupling of splicing and 3′ end processing. We propose in (A) that U2AF 65 bound to the pyrimidine tract of the last 3′ splice site (3′ SS) of a pre-mRNA tethers the 59 kDa subunit of CF Im to the cleavage/polyadenylation site (PA) via an interaction between these two proteins (double-headed arrow). This results in stimulation of 3′ end processing. In (B), PAP recruited to an RNA via the cleavage/polyadenylation complex interacts with U2AF 65 (Vagner et al, 2000) to help to tether this splicing factor to the most proximal 3′ splice site thereby stimulating splicing. The thick black lane corresponds to intronic sequences, while the white rectangle corresponds to the 3′ terminal exon of a pre-mRNA.
We have previously reported another interaction involving the same region of U2AF 65 (17–47) and a component of the 3′ end processing machinery (Vagner et al, 2000). In that case PAP, which associates with the CPSF–CstF–CF Im complex assembled on the 3′ end processing signals, interacts with U2AF 65 to promote its recruitment to 3′ splice sites (Vagner et al, 2000; Figure 7B). The role of this interaction in the stimulatory effect of U2AF 65 on polyadenylation can be excluded since U2AF 65 was not able to stimulate polyadenylation when PAP alone was used in the assay (Figure 6B). Furthermore, U2AF 65 was still able to stimulate polyadenylation in an assay containing CF Im and a deletion mutant of PAP unable to interact with U2AF 65 (Figure 6B). However, we cannot definitively conclude on the eventual role of this interaction in stimulating cleavage. Nevertheless, it is very unlikely that the U2AF 65–PAP interaction is involved in stimulating this step since the recruitment of PAP to cleavage/polyadenylation sites is a late event in the assembly of the cleavage/polyadenylation machinery and since this interaction is not involved in the second step (polyadenylation) of 3′-end processing.
These interactions occur in the context of the definition of the 3′-terminal exon that involves the coordinated recognition of a cleavage/polyadenylation site with the adjacent upstream 3′ splice site (Berget, 1995). This recognition may depend on the intrinsic strength of the processing signals at either end of the 3′-terminal exon. When the cleavage/polyadenylation site is strongly bound by the cleavage/polyadenylation machinery and is present downstream of a weak 3′ splice site, the PAP–U2AF 65 interaction may assist in the recognition of the 3′ splice site (Vagner et al, 2000). In contrast, the interaction between U2AF 65 and CF Im 59 may be involved in helping recognition of a weak cleavage/polyadenylation site by a strong 3′ splice site. However, both interactions could also occur at the same time and mutually stimulate both processes. To discriminate between these possibilities, a detailed study of the interaction between U2AF 65 and both PAP and CF Im 59 will be required.
Interestingly, we have identified a mechanism of 3′ end processing stimulation in vertebrates that involves the recruitment of the basic 3′ end processing machinery as a result of a direct interaction between a regulatory protein bound to a cis-acting enhancer element and a component of this machinery. This kind of mechanism is a common theme in transcription and splicing regulation (Hertel et al, 1997), but has never been demonstrated so far for vertebrate 3′ end processing regulation. However, a precedent might exist in plants. In Arabidopsis thaliana, the FCA RNA-binding protein interacts with the FY 3′ end processing factor to help tether or stabilise the 3′ end processing machinery to an otherwise weak poly(A) site, thereby promoting its selection (Simpson et al, 2003). In that case, another type of interaction between the regulator and the 3′ end processing machinery has been described that involves a WW domain and a proline-rich domain (Simpson et al, 2003), and not SR alternating charge domains such as the ones present in U2AF 65 and CF Im 59.
Regulation at the level of cleavage/polyadenylation has been demonstrated in several eukaryotic cellular or viral pre-mRNAs containing a single polyadenylation signal. Furthermore, when looking at a large number of 3′ expressed sequence tag sequences, several studies demonstrated that 29–50% of human pre-mRNAs displayed two or more polyadenylation signals (Beaudoing and Gautheret, 2001). This indicates that the repertoire of cis-acting elements and trans-acting factors that regulate the use of 3′ end processing signals is far from being complete. Since U2AF 65 binding to the 3′ splice site is one of the earliest steps of spliceosome assembly, the participation of U2AF 65 in the recruitment of the 3′ end processing machinery should occur in a splicing-coupled manner. However, we cannot exclude the possible implication of U2AF 65 bound to polypyrimidine tracts located in 3′-terminal exons in the vicinity of a cleavage/polyadenylation site in the stimulation of 3′ end processing. Indeed, situations have been reported in which U2AF 65 acts independently of its binding to 3′ splice sites. For instance, it has recently been shown that the Drosophila large U2AF subunit (dU2AF50) is able to bind to many intronless mRNAs for which it could regulate nuclear export (Blanchette et al, 2004), although a possible role for dU2AF50 in 3′ end processing was not investigated.
In addition, the ability of an RS-rich region of U2AF 65 to promote 3′ end cleavage raised the possibility that, like U2AF 65, the RS domains of SR proteins might stimulate 3′ end cleavage. SR proteins constitute a family of splicing factors that have diverse roles in constitutive and regulated splicing and that usually interact with exonic sequences (Graveley, 2000). We have indeed shown that several RS domains of SR proteins stimulate 3′ end processing (S Millevoi, M Antoniou and S Vagner, unpublished data) and SR proteins may therefore constitute a novel family of regulators of 3′ end processing. Interestingly, it was recently reported that three members of the SR protein family interact with the RS-like alternating charge domain of CF Im 68 but not CF Im 59 (Dettwiler et al, 2004). This would suggest specific regulatory functions of the various CF Im heterodimers. The CF Im59/25 heterodimer may function in concert with U2AF 65 to link the splicing and 3′ end processing machinery, while the CF Im68/25 heterodimer may be regulated by SR proteins bound to 3′-terminal exons. All recent studies point to a crucial role of the CF Im complex in the coordination of the early assembly of the 3′ end processing machinery. Our study demonstrates that CF Im is not only involved in the basal 3′ end processing process, but constitutes a key target for the regulation of the formation of 3′ ends of vertebrate pre-mRNAs.
Materials and methods
Plasmid constructs
The plasmids used to generate RNA substrates for cleavage/polyadenylation reactions (R17-adenovirus L3 and R17-β-globin) were as described previously (Millevoi et al, 2002). The two plasmids were linearised using EcoRI and XhoI, respectively, for in vitro cleavage assays and with AvaI and SwaI, respectively, for the in vitro polyadenylation assays. Capped, uniformly 32P-labelled RNAs used for cleavage, polyadenylation and UV crosslinking assays were obtained by in vitro transcription of the linearised plasmid with either T3 (R17-adenovirus L3) or T7 (R17-β-globin) RNA polymerase.
Cleavage and polyadenylation assays
Cleavage reactions were as described previously (Boelens et al, 1993). The 32P-labelled RNA (R17-adenovirus L3 or R17-β-globin) substrate was incubated with HeLa cell nuclear extract (prepared according to (Dignam et al, 1983) for 90 min at 30°C in the presence of either purified GST-R17 or the different GST-R17–U2AF 65 recombinant fusion proteins. For the time-course experiment, 20 μl aliquots were withdrawn at the indicated time from a 320 μl reaction mixture.
The in vitro polyadenylation assays using HeLa cell nuclear extracts were performed for 40 min at 30°C as for the cleavage reaction, except that it was carried out without cordycepin and in the presence of 0.7 mM ATP. Reconstituted polyadenylation reactions were performed for 15 min at 37°C as described by Gunderson et al (1997). Analysis and quantification of cleavage/polyadenylation reactions after RNA extraction and resolution on a denaturing 6% polyacrylamide gel was carried out by PhosphorImager (Molecular Dynamics) analysis. All the polyadenylation reactions were carried out in the presence of Mn2+, except the reactions shown in Figure 6A (lanes 1–5) that were carried out in the presence of Mg2+. Cleavage activity was calculated by dividing the amount of upstream cleavage product by the sum of cleavage plus precursor products.
Protein expression and purification
GST pull-down assays
The RS (residues 333–471), RRM (residues 1–160) and RRM-PRO (residues 1–333) domains of CF Im 59 mutants and the RS domain of CF Im 68 were generated by PCR using T7 RNA polymerase promoter-containing oligonucleotides. The various proteins were produced by in vitro transcription/translation using TNT rabbit reticulocyte lysate (Promega) and labelled with 35S-methionine. These experiments were carried out as described by Vagner et al (2000). Briefly, 3 μg of purified GST-R17 fusion proteins were bound to 30 μl of glutathione agarose beads in NETN buffer (20 mM Tris at pH 8.0, 100 mM NaCl, 0.5% NP-40, 0.5 mM EDTA) for 30 min at 4°C followed by three washes with 1 ml of NETN buffer. For each binding reaction, 2–5 μl of translation mixture was used and assays were performed in 200 μl of NETN at 4°C. Beads were then washed five times, treated with 10 μg/ml RNAse A at room temperature for 30 min and washed again. Protein elution was performed by adding SDS loading buffer to the beads. Eluted proteins were resolved by 8% SDS–polyacrylamide gel electrophoresis (PAGE). Gels were fixed, dried and labelled proteins were visualised by fluorography.
Yeast two-hybrid experiments
Yeast two-hybrid assays were performed in the Y190 yeast strain. The Y190 strain was cotransformed with pAS2 and pACTII plasmids. Transformed yeasts were selected on a medium lacking leucine and tryptophane. The transformants were tested for the expression of the lacZ reporter gene by using a β-galactosidase liquid assay (Galacto-light assay, Tropix, Perkin-Elmer).
UV crosslinking
The baculovirus coexpressed CF Im 59 and CF Im 25 proteins were incubated with 32P-labelled RNAs (R17-adenovirus L3 or R17-β-globin) under cleavage conditions for 15 min at room temperature. The reaction mixtures were then irradiated on ice with UV light (254 nm) in a Stratalinker (Stratagene) at 0.4 J/cm2 at 10 cm distance. Then, 50 U of RNAse ONE (Promega) was added and the reaction mixtures incubated for 30 min at 37°C. SDS gel loading buffer was added and the samples boiled for 2 min before fractionation on a 10% SDS–PAGE.
Nitrocellulose filter binding assays
See Supplementary data.
Supplementary Material
Supplementary Figure 1
Acknowledgments
SM and CL were supported by an FEBS long-term fellowship. We thank Alexandra Hembury and Françoise Pujol for their help in the preparation of the GST-R17-RS5 and GST-R17-RS7 proteins. Work in the laboratory of SV was supported by INSERM, Fondation de France, FRM (Equipe FRM, soutenue par la Fondation Recherche Médicale) and the French Ministry of Research (ACI ‘Jeunes Chercheurs'). Work in the laboratory of WK was supported by the University of Basel and the Swiss National Science Foundation.
References
- Antoniou M, Geraghty F, Hurst J, Grosveld F (1998) Efficient 3′ end formation of human β-globin mRNA in vivo requires sequences within the last intron but occurs independently of the splicing reaction. Nucleic Acids Res 26: 721–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauren G, Belikov S, Wieslander L (1998) Transcriptional termination in the Balbiani ring 1 gene is closely coupled to 3′ end formation and excision of the 3′-terminal intron. Genes Dev 12: 2759–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoing E, Gautheret D (2001) Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res 11: 1520–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget SM (1995) Exon recognition in vertebrate splicing. J Biol Chem 270: 2411–2414 [DOI] [PubMed] [Google Scholar]
- Blanchette M, Labourier E, Green RE, Brenner SE, Rio DC (2004) Genome-wide analysis reveals an unexpected function for the Drosophila splicing factor U2AF50 in the nuclear export of intronless mRNAs. Mol Cell 14: 775–786 [DOI] [PubMed] [Google Scholar]
- Boelens WC, Jansen EJ, van Venrooij WJ, Stripecke R, Mattaj IW, Gunderson SI (1993) The human U1 snRNP-specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell 72: 881–892 [DOI] [PubMed] [Google Scholar]
- Brown KM, Gilmartin GM (2003) A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol Cell 12: 1467–1476 [DOI] [PubMed] [Google Scholar]
- Colgan DF, Manley JL (1997) Mechanism and regulation of mRNA polyadenylation. Genes Dev 11: 2755–2766 [DOI] [PubMed] [Google Scholar]
- Cooke C, Alwine JC (1996) The cap and the 3′ splice site similarly affect polyadenylation efficiency. Mol Cell Biol 16: 2579–2584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke C, Alwine JC (2002) Characterization of specific protein–RNA complexes associated with the coupling of polyadenylation and last-intron removal. Mol Cell Biol 22: 4579–4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke C, Hans H, Alwine JC (1999) Utilization of splicing elements and polyadenylation signal elements in the coupling of polyadenylation and last-intron removal. Mol Cell Biol 19: 4971–4979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custodio N, Carmo-Fonseca M, Geraghty F, Pereira HS, Grosveld F, Antoniou M (1999) Inefficient processing impairs release of RNA from the site of transcription. EMBO J 18: 2855–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM (2004) Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein–protein interactions, and subcellular localization. J Biol Chem 279: 35788–35797 [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama-Carvalho M, Krauss RD, Chiang L, Valcarcel J, Green MR, Carmo-Fonseca M (1997) Targeting of U2AF65 to sites of active splicing in the nucleus. J Cell Biol 137: 975–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR (2000) Sorting out the complexity of SR protein functions. RNA 6: 1197–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson SI, Vagner S, Polycarpou-Schwarz M, Mattaj IW (1997) Involvement of the carboxyl terminus of vertebrate poly(A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes Dev 11: 761–773 [DOI] [PubMed] [Google Scholar]
- Hertel KJ, Lynch KW, Maniatis T (1997) Common themes in the function of transcription and splicing enhancers. Curr Opin Cell Biol 9: 350–357 [DOI] [PubMed] [Google Scholar]
- Kaufmann I, Martin G, Friedlein A, Langen H, Keller W (2004) Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J 23: 616–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielkopf CL, Rodionova NA, Green MR, Burley SK (2001) A novel peptide recognition mode revealed by the X-ray structure of a core U2AF35/U2AF65 heterodimer. Cell 106: 595–605 [DOI] [PubMed] [Google Scholar]
- Kim H, Lee Y (2001) Interaction of poly(A) polymerase with the 25-kDa subunit of cleavage factor I. Biochem Biophys Res Commun 289: 513–518 [DOI] [PubMed] [Google Scholar]
- Ko B, Gunderson SI (2002) Identification of new poly(A) polymerase-inhibitory proteins capable of regulating pre-mRNA polyadenylation. J Mol Biol 318: 1189–1206 [DOI] [PubMed] [Google Scholar]
- Maniatis T, Reed R (2002) An extensive network of coupling among gene expression machines. Nature 416: 499–506 [DOI] [PubMed] [Google Scholar]
- Millevoi S, Geraghty F, Idowu B, Tam JL, Antoniou M, Vagner S (2002) A novel function for the U2AF 65 splicing factor in promoting pre-mRNA 3′ end processing. EMBO Rep 3: 869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L, Keller W (1999) mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr Opin Cell Biol 11: 352–357 [DOI] [PubMed] [Google Scholar]
- Nesic D, Cheng J, Maquat LE (1993) Sequences within the last intron function in RNA 3′ end formation in cultured cells. Mol Cell Biol 13: 3359–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesic D, Maquat LE (1994) Upstream introns influence the efficiency of final intron removal and RNA 3′ end formation. Genes Dev 8: 363–375 [DOI] [PubMed] [Google Scholar]
- Nesic D, Zhang J, Maquat LE (1995) Lack of an effect of the efficiency of RNA 3′ end formation on the efficiency of removal of either the final or the penultimate intron in intact cells. Mol Cell Biol 15: 488–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M, Berget SM (1991) Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes Dev 5: 2086–2095 [DOI] [PubMed] [Google Scholar]
- Niwa M, Rose SD, Berget SM (1990) In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev 4: 1552–1559 [DOI] [PubMed] [Google Scholar]
- Philipps D, Celotto AM, Wang QQ, Tarng RS, Graveley BR (2003) Arginine/serine repeats are sufficient to constitute a splicing activation domain. Nucleic Acids Res 31: 6502–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M (2002) Large-scale proteomic analysis of the human spliceosome. Genome Res 12: 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R (2000) Mechanisms of fidelity in pre-mRNA splicing. Curr Opin Cell Biol 12: 340–345 [DOI] [PubMed] [Google Scholar]
- Rudner DZ, Kanaar R, Breger KS, Rio DC (1998) Interaction between subunits of heterodimeric splicing factor U2AF is essential in vivo. Mol Cell Biol 18: 1765–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüegsegger U, Beyer K, Keller W (1996) Purification and characterization of human cleavage factor Im involved in the 3′ end processing of messenger RNA precursors. J Biol Chem 271: 6107–6113 [DOI] [PubMed] [Google Scholar]
- Rüegsegger U, Blank D, Keller W (1998) Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol Cell 1: 243–253 [DOI] [PubMed] [Google Scholar]
- Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C (2003) FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113: 777–787 [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL (2000) Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol 20: 1515–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S, Vagner C, Mattaj IW (2000) The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′ end processing and splicing. Genes Dev 14: 403–413 [PMC free article] [PubMed] [Google Scholar]
- Venkataraman K, Brown KM, Gilmartin GM (2005) Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev 19: 1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E, Rüegsegger U (1999) 3′ End processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev 23: 277–295 [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Steitz JA (1993) Association with terminal exons in pre-mRNAs: a new role for the U1 snRNP? Genes Dev 7: 647–659 [DOI] [PubMed] [Google Scholar]
- Zamore PD, Green MR (1989) Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci USA 86: 9243–9247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Licklider LJ, Gygi SP, Reed R (2002) Comprehensive proteomic analysis of the human spliceosome. Nature 419: 182–185 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1


