Abstract
SPIN90/WISH (SH3 protein interacting with Nck, 90 kDa/Wiskott–Aldrich syndrome protein (WASP) interacting SH3 protein) regulates actin polymerization through its interaction with various actin-regulating proteins. It is highly expressed in the brain, but its role in the nervous system is largely unknown. We report that it is expressed in dendritic spines where it associates with PSD-95. Its overexpression increased the number and length of dendritic filopodia/spines via an N-WASP-independent mechanism, and knock down of its expression with small interfering RNA reduced dendritic spine density. The increase in spinogenesis is accompanied by an increase in synaptogenesis in contacting presynaptic neurons. Interestingly, PSD-95-induced dendritic spinogenesis was completely abolished by knock down of SPIN90/WISH. Finally, in response to chemically induced long-term potentiation, SPIN90/WISH associated with PSD-95 and was redistributed to dendritic spines. Our results suggest that SPIN90/WISH associates with PSD-95, and so becomes localized to dendritic spines where it modulates actin dynamics to control dendritic spinogenesis. They also raise the possibility that SPIN90/WISH is a downstream effector of PSD-95-dependent synaptic remodeling.
Keywords: actin, dendritic spines, PSD-95, SPIN90/WISH, spinogenesis
Introduction
Dendritic spines are the major sites of synapse formation where more than 90% of the excitatory synapses in the central nervous sysytem are generated (Nimchinsky et al, 2002; Tashiro and Yuste, 2003). As the spines are the primary areas of contact of individual axons, changes in their genesis and morphology are considered strong candidates for the cellular mechanism regulating synaptic activity (Tada and Sheng, 2006). Such changes also modify important brain functions such as the formation of long-term memory (Segal and Andersen, 2000; Carlisle and Kennedy, 2005; Halpain et al, 2005; Tada and Sheng, 2006). Actin reorganization is involved in both the formation of dendritic spines during development and their structural plasticity at mature synapses (Matus, 2000; Ethell and Pasquale, 2005). As dendritic spines have an extensive actin cytoskeleton, proteins known to alter the arrangement of actin cytoskeleton, and the mediators of those proteins, have been the primary targets of studies focusing on spine changes (Hayashi and Shirao, 1999; Tashiro et al, 2000; Pak et al, 2001; Penzes et al, 2001; Irie and Yamaguchi, 2002; Hering and Sheng, 2003; Choi et al, 2005). Understanding the mechanisms that control the actin cytoskeleton of dendritic spines may help to reveal the cellular basis of the synaptic plasticity that underlies learning and memory.
The formation and maintenance of dendritic spines require precise targeting and coordinated activation of structural and signaling molecules (Nimchinsky et al, 2002; Yuste and Bonhoeffer, 2004; Tada and Sheng, 2006). Several scaffold proteins, such as GIT1 and intersectin, interact with actin or actin binding proteins in spines (Irie and Yamaguchi, 2002; Zhang et al, 2005), and control the local formation and branching of actin filaments. However, much remains to be determined about the signaling pathways that regulate the formation and pruning of spines. Recent studies show that PSD-95 enhances the maturation of glutamatergic synapses pre- and post-synaptically, and that targeted disruption of PSD-95 alters activity-dependent synaptic plasticity and learning (Migaud et al, 1998; El-Husseini et al, 2000). Although PSD-95 expression increases the number and size of dendritic spines, the underlying signaling pathways are largely unknown.
SPIN90/WISH (SH3 protein interacting with Nck, 90 kDa/Wiskott–Aldrich syndrome protein (WASP) interacting SH3 protein) is a recently cloned protein, originally identified as a binding partner of Nck (Lim et al, 2001) or Ash/Grb2 (Fukuoka et al, 2001). It is highly expressed in many rat tissues including the brain, heart, testis, and skeletal muscle (Fukuoka et al, 2001; Lim et al, 2001, see also Supplementary Figure 1). It contains an N-terminal Src homology 3 (SH3) domain, three central proline-rich domains (PRDs), and a hydrophobic C-terminus, and each of these domains is a potential site of protein–protein interaction (Lim et al, 2001, 2003). SPIN90/WISH activates the Arp2/3 complex by N-WASP-dependent as well as N-WASP-independent mechanisms, inducing the rapid actin polymerization that is required for microspike formation (Fukuoka et al, 2001; Kim et al, 2006). It interacts with N-WASP via its N-terminal SH3 domain, but it is not known how it regulates actin polymerization in an N-WASP-independent manner. We have recently shown that it associates with the Arp2/3 complex via its C-terminal A-like domain (amino acids 336–407), and that this A-like domain contains conserved arginine, tryptophan, and hydrophobic residues essential for strong activation of the Arp2/3 complex (Kim et al, 2006). It also binds directly to G-actin through its verprolin homology (VPH)-like domain, so bringing about Arp2/3-induced actin polymerization in vitro (Kim et al, 2006). These results strongly suggest that the SPIN90/WISH C-terminal hydrophobic region is responsible for inducing actin polymerization in the N-WASP-independent pathway (Kim et al, 2006).
We recently found that SPIN90/WISH is highly expressed in presynaptic terminals where it interacts with dynamin I and participates in synaptic vesicle endocytosis (Kim et al, 2005). We now report that it is also highly expressed in postsynaptic dendritic spines. We have identified PSD-95, a postsynaptic synaptic protein, as a new interaction partner of SPIN90/WISH. Overexpression of SPIN90/WISH resulted in increased length and density of dendritic filopodia and a higher density of spines. As SH3-deleted SPIN90/WISH, which cannot interact with N-WASP, retained its ability to increase the density of dendritic protrusions, it appears to regulate dendritic spine formation in an N-WASP-independent manner. We also report that the increase in dendritic spinogenesis by PSD-95 was completely abolished by knock down of SPIN90/WISH, thus strongly suggesting that SPIN90/WISH is a downstream effector of PSD-95. We observed that SPIN90/WISH translocated from the dendritic shaft into the dendritic spines during chemically induced long-term potentiation (LTP) and that this required it to associate with PSD-95. This raises the possibility that SPIN90/WISH may not only regulate dendritic spinogenesis in developing neurons but also contribute to PSD-95-dependent types of synaptic remodeling in mature neurons.
Results
SPIN90/WISH localizes to dendritic spines in cultured neurons and interacts with PSD-95
To determine the location of SPIN90/WISH in cultured hippocampal neurons, we stained it with a specific SPIN90/WISH antibody and a fluorescent F-actin probe, Texas-Red phalloidin. SPIN90/WISH and actin showed considerable colocalization in dendritic spines (Figure 1A). As SPIN90/WISH contains a PRD domain, it could interact with an SH3 domain-containing protein in the dendritic spines. PSD-95 is a key scaffolding protein of postsynaptic densities (PSDs) and has an SH3 domain. We therefore tested whether SPIN90/WISH interacts with PSD-95. Brain lysates were immunoprecipitated with an SPIN90/WISH antibody and sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gels were silver-stained. After micro-liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis, PSD-95 was identified as a binding partner (Figure 1B). In vivo immunoprecipitation assays confirmed that PSD-95 indeed binds to SPIN90/WISH whereas another MAGUK protein, SAP-97 does not (Figure 1C). Immunocytochemistry showed that SPIN90/WISH is found in dendritic spines, where it colocalizes with PSD-95 (Figure 1D). Exogenously expressed SPIN90/WISH and PSD-95 also colocalize in the dendritic spines of hippocampal neurons (Figure 1E).
Figure 1.
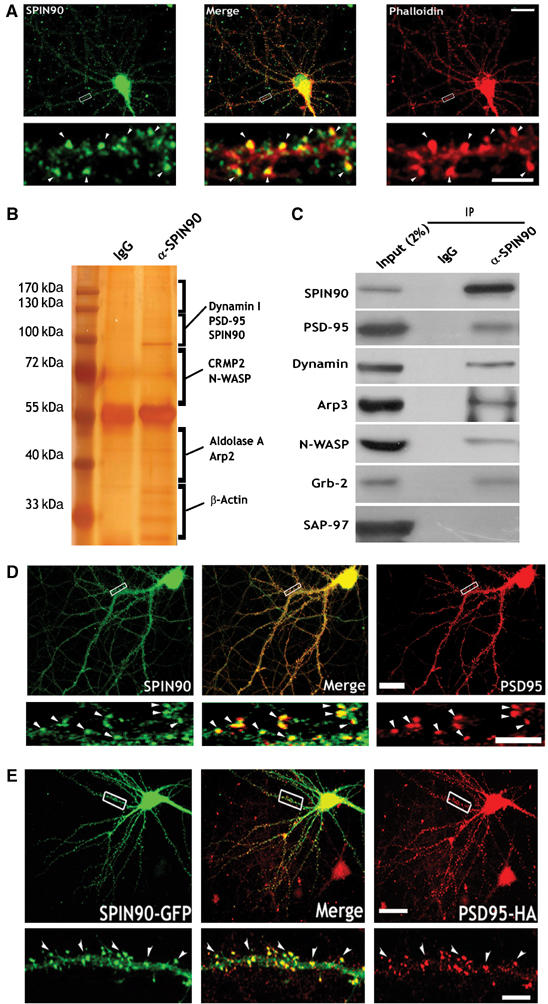
SPIN90/WISH is expressed in dendritic spines and colocalizes with actin and PSD-95. (A) Colocalization of SPIN90/WISH and actin in cultured hippocampal neurons. Neurons at 17DIV were stained with anti-SPIN90/WISH antibody (green) and phalloidin (red). A high-magnification view of the region enclosed by the rectangle is shown below each image. Scale bars, low magnification: 20 μm; high magnification: 5 μm. (B) Brain lysates were immunoprecipitated with anti-SPIN90/WISH antibody, and SDS–PAGE gels were silver stained. Protein bands were excised from the stained gels, analyzed by micro-LC-MS/MS, and identified by a protein database search. Proteins that are unknown or not identified clearly yet were not shown. (C) Rat brain lysates were immunoprecipitated (IP) with anti-SPIN90/WISH antibody and immunoblotted (IB) with the indicated antibodies. IgG: normal rabbit serum. (D) Colocalization of endogenously expressed SPIN90/WISH and PSD-95. Neurons at 17DIV were doubly immunostained with SPIN90/WISH antibody (green) and PSD-95 antibody (red). Arrowheads indicate the dendritic spine regions in which colocalization is readily seen. Scale bars, as before. (E) Colocalization of overexpressed GFP-SPIN90/WISH and PSD-95-HA. Neurons were transfected with GFP-SPIN90/WISH and PSD-95-HA, fixed at 17DIV, and immunostained with GFP antibody for SPIN90/WISH and HA antibody for PSD-95 (red). High-magnification views are of the regions enclosed in rectangles. Scales as before.
SPIN90/WISH binds to PSD-95 via its SH3 domain in vitro
To map the binding site of SPIN90/WISH that interacts with PSD95, we carried out a series of GST pull-down assays (Figure 2A). PSD-95 interacted with the N-terminal region of SPIN90/WISH and with its PRD domain but not with the SPIN90/WISH C-terminus or SPIN90/WISH lacking its PRD domain (SPIN90/WISH-ΔPRD, Figure 2C). Evidently, the PRD region of SPIN90/WISH binds to the SH3 domain of PSD-95. Indeed, full-length PSD-95 or its SH3 domain interacted with SPIN90/WISH, but PSD-95 lacking the SH3 domain did not (Figure 2B and D).
Figure 2.
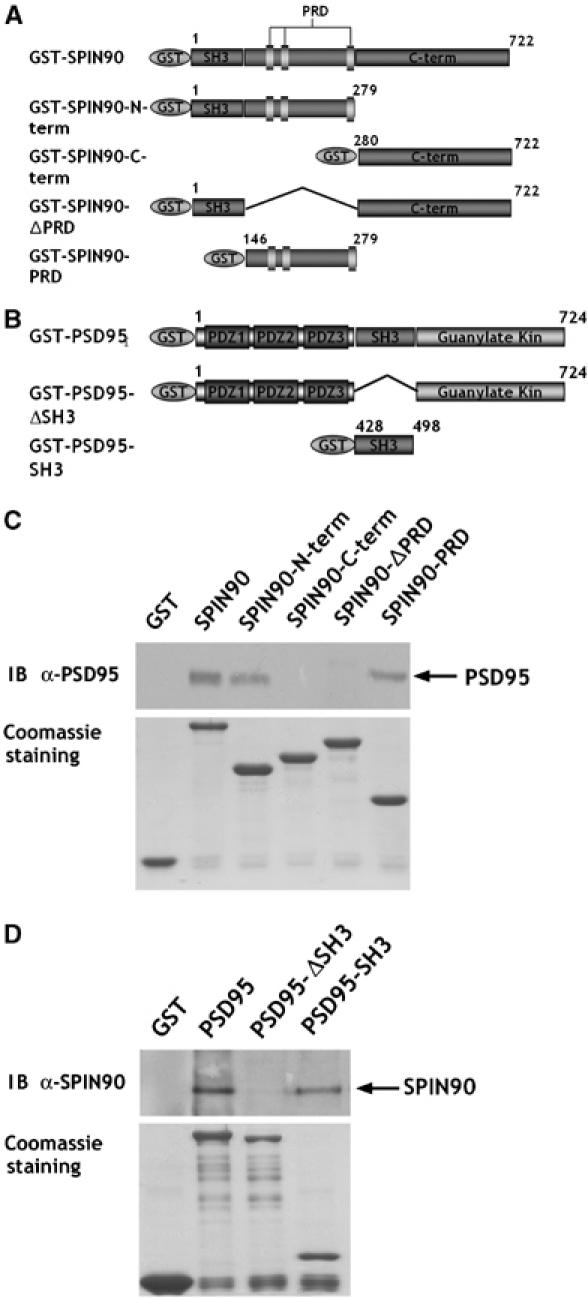
The PRD of SPIN90/WISH interacts with the SH3 domain of PSD-95 in vitro. (A, B) Schematic diagrams of the series of truncated GST-SPIN90/WISH and GST-PSD-95 constructs used in GST pull-down assays. (C) GST alone, GST-tagged full-length SPIN90/WISH or truncated variants were expressed in BL-21, then immobilized on glutathione-Sepharose beads and incubated with rat brain lysates. Bound proteins were analyzed by SDS–PAGE and immunoblotted with PSD-95 antibody. (D) After expression in BL-21, the GST-PSD-95 and truncated variants were immunoblotted with SPIN90/WISH antibody.
Overexpression of SPIN90/WISH induces the formation of dendritic filopodia by an N-WASP-independent mechanism
Dendritic filopodia are actin-rich structures that are directly controlled by alterations of actin dynamics (Ziv and Smith, 1996; Yuste and Bonhoeffer, 2004). The dendritic filopodia of developing neurons are responsible for initiating spine formation following contact with a nearby axon (Ziv and Smith, 1996; Yuste and Bonhoeffer, 2004). As SPIN90/WISH induces actin polymerization and exists in high concentration in dendritic spines, we tested whether it plays a role in the formation of dendritic filopodia in developing neurons. Hippocampal neurons transfected with green fluorescent protein (GFP)-tagged SPIN90/WISH had a higher density of dendritic filopodia than the GFP control (0.25±0.01, n=8 for SPIN90/WISH; 0.14±0.01, n=9 for control, P<0.01, Figure 3A–C), whereas overexpression of SPIN90/WISH-ΔPRD or SPIN90/WISH-PRD had no effect on filopodial density (0.13±0.01, n=11 for SPIN90/WISH-ΔPRD, P=0.47; 0.14±0.01, n=9 for SPIN90/WISH-PRD, P=0.90, Figure 3A–C), suggesting that recruitment of SPIN90/WISH via its interaction with PSD-95 to the site where dendritic filopodia form is required for its effect on filopodial density. Interestingly, overexpression of SPIN90/WISH-ΔSH3, which cannot interact with N-WASP, also increased filopodial density (0.22±0.01, n=10, P<0.01, Figure 3C) indicating that interaction of SPIN90/WISH with N-WASP is not necessary for the induction of dendritic filopodia. Expression of SPIN90/WISH also resulted in an increase of filopodial length whereas the other constructs had no significant effect (Figure 3D).
Figure 3.
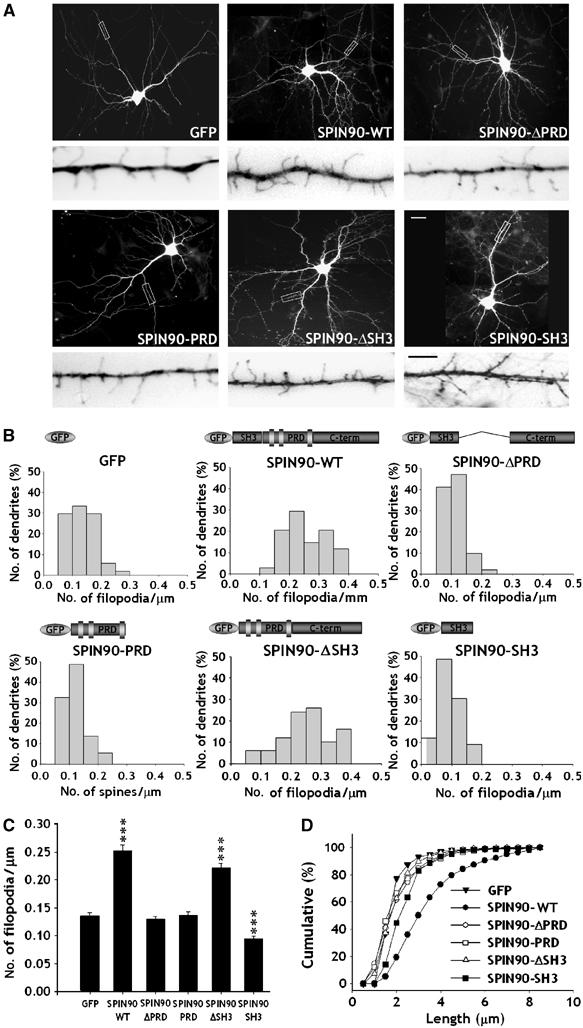
Overexpression of SPIN90/WISH increases the number and length of dendritic filopodia in early hippocampal neurons. Neurons were transfected with various constructs, fixed at 7DIV and immunostained with GFP antibody. (A) GFP fluorescence images of the transfected neurons. High-magnification views are of the regions enclosed in rectangles. Images are inverted for clarity. Scale bars, low magnification: 20 μm; high magnification: 5 μm (B, C) Histograms of the number of dendritic filopodia per μm in the transfected neurons (B); comparison of average numbers of filopodia (C). Data are means±s.e. (GFP, n=9; SPIN90/WISH, n=8; SPIN90/WISH-ΔPRD, n=11; SPIN90/WISH-PRD, n=9; SPIN90/WISH-ΔSH3, n=10; SPIN90/WISH-SH3, n=8; ***P<0.01, significantly different from GFP alone by ANOVA and Tukey's HSD post hoc test). (D) Cumulative frequency distribution of dendritic filopodial length in the transfected neurons.
Overexpression of SPIN90/WISH induces dendritic spine formation by an N-WASP-independent mechanism
We next investigated the effect of SPIN90/WISH on the dendritic spines of neurons at later developmental stages. The average density of spines proved to be higher in cells overexpressing full-length SPIN90/WISH (0.79±0.04, n=8, P<0.01) than in the GFP control (0.41±0.02, n=6, Figure 4A–C). SPIN90/WISH-ΔPRD or SPIN90/WISH-PRD failed to have any effect on the density of dendritic spines (0.43±0.04, n=7 for SPIN90/WISH-ΔPRD, P=0.77; 0.40±0.01, n=6 for SPIN90/WISH-PRD, P=0.58; Figure 4A–C), suggesting that recruitment of SPIN90/WISH to dendritic spines via its interaction with PSD-95 is required for its effect on dendritic spine formation. As in the case of dendritic filopodia, expression of SPIN90/WISH-ΔSH3, which lacks an N-WASP binding site, led to an increase in spine density (0.62±0.04, n=7, P<0.01), whereas SPIN90/WISH-SH3 expression caused a decrease in spine density (0.30±0.01, n=7, P<0.01; Figure 4A–C). These results confirm the idea that although binding of SPIN90/WISH with N-WASP via SH3–PRD interaction can induce actin polymerization (Fukuoka et al, 2001), it is not required for the induction of dendritic spines; instead, an N-WASP-independent mechanism is mainly responsible. The length of dendritic spines was decreased by expression of SPIN90/WISH-SH3, not by the other constructs (Figure 4D).
Figure 4.
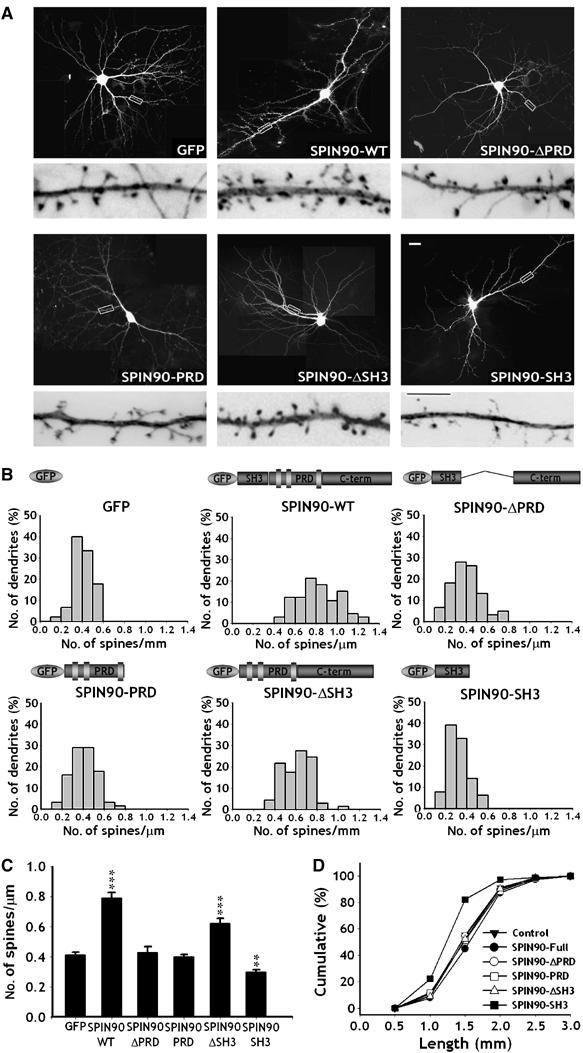
Overexpression of SPIN90/WISH increases the density and length of dendritic spines in mature hippocampal neurons. Neurons were transfected and imaged as in Figure 3. (A) GFP fluorescence images of the transfected neurons. High-magnification views are of the regions enclosed in rectangles. Images are inverted for clarity. Scale bars, low magnification: 20 μm; high magnification: 5 μm. (B, C) Histograms of the number of dendritic spines per μm in the transfected neurons (B); comparison of the average numbers of spines (C). Data are means±s.e. (GFP, n=6; SPIN90/WISH, n=8; SPIN90/WISH-ΔPRD, n=7; SPIN90/WISH-PRD, n=6; GFP-SPIN90/WISH-ΔSH3, n=7; GFP-SPIN90/WISH-SH3, n=7; ***P<0.01 and **P<0.05, significantly different from the GFP alone by ANOVA and Tukey's HSD post hoc test). SPIN90-ΔSH3, which lacks an N-WASP binding site, increased spine density like full-length of SPIN90/WISH, indicating an N-WASP-independent mechanism. (D) Cumulative frequency distribution of dendritic spine lengths in the transfected neurons.
The C-terminal A-like domain is responsible for the effect of SPIN90/WISH on spinogenesis
SPIN90/WISH contains an A-like domain, an Arp2/3 binding region, at its C-terminus and increased Arp2/3-induced actin polymerization in vitro (Kim et al, 2006). We tested whether this A-like domain is the effector domain of SPIN90/WISH and whether deletion of this domain had a dominant-negative effect preventing endogenous SPIN90/WISH from interacting with PSD-95. Figure 5 shows that the average density of spines was lower in cells overexpressing SPIN90/WISH-ΔALD than in the GFP control whereas SPIN90/WISH-ALD had no effect on the density of dendritic spines (0.41±0.01, n=7 for GFP control; 0.32±0.01, n=8 for SPIN90-ΔALD-GFP; 0.39±0.01, n=6 for SPIN90/WISH-ALD, P<0.05), suggesting that A-like domain is responsible for the effect of SPIN90/WISH on spine formation.
Figure 5.
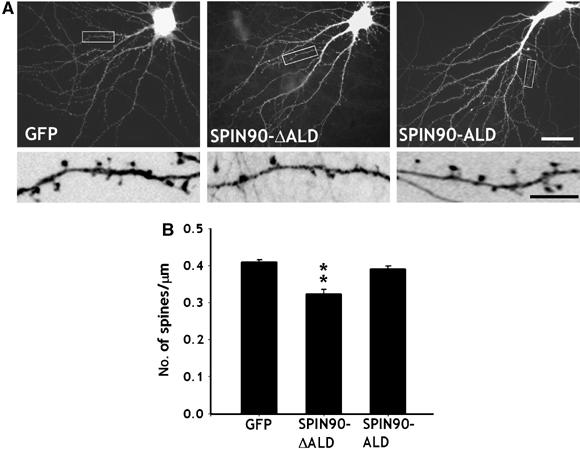
The C-terminal A-like domain is responsible for the effect of SPIN90/WISH on spinogenesis. (A, B) Neurons were transfected with GFP alone, GFP-SPIN90/WISH-ΔALD or GFP-SPIN90/WISH-ALD and visualized at 17DIV by immunofluorescence staining for GFP. High-magnification views are of the regions enclosed in rectangles. Images are inverted for clarity. The average number of spines per μm was analyzed (0.41±0.01, n=7 for GFP control; 0.32±0.01, n=8 for SPIN90-ΔALD-GFP; 0.39±0.01, n=6 for SPIN90/WISH-ALD; **P<0.05 by ANOVA and Tukey's HSD post hoc test). Scale bars, low magnification: 20 μm, high magnification: 5 μm.
SPIN90/WISH expression increases the number of synapses
The formation of dendritic spines in neurons is correlated with synapse formation (Yuste and Bonhoeffer, 2004; Ethell and Pasquale, 2005). We therefore assessed whether overexpression of SPIN90/WISH altered the number of synapses in contact with the dendritic spines of neurons transfected with SPIN90/WISH (Figure 6A–D). When we analyzed synaptobrevin 2 immunoreactivity that was juxtaposed to or merged with GFP-positive dendritic spines, the number of synapses contacting dendritic spines was significantly increased in SPIN90/WISH-transfected neurons (0.51±0.01, n=6 for SPIN90/WISH; 0.26±0.01, n=6 for control, P<0.01). Expression of SPIN90/WISH-ΔPRD or SPIN90/WISH-PRD failed to affect synapse formation (0.26±0.02, n=7 for SPIN90/WISH-ΔPRD, P=0.88; 0.28±0.01, n=6 for SPIN90/WISH PRD, P=0.25; Figure 6E). These results indicate that the increase in spinogenesis induced by SPIN90/WISH is accompanied by an increase in synaptogenesis in contacting presynaptic neurons.
Figure 6.
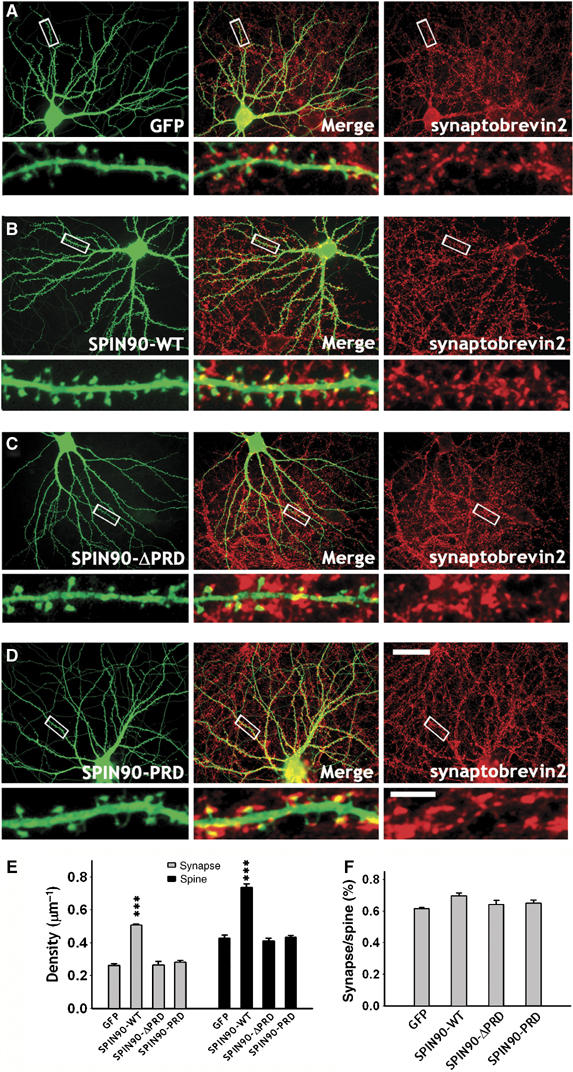
SPIN90/WISH expression increases the number of synapses as well as the density of dendritic spines. (A–D) Neurons were transfected with various GFP-tagged SPIN90/WISH variants and immunostained using GFP (green) and synaptobrevin 2 (red) antibodies. Middle panels are merged images. High-magnification views of the regions enclosed by rectangles are shown below each image. Scale bars, low magnification:, 30 μm, high magnification: 5 μm. (E) Quantification of the number of synapses (gray bars) that contact GFP-positive dendritic spines (black bars). Data are means±s.e. (GFP, n=6; SPIN90/WISH, n=6; SPIN90/WISH-ΔPRD, n=7; SPIN90/WISH-PRD, n=6; ***P<0.01 by ANOVA and Tukey's HSD post hoc test). (F) Correlation between spinogenesis and synaptogenesis. The ratio of synapse density to spine density is displayed.
Knock down of endogenous SPIN90/WISH by small interfering RNA inhibits dendritic spinogenesis
To assess the function of endogenously expressed SPIN90/WISH, we used small-interfering RNA (siRNA) to knock down SPIN90/WISH expression. Suppression of SPIN90/WISH expression by siRNA in Rat-1 fibroblasts was confirmed by immunoblotting (Figure 7A and B). The efficiency of siRNA was further established by immunofluorescence staining of SPIN90/WISH in the hippocampal neurons (Figure 7C). When endogenous SPIN90/WISH was knocked down by siRNA treatment, the density of dendritic spines was reduced (0.41±0.02, n=8 for psiRNA-virgin; 0.20±0.02, n=7 for psiRNA-SPIN90/WISH, P<0.01; Figure 7D and E). In addition, the mean spine length of the psiRNA-SPIN90/WISH-transfected neurons was ∼16% less than that of neurons expressing control vector (1.80±0.06, n=8 for psiRNA-virgin; 1.51±0.1, n=7 for psiRNA-SPIN90/WISH, P=0.02; Figure 7F). This demonstrates that SPIN90/WISH plays a role in neuronal spine formation.
Figure 7.
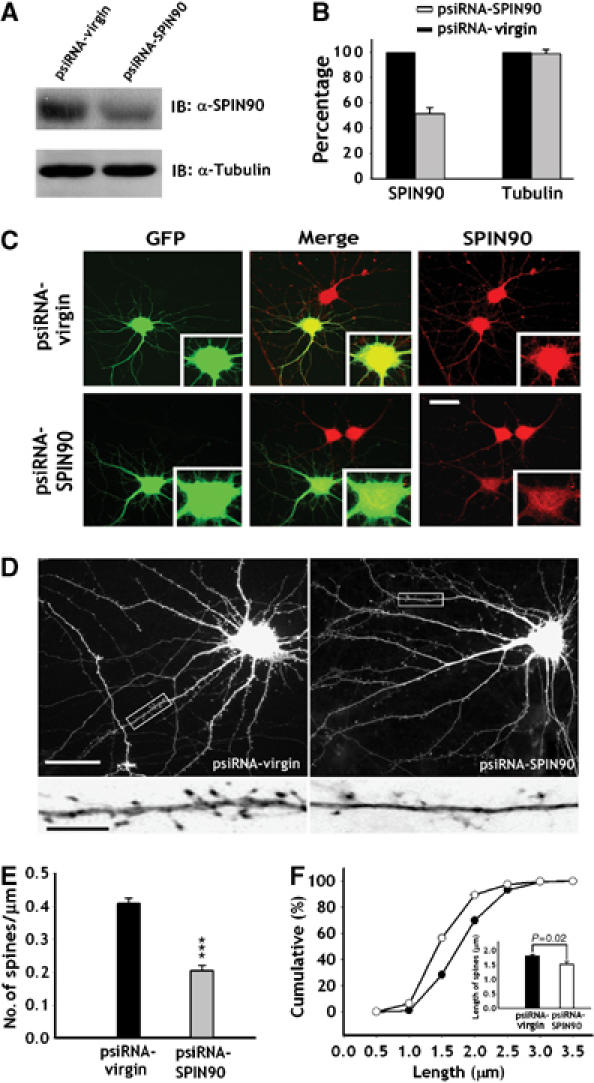
Knock down of endogenous SPIN90/WISH decreases dendritic spine density and length. (A, B) Rat-1 fibroblasts were transfected with psiRNA-virgin and psiRNA-SPIN90/WISH, and lysates were immunoblotted with SPIN90/WISH antibody. Relative band intensities are displayed as histograms. (C) Representative images of SPIN90/WISH knockdown by siRNA. Neurons that had been transfected with psiRNA-SPIN90/WISH were immunostained with GFP antibody (green) and SPIN90/WISH antibody (red). The cell body areas are magnified to show the effect of knock down. Scale bar, 30 μm. (D) GFP fluorescence images of neurons transfected with empty or SPIN90/WISH siRNA vector. The high-magnification views are of the regions enclosed in rectangles. Scale bars, low magnification: 20 μm; high magnification: 5 μm. (E) Analysis of the effect of SPIN90/WISH-siRNA on spine density. Data are means±s.e. (psiRNA-virgin, n=8; psiRNA-SPIN90/WISH, n=7; ***P<0.01, significantly different from the virgin control, Student's t-test). (F) Cumulative frequency distribution of dendritic spine lengths in neurons transfected with psiRNA-virgin (open circles) and psiRNA-SPIN90/WISH (solid circles). Inset gives mean spine lengths.
The effect of PSD-95 on dendritic spinogenesis is abolished by knock down of SPIN90/WISH
Previous studies have shown that overexpression of PSD-95 leads to an increase in spine density as well as in the number of synapses (Migaud et al, 1998; El-Husseini et al, 2000). The fact that SPIN90/WISH interacts with PSD-95, and that overexpression of SPIN90/WISH leads to an increase of spinogenesis, raised the possibility that SPIN90/WISH is a downstream effector of PSD-95-induced spinogenesis. To test this possibility, we overexpressed PSD-95 in a SPIN90/WISH knock down background. In agreement with the previous studies, overexpression of PSD-95 alone resulted in an increase of spine density (0.54±0.02, n=7 for PSD-95; 0.41±0.02, n=6 for control, P<0.01; Figure 8A and B). This effect was completely abolished in neurons in which expression of SPIN90/WISH was knocked down by siRNA (0.26±0.02, n=7 for psiRNA-SPIN90/WISH; 0.50±0.01, n=6 for psiRNA-virgin, P<0.01; Figure 8C and D), suggesting that the stimulatory effect of PSD-95 on spine formation is mediated by SPIN90/WISH.
Figure 8.
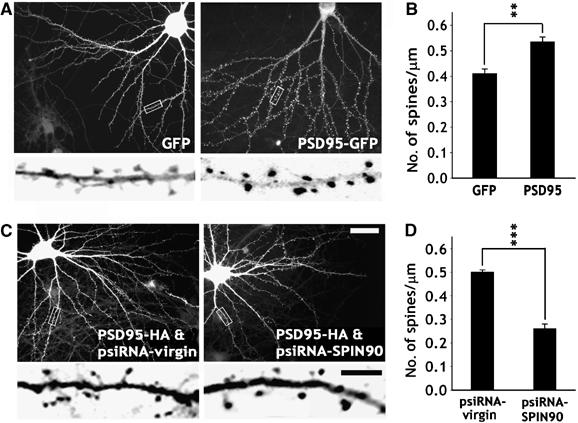
The effect of PSD-95 on dendritic spinogenesis is mediated by SPIN90/WISH. (A, B) Neurons were transfected with GFP alone or PSD-95-GFP and visualized at 17DIV by immunoassaying for GFP. High-magnification views are of the regions enclosed in rectangles. The average number of spines per μm was analyzed (GFP, n=5; PSD-95-GFP, n=8; **P<0.05, Student's t-test). Scale bars, low magnification: 20 μm, high magnification: 5 μm (C, D) Neurons were cotransfected with PSD-95-HA and psiRNA-virgin, or with PSD-95-HA and psiRNA-SPIN90/WISH, and immunostained at 17DIV with HA and GFP antibodies. Only GFP images are shown. High-magnification views are of the regions enclosed in rectangles. Average number of spines per μm are presented (psiRNA-virgin/PSD-95-HA, n=6; psiRNA-SPIN90/WISH/PSD-95-HA, n=7; ***P<0.01, Student's t-test).
SPIN90/WISH is enriched in dendritic spines and translocates in response to chemically induced LTP
Spines undergo activity-dependent morphological changes that include the creation of new spines or splitting of existing ones (Segal and Andersen, 2000; Carlisle and Kennedy, 2005; Halpain et al, 2005; Tada and Sheng, 2006). PSD-95 is also known to mimic LTP and so to be involved in synaptic plasticity and learning (Migaud et al, 1998; Beique and Andrade, 2003; Stein et al, 2003; Ehrlich and Malinow, 2004; Yao et al, 2004). Because SPIN90/WISH binds to PSD-95 and acts as a downstream effector of PSD-95-induced spinogenesis, we asked whether the distribution of SPIN90/WISH changes in response to synaptic activity. To ensure that a very large proportion of the synapses and spines examined had been potentiated, chemical LTP was induced (Hosokawa et al, 1995; Zakharenko et al, 2001). After challenging with chemical LTP protocol, the number of synapses contacting dendritic spines increased, suggesting that LTP had indeed been induced (Supplementary Figure 2). When chemical LTP was induced, the fluorescence intensities of spines that were transfected with GFP full-length SPIN90/WISH increased dramatically (Figure 9A and B). Furthermore, the ratio of fluorescence intensities in dendritic spines and shafts (Fspine/Fshaft) increased, indicating that chemical LTP causes a redistribution of SPIN90/WISH from dendritic shafts to spines (Figure 9D). SPIN90/WISH-ΔPRD did not produce either of these effects (Figure 9C), suggesting that the translocation of SPIN90/WISH is mediated by interaction with PSD-95. We confirmed this notion by showing that coexpression of SPIN90/WISH and PSD-95-SH3 caused no change in the fluorescence of the dendritic spines or in the ratio Fspine/Fshaft after induction of chemical LTP (Figure 9C and D). Taken together, our results suggest that as a result of associating with PSD-95, SPIN90/WISH undergoes activity-dependent translocation from dendritic shafts to dendritic spines.
Figure 9.
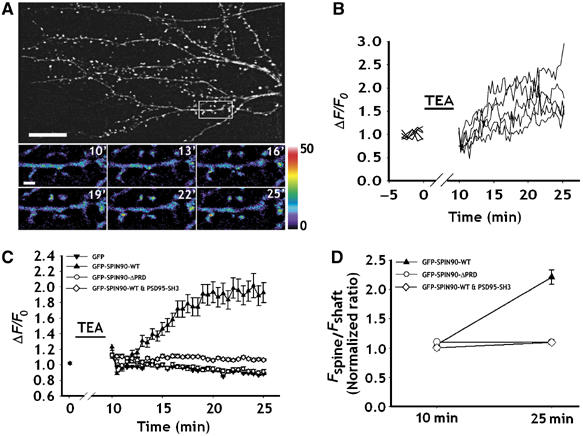
SPIN90/WISH is redistributed into dendritic spines by chemically induced LTP. Neurons were transfected with various GFP-tagged SPIN90/WISH constructs and chemical LTP was induced. Time-lapse images were acquired every 30 s for 15 min after TEA treatment. (A) Representative fluorescence images of neurons that underwent chemical LTP. High-magnification views are of the dendritic spine regions enclosed in rectangles. The color scale gives fluorescence intensities in arbitrary fluorescence units. Scale bar, 1 μm. (B) The curves represent the changes in fluorescence intensity of each dendritic spine in (A). TEA indicates the time of application of medium inducing chemical LTP. (C) Quantitative analysis of the normalized fluorescence intensities of the GFP signal in the dendritic spines. F0 indicates average fluorescence intensity before TEA treatment. Net fluorescence changes (ΔF) and ΔF/F0 values were obtained as described in Materials and methods. (D) The ratios of fluorescence intensities of dendritic spines over shafts (Fspine/Fshaft). Black triangles: ratios in GFP-SPIN90/WISH-transfected neurons; white diamonds: ratios in GFP-SPIN90/WISH-ΔPRD-transfected neurons. Ten minutes refers to immediately after TEA treatment and 25 min indicates the end of time-lapse recording. Data are means±s.e.
Discussion
SPIN90/WISH was originally identified as a NCK binding protein, and its characterization has mainly focused on the regulation of actin polymerization in non-neuronal cells (Kim et al, 2006). However, as it is a binding partner of N-WASP (Fukuoka et al, 2001), dynamin (Kim et al, 2005), and other types of protein (Lim et al, 2003), it is a strong candidate for an actin modulator in neuronal systems. Although its role in neurons is not fully understood, the complexity of its binding partners and its high level of expression in the brain make this inference plausible.
Dendritic spines and filopodia are rich in F-actin, and regulation of the actin cytoskeleton controls the formation, motility, and stability of filopodia as well as the maturation of spines (Ziv and Smith, 1996; Halpain, 2000; Matus, 2000; Yuste and Bonhoeffer, 2004; Ethell and Pasquale, 2005; Tada and Sheng, 2006). Overexpression of SPIN90/WISH in neurons caused significant increases of filopodial density and length and, subsequently, of spine density. SPIN90/WISH-mediated modulation of the actin cytoskeleton has been suggested to involve N-WASP-dependent as well as N-WASP-independent mechanisms (Fukuoka et al, 2001; Kim et al, 2006). SPIN90/WISH strongly enhanced N-WASP-induced activation of the Arp2/3 complex independent of Cdc42 (Kim et al, 2006). However, it turned out that SPIN90/WISH could activate actin polymerization even in N-WASP-depleted extracts, suggesting that SPIN90/WISH can activate the Arp2/3 complex through both N-WASP-dependent and -independent pathways without Cdc42 (Fukuoka et al, 2001). In a separate biochemical study, we found that the C-terminus of SPIN90/WISH contains a VPH-like domain (amino acids 527–549), which is known to bind G-actin and an A-like domain (amino acids 336–407), which is a binding site for the Arp2/3 complex (Kim et al, 2006). Indeed, SPIN90/WISH polymerizes actin in vitro and induces actin comet formation in non-neuronal cells (Kim et al, 2006). Hence, SPIN90/WISH regulates actin polymerization in an N-WASP-independent manner via its C-terminal hydrophobic region. We showed above that SPIN90/WISH induces dendritic filopodia and spine formation via an N-WASP-independent mechanism, as SPIN90/WISH lacking its SH3 domain (the N-WASP-interacting region) still increased the density of dendritic filopodia and spines. However, we cannot rule out the possibility that an N-WASP-dependent pathway makes some contribution to the effect of SPIN90/WISH, as the increase in the density of filopodia and spines in response to SPIN90/WISH-ΔSH3 was not fully comparable to that caused by full-length SPIN90/WISH (87.9% of the full-length effect on filopodia, 78.7% of the full-length effect on spines, Figures 3C and 4C). Moreover, expression of the SPIN90/WISH-SH3 domain resulted in a small but significant decrease in the density of dendritic filopodia and spines (30.5% decrease in filopodial density, 27.5% decrease in spine density). The bulk of the effect of SPIN90/WISH on dendritic filopodia and spine formation is, however, clearly mediated by an N-WASP-independent mechanism.
We showed that SPIN90/WISH binds to PSD-95, a well-known scaffolding protein in PSDs. Indeed, micro-LC-MS/MS analysis showed that PSD-95 is a major postsynaptic binding partner of SPIN90/WISH-PRD in brain lysates (Figure 1B and C). PSD-95 is known to enhance the maturation of glutamatergic synapses; its overexpression increases the number and size of dendritic spines as well as the number of synapses (Migaud et al, 1998; El-Husseini et al, 2000). Its effect is related to that of LTP (Migaud et al, 1998; Beique and Andrade, 2003; Stein et al, 2003; Ehrlich and Malinow, 2004; Yao et al, 2004), and targeted disruption of PSD-95 alters activity-dependent synaptic plasticity and learning (Migaud et al, 1998; El-Husseini et al, 2000). It does not, however, interact directly with the actin cytoskeleton, and the mechanism by which it modulates dendritic morphogenesis is largely unknown. Recent studies show that SPAR (Pak et al, 2001), Kalirin-7 (Penzes et al, 2001), Drebrin (Takahashi et al, 2003), and IRSp53 (Choi et al, 2005) interact with PSD-95 and regulate dendritic spine morphogenesis. SPAR is a RapGAP and its overexpression causes spine enlargement and morphological changes (Pak et al, 2001). Drebrin causes synaptic targeting of PSD-95 and regulates filopodia/spine maturation (Takahashi et al, 2003). Kalirin-7 is a Rho-GEF, and IRSp53 is a downstream effector of Rac1 and Cdc42. Their overexpression increases the density of dendritic spines and siRNA downregulation of IRSp53 decreases it (Penzes et al, 2001; Choi et al, 2005). The effects of SPIN90/WISH on spines differ from the above proteins; for instance, SPIN90/WISH affects spine formation without apparent effect on head enlargement or spine maturation, and it specifically interacts with PSD-95, leading to an increase of spinogenesis whereas the above proteins also interact with other PSD-95/Discs large/zona occludens-1(PDZ) containing proteins such as SAP-97 or Chapsyn-110. Taken together, these findings suggest that PSD-95 interacts with many downstream partners that could contribute to multiple distinct mechanisms of PSD-95-induced spine morphogenesis.
Actin probably plays a central role in the morphological changes of spines that occur in response to a variety of neural events, including the evocation of LTP (Matus, 2000; Segal and Andersen, 2000; Yuste and Bonhoeffer, 2001; Nimchinsky et al, 2002; Ethell and Pasquale, 2005; Tada and Sheng, 2006). The stimuli that produce the various forms of LTP cause rapid local increments in the extensions of filopodia and formation of new spines at the sites of stimulation (Engert and Bonhoeffer, 1999; Maletic-Savatic et al, 1999; Yuste and Bonhoeffer, 2004). We showed that SPIN90/WISH translocates from dendritic shafts to spines in response to chemically induced LTP, and that this effect requires interaction of SPIN90/WISH with PSD-95. This implies that SPIN90/WISH not only regulates dendritic spinogenesis in developing neurons but also contributes to synaptic remodeling in mature neurons. As other proteins also modulate spine morphology, further investigation is needed to see how the different mechanisms are integrated to control dendritic spinogenesis and activity-dependent synaptic plasticity.
Materials and methods
Antibodies and plasmid constructs
Anti-SPIN90/WISH antibody (Lim et al, 2001) (rabbit polyclonal or mouse polyclonal) and anti-PSD95 antibody (Choi et al, 2005) (rabbit polyclonal, kindly provided by Dr Eoonjun Kim of the KAIST, Daejon, Korea) have been previously described. The following additional antibodies were used: anti-GFP (rabbit polyclonal), anti-SAP-97 (rabbit polyclonal), ant-N-WASP (rabbit polyclonal) and anti-Grb2 (mouse monoclonal) (all from Abcam, Cambridge, UK), anti-dynamin I (Affinity Bioreagents, Golden, CO), anti-HA (rat monoclonal, Roche, Penzberg, Germany), anti-tubulin (Sigma, St Louis, MO), and Oregon-Green conjugated goat-anti-rabbit, Oregon-Green conjugated goat-anti-mouse, Texas-Red-conjugated goat-anti-rat (Invitrogen, Carlsbad, CA). Full-length SPIN90/WISH, as well as SPIN90/WISH ΔPRD, PRD, ΔSH3, and SH3 were subcloned in-frame into vector pEGFP (Clontech, Palo Alto, CA). Full-length, ΔPRD, PRD, and the N-terminus and C-terminus of SPIN90/WISH were subcloned in-frame into pGEX4T-1 (Amersham Bioscience, Arlington, IL), as previously described (Lim et al, 2001). Full-length PSD-95, and ΔSH3, SH3 of PSD-95, were amplified by PCR and subcloned into pGEX4T-1 for the GST pull-down assays.
Culture and transfection of hippocampal neurons
Cultured hippocampal neurons were prepared from embryonic day 18 fetal Sprague–Dawley rats, as described (Chang and De Camilli, 2001). They were plated on poly-D-lysine-coated 18 mm glass coverslips and grown in Neurobasal medium (Invitrogen) supplemented with 2% B-27 and 0.5 mM L-glutamine. The neurons were transfected at 5 or 14DIV by the calcium-phosphate method (Chang and De Camilli, 2001). Briefly, 8–10 μg of cDNA and 7.5 μl of 2 M CaCl2 were mixed in distilled water to a total volume of 75 μl, and same volume of 2 × BBS was added. The cell culture medium was completely replaced by transfection medium (MEM, 1 mM pyruvate, 0.6% glucose, 10 mM glutamine, and 10 mM N-2-hydroxyl piperazine-N′-2-ethane sulfonic acid (HEPES), pH 7.65), and the cDNA mixture was added to the cells, which were then incubated in a 5% CO2 incubator for 90 min. They were washed twice with transfection medium (pH 7.35) and then returned to the original culture medium.
Immunocytochemistry
Immunocytochemistry was performed as previously described (Chang and De Camilli, 2001). Briefly, cells were fixed in 4% paraformaldehyde/4% sucrose/phosphate-buffered saline (PBS) for 15 min, washed 2 × 5 min in PBS, and permeabilized for 5 min in 0.25% Triton X-100/PBS. They were blocked for 30 min in 10% bovine serum albumin (BSA)/PBS at 37°C and incubated in 3% BSA/PBS/primary antibody for 2 h at 37°C, or overnight at 4°C, and then washed 6 × 2 min in PBS, followed by an additional incubation for 45 min at 37°C in secondary antibody/3% BSA/PBS.
Co-immunoprecipitation and immunoblotting
Adult rat brains were washed with cold PBS and homogenized in modified radio-immunoprecipitation assay (RIPA) buffer (50 mM Tris–HCl, pH 7.5, 5 mM ethylene diaminetetra acetic acid, 150 mM NaCl, 1% NP-40, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 mM leupeptin, 1.5 mM pepstatin, and 1 mM aprotinin) with a Teflon glass-homogenizer. The extract was then clarified by centrifugation at 100 000 g for 1 h, and the supernatant protein concentration was determined with a Bradford Protein assay Reagent kit (Bio-Rad, Hercules, CA). Samples containing 5 mg of total protein were immunoprecipitated for 2 h, following by an additional 1 h of incubation at 4°C with protein A-Sepharose beads (Amersham Biosciences). The precipitates were extensively washed with RIPA buffer, fractionated by SDS–PAGE, and transferred to a polyvinylidine diflouride membrane (Bio-Rad). The membrane was blocked with 5% skim milk TBST (10 mM Tris–HCl, 100 mM NaCl, and 0.1% Tween-10, pH 7.5) for 1 h, washed, and probed with primary antibody for 1 h at room temperature. After extensive washing in TBST, the membrane was incubated with Horseradish peroxidase-conjugated secondary antibody (Jackson Immuno Research Laboratories, West Grove, PA). Proteins were visualized with enhanced chemiluminescence reagent (Amersham Biosciences). The images of the bands were scanned with a GS710 densitometer (Bio-Rad), and quantified with Science Lab Image software.
GST pull-down assay
The various plasmids were transformed into Escherichia coli BL-21 and the cells were cultured in LB medium supplemented with ampicillin. After overnight induction with 0.5 mM IPTG at 25°C, the cells were sonicated in lysis buffer (1% Triton X-100, 0.5% sodium deoxy-cholate, 20 mM Tris, pH 8.0, 150 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 0.1 mM PMSF). The sonicates were centrifuged for 15 min at 12 000 r.p.m., and the resulting supernatants incubated with glutathione-Agarose-4B beads (Amersham Biosciences) at 4°C for 30 min. After three washes with lysis buffer, the beads were incubated with brain homogenate in lysis buffer for 2 h at 4°C, washed extensively with lysis buffer and analyzed by SDS–PAGE and immunoblotting.
In-gel digestion and peptide sample preparation
The SDS–polyacrylamide gels were silver-stained and protein bands were excised from the stained gel. The resulting samples were washed three times with 1:1 (v/v) solution of acetonitrile/deionized water for 10 min, dehydrated with 100% acetonitrile, washed with 1:1 (v/v) solution of 100% acetonitrile: 100 mM ammonium bicarbonate and dried using a Speed-Vac. Then samples were reduced with 10 mM Tris(2-carboxyethyl)phosphine hydrochloride in 0.1 M ammonium bicarbonate at 56°C for 45 min and alkylated with 55 mM iodoacetamide in 0.1 M ammonium bicarbonate at room temperature for 30 min. The above washing step was repeated on the alkylated samples, which were dried, soaked in sequencing-grade trypsin solution (500 ng) on ice for 45 min and immersed in 100 μl of 50 mM ammonium bicarbonate (pH 8.0) at 37°C for 14–18 h. The resulting peptides were extracted sequentially by agitation for 20 min with 45% acetonitrile in 20 mM ammonium bicarbonate, 45% acetonitrile in 0.5% trifluoroacetic acid (TFA) and 75% acetonitrile in 0.25% TFA. The extracts containing tryptic peptides were pooled and evaporated under vacuum.
Micro-LC-MS/MS analysis and protein database search
In gel digested proteins were loaded onto fused silica capillary columns (100 μm i.d., 360 μm o.d.) containing 8 cm of 5 μm particle size Aqua C18 reverse-phase column material. The columns were placed in line with an Agilent HP 1100 quaternary LC pump, and a splitter system was used to achieve a flow rate of 250 nl/min. Buffer A (5% acetonitrile and 0.1% formic acid) and buffer B (80% acetonitrile and 0.1% formic acid) were used to make a 90-min gradient. The gradient profile started with 5 min of 100% buffer A, followed by a 60-min gradient from 0 to 55% buffer B, a 25-min gradient from 55 to 100% buffer B, and a 5-min gradient of 100% buffer B. Eluted peptides were directly electrosprayed into an LTQ linear ion trap mass spectrometer (ThermoFinnigan, Palo Alto, CA) by applying 2.3 kV of DC voltage. Data-dependent scans consisting of one full MS scan (400–1400 m/z) and five data-dependent MS/MS scans were used to generate MS/MS spectra of the eluted peptides. A normalized collision energy of 35% was used throughout the data acquisition. MS/MS spectra were searched against an NCBI rat protein sequence database using Bioworks Ver. 3.1 and Sequest Cluster System (14 nodes). DTASelect was used to filter the search results and the following Xcorr values were applied to different charge states of peptides: 1.8 for singly charged peptides, 2.2 for doubly charged peptides, and 3.2 for triply charged peptides. Manual assignments of fragment ions in each MS/MS spectrum were performed to confirm the protein database search results.
RNA interference
siRNA for SPIN90/WISH was designed from nucleotides 1771–1791 of the rat SPIN90/WISH cDNA sequence (GenBank accession number XP_238555). Complementary oligonucleotides (66 nt) were synthesized separately, with the addition of an Acc65I site at the 5′ end and a HindIII site at the 3′ end. The forward primer sequence was 5′-GTACCTCGATCCTGTGCGTATCTTCAGATCAA GAGTCTGAAGATACGCACAGGATCTTTTTGGAAA- 3′, where the underlined letters represent the SPIN90/WISH siRNA target sequence. The annealed 66 bp cDNA fragment of SPIN90/WISH siRNA was cloned into the Acc65I–HindIII sites of the vector, psiRNA-hH1GFPzeo G2 (InvivoGen, San Diego, CA). The efficiency of siRNA was tested on Rat-1 cells of rat fibroblast origin, and confirmed in neurons as previously described (Kim et al, 2005).
Image acquisition and data analysis
Images were obtained with an Olympus IX-71 inverted microscope (Olympus Optical, Tokyo, Japan) with a 40 × , 1.0. NA or a 60 × 1.4 NA oil lens using a CoolSNAP-Hq CCD camera (Roper Scientific, Tucson, AZ) driven by MetaMorph imaging software (Universal Imaging Corporation, West Chester, PA) with a GFP optimized filter set (Omega Optical, Brattleboro, VT). Analysis and quantification of data were performed with MetaMorph software and statistical analyses were performed using SigmaStat (Systat Software, Point Richmond, CA). For multiple conditions, we compared means by analysis of variance (ANOVA). All data found to be significant by ANOVA were compared with Tukey's honestly significant difference (HSD) or Fisher's least significant difference (depending on the number of groups) post hoc test to reveal statistically different groups. Statistical differences between two conditions were determined using Student's t-test. Data are presented as means±s.e.
Live-cell imaging
For live-cell imaging of translocation of SPIN90/WISH to the dendritic spines in response to chemical LTP, hippocampal neurons were transfected with GFP-tagged full-length and truncated forms of SPIN90/WISH. Immediately before image acquisition, cells were mounted in a temperature-controlled perfusion chamber (Chamlide AC-S-10, LCI, Seoul, Korea) on the stage of an Olympus IX-71 microscope and perfused at 30°C with Tyrode's solution (119 mM NaCl, 2.5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 25 mM HEPES, pH 7.4, 30 mM glucose) using an inline heating system (IHS-101, LCI). Control images were acquired every 30 s for 3 min using a CoolSNAP-Hq CCD camera driven by MetaMorph Imaging software with a GFP optimized filter set, and chemical LTP was induced by exposing the cells to modified artificial cerebrospinal fluid solution (123 mM NaCl, 5 mM KCl, 1.25 mM KH2PO4, 24 mM NaHCO3, 0.1 mM MgCl2, 5 mM CaCl2, 10 mM D-glucose, and 25 mM tetraethylammonium chloride, pH 7.2) for 10 min (Hosokawa et al, 1995; Zakharenko et al, 2001). The cells were then continuously perfused with Tyrode's solution and time-lapse images were acquired every 30 s for 15 min. The fluorescence intensity of individual spines was measured by averaging a selected area of pixel intensities using MetaMorph software. Net fluorescence changes (ΔF) were obtained for individual spines by subtracting the average of the intensities of the first six frames (F0) from the intensity of each frame (Ft). They were then divided by F0 to yield ΔF/F0 values. Analysis and quantification of data were performed using MetaMorph software.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Acknowledgments
This research was supported by a grant from the Brain Research Center of the 21st Century Frontier Research Program (M103KV010009-06K2201-00910) to SC funded by the Ministry of Science and Technology, Republic of Korea.
References
- Beique J, Andrade R (2003) PSD-95 regulates synaptic transmission and plasticity in rat cerebral cortex. J Physiol 546: 859–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle H, Kennedy M (2005) Spine architecture and synaptic plasticity. Trends Neurosci 28: 182–187 [DOI] [PubMed] [Google Scholar]
- Chang S, De Camilli P (2001) Glutamate regulates actin-based motility in axonal filopodia. Nat Neurosci 4: 787–793 [DOI] [PubMed] [Google Scholar]
- Choi J, Ko J, Racz B, Burette A, Lee JR, Kim S, Na M, Lee HW, Kim K, Weinberg RJ, Kim E (2005) Regulation of dendritic spine morphogenesis by insulin receptor substrate 53, a downstream effector of Rac1 and Cdc42 small GTPases. J Neurosci 25: 869–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich I, Malinow R (2004) Postsynaptic density 95 controls AMPA receptor incorporation during long-term potentiation and experience-driven synaptic plasticity. J Neurosci 24: 916–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS (2000) PSD-95 involvement in maturation of excitatory synapses. Science 290: 1364–1368 [PubMed] [Google Scholar]
- Engert F, Bonhoeffer T (1999) Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature 399: 19–21 [DOI] [PubMed] [Google Scholar]
- Ethell I, Pasquale E (2005) Molecular mechanisms of dendritic spine development and remodeling. Prog Neurobiol 75: 161–205 [DOI] [PubMed] [Google Scholar]
- Fukuoka M, Suetsugu S, Miki H, Fukami K, Endo T, Takenawa T (2001) A novel neural Wiskott-Aldrich syndrome protein (N-WASP) binding protein, WISH, induces Arp2/3 complex activation independent of Cdc42. J Cell Biol 152: 471–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpain S (2000) Actin and the agile spine: how and why do dendritic spines dance? Trends Neurosci 23: 141–146 [DOI] [PubMed] [Google Scholar]
- Halpain S, Spencer K, Graber S (2005) Dynamics and pathology of dendritic spines. Prog Brain Res 147: 29–37 [DOI] [PubMed] [Google Scholar]
- Hayashi K, Shirao T (1999) Change in the shape of dendritic spines caused by overexpression of drebrin in cultured cortical neurons. J Neurosci 19: 3918–3925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hering H, Sheng M (2003) Activity-dependent redistribution and essential role of cortactin in dendritic spine morphogenesis. J Neurosci 23: 11759–11769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa T, Rusakov DA, Bliss TV, Fine A (1995) Repeated confocal imaging of individual dendritic spines in the living hippocampal slice: evidence for changes in length and orientation associated with chemically induced LTP. J Neurosci 15: 5560–5573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie F, Yamaguchi Y (2002) EphB receptors regulate dendritic spine development via intersectin, Cdc42 and N-WASP. Nat Neurosci 5: 1117–1118 [DOI] [PubMed] [Google Scholar]
- Kim DJ, Kim SH, Lim CS, Choi KY, Park CS, Sung BH, Yeo MG, Chang S, Kim JK, Song WK (2006) Interaction of SPIN90 with the Arp2/3 complex mediates lamellipodia and actin comet tail formation. J Biol Chem 25: 9515–9523 [DOI] [PubMed] [Google Scholar]
- Kim Y, Kim S, Lee S, Kim SH, Park ZY, Song WK, Chang S (2005) Interaction of SPIN90 with dynamin I and its participation in synaptic vesicle endocytosis. J Neurosci 25: 9515–9523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CS, Kim SH, Jung JG, Kim JK, Song WK (2003) Regulation of SPIN90 phosphorylation and interaction with Nck by ERK and cell adhesion. J Biol Chem 278: 52116–52123 [DOI] [PubMed] [Google Scholar]
- Lim CS, Park ES, Kim DJ, Song YH, Eom SH, Chun JS, Kim JH, Kim JK, Park D, Song WK (2001) SPIN90 (SH3 protein interacting with Nck, 90 kDa), an adaptor protein that is developmentally regulated during cardiac myocyte differentiation. J Biol Chem 276: 12871–12878 [DOI] [PubMed] [Google Scholar]
- Maletic-Savatic M, Malinow R, Svoboda K (1999) Rapid dendritic morphogenesis in CA1 hippocampal dendrites induced by synaptic activity. Science 283: 1923–1927 [DOI] [PubMed] [Google Scholar]
- Matus A (2000) Actin-based plasticity in dendritic spines. Science 290: 754–758 [DOI] [PubMed] [Google Scholar]
- Migaud M, Charlesworth P, Dempster M, Webster LC, Watabe AM, Makhinson M, He Y, Ramsay MF, Morris RG, Morrison JH, O'Dell TJ, Grant SG (1998) Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. Nature 396: 433–439 [DOI] [PubMed] [Google Scholar]
- Nimchinsky E, Sabatini B, Svoboda K (2002) Structure and function of dendritic spines. Annu Rev Physiol 64: 313–353 [DOI] [PubMed] [Google Scholar]
- Pak DT, Yang S, Rudolph-Correia S, Kim E, Sheng M (2001) Regulation of dendritic spine morphology by SPAR, a PSD-95-associated RapGAP. Neuron 31: 289–303 [DOI] [PubMed] [Google Scholar]
- Penzes P, Johnson RC, Sattler R, Zhang X, Huganir RL, Kambampati V, Mains RE, Eipper BA (2001) The neuronal Rho-GEF Kalirin-7 interacts with PDZ domain-containing proteins and regulates dendritic morphogenesis. Neuron 29: 229–242 [DOI] [PubMed] [Google Scholar]
- Segal M, Andersen P (2000) Dendritic spines shaped by synaptic activity. Curr Opin Neurobiol 10: 582–586 [DOI] [PubMed] [Google Scholar]
- Stein V, House DR, Bredt DS, Nicoll RA (2003) Postsynaptic density-95 mimics and occludes hippocampal long-term potentiation and enhances long-term depression. J Neurosci 23: 5503–5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T, Sheng M (2006) Molecular mechanisms of dendritic spine morphogenesis. Curr Opin Neurobiol 16: 95–101 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Sekino Y, Tanaka S, Mizui T, Kishi S, Shirao T (2003) Drebrin-dependent actin clustering in dendritic filopodia governs synaptic targeting of postsynaptic density-95 and dendritic spine morphogenesis. J Neurosci 23: 6586–6595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro A, Minden A, Yuste R (2000) Regulation of dendritic spine morphology by the rho family of small GTPases: antagonistic roles of Rac and Rho. Cereb Cortex 10: 927–938 [DOI] [PubMed] [Google Scholar]
- Tashiro A, Yuste R (2003) Structure and molecular organization of dendritic spines. Histol Histopathol 18: 617–634 [DOI] [PubMed] [Google Scholar]
- Yao WD, Gainetdinov RR, Arbuckle MI, Sotnikova TD, Cyr M, Beaulieu JM, Torres GE, Grant SG, Caron MG (2004) Identification of PSD-95 as a regulator of dopamine-mediated synaptic and behavioral plasticity. Neuron 41: 625–638 [DOI] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T (2001) Morphological changes in dendritic spines associated with long-term synaptic plasticity. Annu Rev Neurosci 24: 1071–1089 [DOI] [PubMed] [Google Scholar]
- Yuste R, Bonhoeffer T (2004) Genesis of dendritic spines: insights from ultrastructural and imaging studies. Nat Rev Neurosci 5: 24–34 [DOI] [PubMed] [Google Scholar]
- Zakharenko S, Zablow L, Siegelbaum S (2001) Visualization of changes in presynaptic function during long-term synaptic plasticity. Nat Neurosci 4: 711–717 [DOI] [PubMed] [Google Scholar]
- Zhang H, Webb DJ, Asmussen H, Niu S, Horwitz AF (2005) A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J Neurosci 25: 3379–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv NE, Smith SJ (1996) Evidence for a role of dendritic filopodia in synaptogenesis and spine formation. Neuron 17: 91–102 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2


