Abstract
The trimeric HIV/SIV envelope glycoprotein, gp160, is cleaved to noncovalently associated fragments, gp120 and gp41. Binding of gp120 to viral receptors leads to large structural rearrangements in both fragments. The unliganded gp120 core has a disordered β3–β5 loop, which reconfigures upon CD4 binding into an ordered, extended strand. Molecular modeling suggests that residues in this loop may contact gp41. We show here that deletions in the β3–β5 loop of HIV-1 gp120 weaken the binding of CD4 and prevent formation of the epitope for monoclonal antibody (mAb) 17b (which recognizes the coreceptor site). Formation of an encounter complex with CD4 binding and interactions of gp120 with mAbs b12 and 2G12 are not affected by these deletions. Thus, deleting the β3–β5 loop blocks the gp120 conformational change and may offer a strategy for design of restrained immunogens. Moreover, mutations in the SIV β3–β5 loop lead to greater spontaneous dissociation of gp120 from cell-associated trimers. We suggest that the CD4-induced rearrangement of this loop releases structural constraints on gp41 and thus potentiates its fusion activity.
Keywords: CD4, envelope, gp120, HIV
Introduction
Infection by enveloped viruses, such as influenza virus and HIV/SIV, begins with fusion of viral and cellular membranes. The membrane fusion reaction is facilitated by the viral envelope glycoproteins (Harrison, 2005). The HIV or SIV envelope glycoprotein is synthesized as a precursor, gp160, and cleaved by a furin-like protease after trimerization (Allan et al, 1985; Veronese et al, 1985). The two fragments produced by the cleavage, gp120 and gp41, form a noncovalently associated heterodimer, three of which make up the mature viral spike (Center et al, 2001, 2002; Zhu et al, 2003). The interaction between gp120 and gp41 is relatively weak, and gp120 spontaneously dissociates from mature virions (shedding), especially in some laboratory-adapted strains (Moore et al, 1990). Sequential binding of gp120 to viral receptor CD4 and coreceptor (e.g. CCR5 or CXCR4) triggers a cascade of conformational changes within both gp120 and gp41 (Wyatt and Sodroski, 1998; Harrison, 2005). It is generally believed that refolding of gp41 pulls viral and cellular membranes together (Chan et al, 1997; Weissenhorn et al, 1997). Changes in gp120 that accompany CD4 and coreceptor binding must somehow liberate gp41 to undergo the fusion-inducing, refolding process, either by dissociating from it or by releasing conformational constraints.
CD4 binding is the first step in viral entry. A large body of evidence derived from biochemical and thermodynamic experiments supports the notion that some major conformational changes take place in gp120 upon binding to CD4 (Sattentau and Moore, 1991; Sattentau et al, 1993; Myszka et al, 2000). CD4 binding has a number of consequences. It leads to formation of the coreceptor binding site (Wu et al, 1996; Trkola et al, 1996a; Rizzuto et al, 1998), to formation of binding sites (epitopes) for the so-called CD4i (CD4 induced) antibodies, such as 17b and 48d (Thali et al, 1993; Sullivan et al, 1998), to enhanced exposure of the V1V2 and V3 variable loops in the context of the gp160 trimer (Wyatt et al, 1995), to exposure of the gp41 HR1 region in the trimer (Furuta et al, 1998; Gallo et al, 2004), and to dissociation (shedding) of gp120 from gp41, especially in some laboratory-adapted strains (Moore et al, 1990, 1992). Crystal structures of gp120 in both CD4-bound and unliganded conformations have recently allowed us to visualize these changes in considerable detail (Kwong et al, 1998; Chen et al, 2005b).
Fragments of gp120 and gp41, truncated to facilitate structural studies, have been crystallized and have yielded atomic structures. The gp120 ‘core'—that is, gp120 stripped of nonessential variable regions and also of N- and C-terminal segments—has been crystallized in two forms: in an unliganded and fully glycosylated state (from SIV mac32H) (Chen et al, 2005b) and in a CD4-bound form (from HIV-1 HXBc2 and YU2) in complex with the Fab from monoclonal antibody (mAb) 17b (Kwong et al, 1998, 2000). The latter is the receptor-induced conformation. It has been described as two closely associated domains, named ‘inner' and ‘outer' (Kwong et al, 1998). The relative orientations of the two domains are fixed by a four-strand β-sheet, termed the ‘bridging sheet', which is formed by two β hairpins, the base of the V1V2 variable loop from the inner domain and a hairpin that projects from the outer domain. CD4 makes direct contacts with both domains as well as with the bridging sheet. The bridging sheet and the V3 variable loop together contribute to the binding site for coreceptor (Rizzuto et al, 1998; Rizzuto and Sodroski, 2000). Comparison of the liganded and unliganded core structures shows that CD4 binding results in large rearrangements of the inner domain as well as formation of the bridging sheet (Chen et al, 2005b). The latter constitutes at least part of the binding sites for both coreceptor and antibody 17b. These structural rearrangements are consistent with the observation that unusually large negative entropy changes accompany CD4 binding (Myszka et al, 2000).
The inner-domain rearrangements are extensive: each secondary structural element shifts around independently, so that the conformational change cannot be described as a rigid-body motion of the whole domain. For example, the relative displacements of the V1V2 stem (yellow in Figure 1A and B) and the three-stranded sheet (light blue in Figure 1) are such that the loop that connects them (joining strands β3 and β5, and hence designated the ‘β3–β5 loop'), which is disordered in the unliganded structure, becomes extended and ordered in the liganded conformation (compare Figure 1A and B). We have suggested that the β3–β5 loop interacts with gp41 in the trimer (explaining why it is disordered on the free gp120 subunit) and that when it is drawn into a different structure during the gp120 conformational change accompanying CD4 binding, it dissociates from gp41. In a model for the unliganded trimer, described in Chen et al (2005b), this loop is indeed in a position to contact gp41. It may interact with the C–C loop and HR1 of gp41, which are implicated by mutagenesis studies in direct contacts with gp120 (Maerz et al, 2001; York and Nunberg, 2004; Jacobs et al, 2005).
Figure 1.
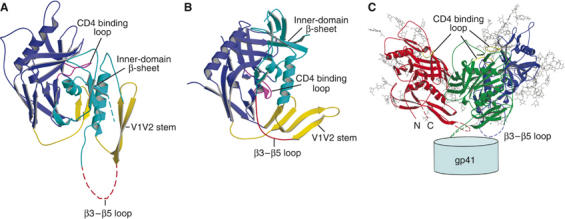
Location of the β3–β5 loop in the gp120 structures. (A) The structure of the unliganded SIV gp120 core; the disordered segment (residues 220–228; the β3–β5 loop) is evident as a dashed, purple line. The β3–β5 loop connects the inner-domain β-sheet in cyan and the V1V2 stem in red. (B) The structure of CD4-bound HIV gp120 core; the β3–β5 loop is a well-ordered strand shown in purple. (C) Proposed gp120 trimer structure, based on the unliganded gp120 core structure (Chen et al, 2005b). Note that the three β3–β5 loops from the three gp120 subunits, shown in red, green and blue, respectively, project around the three-fold axis towards gp41.
The initial and final residues of the β3–β5 loop are spatially adjacent in the unliganded conformation of gp120, but 26 Å apart in the CD4-bound state (Kwong et al, 1998; Chen et al, 2005b). Deletion of the loop should therefore not destabilize the unliganded conformation of free gp120 but should inhibit or weaken the interaction of gp120 with CD4 by preventing the complete conformational change. Binding of CD4 and gp120 is a two-stage process—rapid formation of an initial encounter complex is followed by a slower rearrangement to a tightly docked association (Zhang et al, 2001). We show here that deletions in the β3–β5 loop of HIV-1 gp120 indeed weaken the interaction with CD4 and block formation of the mAb 17b epitope. Thus, they probably also block formation of the coreceptor binding site. The deletions selectively affect the slow step of CD4 binding. They do not affect formation of an encounter complex with CD4, nor do they alter interaction of gp120 with mAbs b12 and 2G12. In addition, point mutations in the β3–β5 loop lead to greater spontaneous dissociation of gp120 from cell-associated gp120/gp41 trimers while not diminishing the capacity of the envelope protein to mediate cell–cell fusion. These findings suggest that CD4-induced rearrangement of the β3–β5 loop may release structural constraints on gp41 and thus potentiate its fusion activity. Deleting the β3–β5 loop may represent a simple strategy for producing a gp120 immunogen locked in its prefusion conformation.
Results
Conservation of the β3–β5 loop in the inner domain
The β3–β5 loop (residues 220–228 in SIVmac 32H) connecting the V1V2 stem and the inner-domain β-sheet is in conserved region C2. It immediately follows a conserved cysteine in the protein sequence, so the sequence alignment in this region is indisputable. Table I shows that the β3–β5 loop is highly conserved in strains among HIV-1/SIVcpz (residues 206–214, HXBc2 numbering) and among HIV-2/SIV, but differs between these two groups. The only invariant residue among all viruses is a lysine at the second position of the loop. A similar pattern of conservation is also true for the C–C loop and HR1 of gp41, as expected for interacting segments that must coevolve to maintain optimal fit (Douglas et al, 1997; Leitner T, 2003, also see Table I).
Table 1.
The sequences of β3–β5 loop and gp41 C–C loop from various HIV/SIV strains
| HIV/SIV strain | Loop sequence | gp41 C–C loop sequence |
|---|---|---|
| HIV-1.A.92UG037.8 | PKVTFEPIP | RDQQLLGIWGCSGKLICPTNV |
| HIV-1.B.AR.99.ARMA132 | PKVSFEPIP | KDQQLLGIWGCSGKLICTTAV |
| HIV-1.C.BI.91.BU910112 | PKVSFDPIP | KDQQLLGIWGCSGKLICTTAV |
| HIV-1.D.CD.84.84ZR085 | PKVSFEPIP | KDQQLLGIWGCSGKHICTTTV |
| HIV-1.F1.BE.93.VI850 | PKVSWDPIP | KDQQLLGIWGCSGKLICTTNV |
| HIV-1.G.BE.96.DRCBL | PKVTFEPIP | KDQQLLGIWGCSGKLICTTNV |
| HIV-1.H.BE.93.VI991 | PKVSFEPIP | KDQQLLGIWGCSGKLICTTNV |
| HIV-1.J.SE.94.SE7022 | PKVSFQPIP | KDQQLLGIWGCSGKLICTTNV |
| HIV-1.K.CM.96.MP535 | PKVTFEPIP | KDQQLLGIWGCSGKLICTTNV |
| HIV-1.AE.TH.90.CM240 | PKISFDPIP | KDQKFLGLWGCSGKIICTTAV |
| HIV-1.N.CM.-.YBF106 | PKTTFEPIP | RDQQILSLWGCSGKTICYTTV |
| HIV-1.O.FR.92.VAU | PKVSFEPIP | QNQQLLNLWGCKNRLICYTSV |
| CPZ.CD.-.ANT | EKSTFEPIP | RDQQLLSLWGCADKVTCHTTV |
| HIV-2 H2A.CI.88.UC2 | DKHYWDSMK | KDQAQLNSWGCTFRQVCHTTV |
| HIV-2 H2B.FR.-.97244 | DKHYWDSLR | KDQAQLNSWGCAFRQVCHTTV |
| SIV MAC32H | DKHYWDTIR | KDQAQLNAWGCAFRQVCHTTV |
| SIV MAC.US.-.239 | DKHYWDAIR | KDQAQLNAWGCAFRQVCHTTV |
| SIV SMM.SL.92.SL92B | DKHYWDAIR | KDQAQLNSWGCAFRQVCHTTV |
Production of gp120 core proteins with loop deletions
The β3–β5 loop is flexible (hence disordered) in the unligand gp120 structure, but extends into a well-ordered strand upon CD4 binding. Deletion in the β3–β5 loop will hinder movement of at least two structural elements, the V1V2 stem and the inner-domain β-sheet, into their CD4-induced positions, and thus will lock gp120 in the unliganded conformation. We have designed loop deletions in the SIV gp120 core based on our crystal structure of the unliganded protein. Because the two ordered residues (219 and 229 in SIV) that flank the β3–β5 loop are about 15 Å apart (Cα positions) but might tolerate some degree of flexibility, we generated two constructs: one with deletion of five residues in the middle of the loop and another with the entire 9-residue loop replaced by two glycine residues. When introduced into insect cells, both constructs yielded secreted gp120 core proteins that could be purified using a 17A11 antibody column. The antibody 17A11 recognizes a conformation-dependent epitope close to the coreceptor binding site (Edinger et al, 2000). A monodisperse protein preparation can be obtained by further purification using size-exclusion chromatography (data not shown). These observations indicate that the deletions in the β3–β5 loop do not affect protein folding.
We then went on to make similar constructs for HIV-1, to take advantage of the many well-studied reagents available for characterizing HIV-1 gp120. We chose a primary isolate, 92UG037.8, from clade A (Figure 2A). The wild-type gp120 core (here designated HIV92ug) was generated as described previously (Chen et al, 2005a) and a His-tag was added to the N-terminus for purification. This HIV-1 gp120 core still contains 17 N-linked glycosylation sites. Construct HIV92ugD5 has five residues deleted in the middle of the β3–β5 loop and HIV92ugD9GG contains a short linker GG replacing the entire β3–β5 loop. The proteins were expressed in insect cells and purified by metal-chelate affinity chromatography with Ni-NTA agarose resin from cell supernatants, followed by gel filtration. All three proteins elute from a Superdex 200 column as a sharp peak, with a size corresponding to a 60-kDa globular protein (Figure 2B). It is consistent with the calculated mass for the monomeric gp120 core that contains a polypeptide chain of about 36 kDa and 17 N-linked glycans. The proteins migrate as a single, but diffuse protein band with an average molecular mass of 64 kDa, as expected for a heavily glycosylated species.
Figure 2.
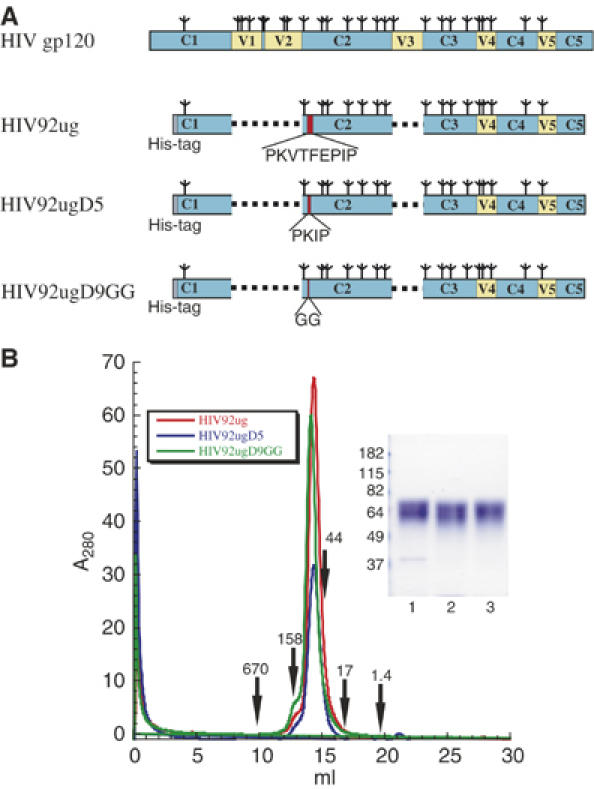
Protein production of HIV-1 gp120 core and its loop-deletion variants. (A) Schematic representations of HIV-1 gp120 protein and its loop-deletion variants. HIV gp120, the surface subunit of the envelope glycoprotein; N-linked glycans are represented by tree-like symbols and various segments of gp120 are designated as follows: C1–C5 in light blue, conserved regions 1–5; V1–V5 in yellow, variable regions 1–5. HIV92ug, HIV-1 gp120 core, the protein is truncated the same way as described in Chen et al (2005a) for crystallographic studies. HIV92ugD5, gp120 core with five residues deleted from the β3–β5 loop. HIV92ugD9GG, gp120 core with the entire β3–β5 loop replaced by a short linker GG. The location of the β3–β5 loop is highlighted in red. The actual residues in the loop are shown beneath each construct. All the gp120 core proteins have a His-tag (in gray) at the N-terminus to facilitate protein purification. (B) HIV-1 gp120 core protein and its loop-deletion variants were purified from supernatants of insect cell culture, and then resolved by gel-filtration chromatography using a Superdex 200 column. The traces are shown in red for HIV92ug, blue for HIV92ugD5 and green for HIVug92D9GG. The apparent molecular masses were calculated based on a standard curve using the following known standards: thyoglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa) and vitamin B12 (1.4 kDa). Peak fractions were pooled and analyzed by Commassie-stained sodium dodecyl sulfate–polyacrylamide gel electrophoresis (inset). Lane 1, HIV92ug; lane 2, HIV92ugD5; lane 3, HIV92ugD9GG.
We examined binding of these proteins to some well-characterized mAbs by surface plasmon resonance (SPR) biosensor analysis. mAb 2G12 is a broadly neutralizing antibody that recognizes a glycan- and conformation-dependent epitope in the outer domain of gp120 (Trkola et al, 1996b). To measure the binding kinetics of various gp120 core proteins to 2G12, the intact IgG was immobilized on a CM5 chip, and gp120, at different molar concentrations, was passed over the surface of the chip. In Figure 3, the sensorgrams for binding of 2G12 to the HIV92ug protein and its two loop deletion variants, HIV92ugD5 and HIV92ugD9GG, are almost identical. The data were analyzed with a 1:1 Langmuir binding model; the kinetic binding constants are listed in Table II. The on- and off-rate constants and the Kd are essentially identical for all three proteins, indicating that the loop deletions did not affect the conformation of the 2G12 epitope. We also tested binding of the three proteins to another broadly neutralizing antibody, b12, which recognizes an epitope that overlaps the CD4 binding site (CD4 BS) (Burton et al, 1994). Among all CD4 BS antibodies, b12 is the only one with potent, broadly neutralizing activity. Unlike many other CD4 BS antibodies, its association with gp120 does not appear to require a large, entropically costly conformational change (Kwong et al, 2002). The sensorgrams for b12 binding are very similar for all three proteins (not shown), and the kinetic data, derived from a 1:1 Langmuir binding model, are summarized in Table II. Although b12 binds to the gp120 core of this particular HIV-1 strain from clade A with relatively low affinity (Kd=1.42 μM), the rate constants for two deletion variants, HIV92ugD5 and HIV92ugD9GG, do not differ significantly (within a factor of 1.6) from that of the wild-type core, HIV92ug. We conclude from the antibody binding studies and from the data described above that the deletions introduced into the β3–β5 loop do not have deleterious effects on the conformation or stability of gp120.
Figure 3.
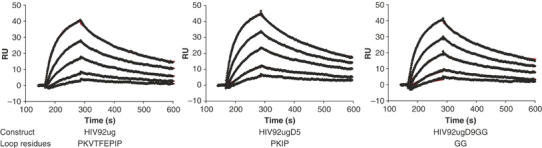
Kinetic analysis of binding of mAb 2G12 to HIV-1 gp120 core proteins. mAb 2G12 was immobilized on a CM-5 chip, and gp120 at various concentrations was passed over the chip surface as described in Materials and methods. All injections were carried out in duplicate, which gave essentially identical results. Binding kinetics were evaluated using BiaEvaluation software (Biacore) using a 1:1 Langmuir binding model. The recorded sensorgrams are shown in black (one of the duplicates) and the calculated curves in red. The residues in the β3–β5 loop are also shown for each protein at the bottom.
Table 2.
Rate constants for binding of gp120 core and its loop-deletion variants with the broadly neutralizing antibodies
| Antibody | Construct | ka (1/Ms) | kd (1/s) | Kd (M) |
|---|---|---|---|---|
| 2G12 | HIV92ug | 1.85E4 | 5.03E-3 | 2.72E-7 |
| HIV92ugD5 | 2.01E4 | 4.89E-3 | 2.44E-7 | |
| HIV92ugD9GG | 2.10E4 | 4.58E-3 | 2.18E-7 | |
| b12 | HIV92ug | 1.02E4 | 1.45E-2 | 1.42E-6 |
| HIV92ugD5 | 1.60E4 | 1.78E-2 | 1.11E-6 | |
| HIV92ugD9GG | 7.75E3 | 1.97E-2 | 2.54E-6 |
CD4 binding to HIV-1 gp120 core proteins
The large-amplitude structural rearrangements in gp120 that accompany CD4 binding are primarily in the inner domain. We have proposed that the initial contact of CD4 with gp120 might be with the ‘CD4-binding loop' in the outer domain (Chen et al, 2005b), which will be unaffected by deletion of the β3–β5 loop. Thus, on this basis, we expect that in a two-step kinetic analysis of CD4-gp120 binding, encounter-complex formation might be insensitive to β3–β5 loop deletion, whereas the slower, tight-binding step might be strongly retarded. We have therefore analyzed kinetics of CD4 binding to HIV92ug and its loop deletion variants, HIV92ugD5 and HIV92ugD9GG, using SPR. Soluble, four-domain CD4 was immobilized on the chip, and solutions of gp120 at various concentrations were allowed to flow over the surface. As shown in Figure 4, all three proteins had similar rate constants for the initial, rapid encounter step (both kon and koff), but quite different rate constants for the tight-binding step. Both loop-deletion variants had association rate constants three to four times lower than did wild-type gp120, and dissociation rate constants 15–20 times greater. The equilibrium dissociation constants (at 25°C) derived from these data are 13.9 nM for wild-type HIV92ug, 228 nM for HIV92ugD5, and 1.0 μM for HIV92ugD9GG. The same experiment was also carried out at 10°C with similar results.
Figure 4.
Binding of soluble CD4 to HIV-1 gp120 core proteins. Soluble four-domain CD4 was immobilized on a CM-5 chip, and gp120 at various concentrations was passed over the chip surface. All injections were carried out in duplicate, which gave essentially identical results. Binding kinetics was evaluated using BiaEvaluation software (Biacore) using a two-step binding model. The recorded sensorgrams are shown in black (one of the duplicates) and the calculated curves in red. The residues in the β3–β5 loop are also shown for each protein at the bottom. The two-step model includes formation of the encounter complex [gp120/CD4]* and, after conformational changes, formation of docking complex gp120/CD4. Rate constants and equilibrium dissociation constants derived from the fits are also summarized.
mAb 17b binding site is not formed when the β3–β5 loop is deleted
If the β3–β5 loop deletion prevents the conformational changes required for CD4 docking, including translocation of V1V2 stem, then the loop-deleted gp120 should be unable to close up the bridging sheet and hence unable to display the coreceptor binding site or epitopes for CD4i antibodies such as 17b. We examined by SPR biosensor analysis binding of the three gp120 core proteins to mAb 17b in presence or absence of soluble CD4. We immobilized 17b IgG molecules on a CM5 chip and passed various gp120 proteins over the chip surface with or without preincubation with two-domain soluble CD4. Wild-type gp120 core (HIV92ug) does not bind to the 17b surface in the absence of CD4 (Figure 5; sensorgram in cyan) but does so in complex with soluble CD4 (sensorgram in black), in agreement with published results (Edwards et al, 2001; Zhang et al, 2001). In contrast, the two variants with deletions in the β3–β5 loop (HIV92ugD5 and HIV92ugD9GG) fail to bind the 17b surface, regardless of whether they have been preincubated with CD4 or not. Shortening the β3–β5 loop thus blocks formation of the 17b epitope.
Figure 5.
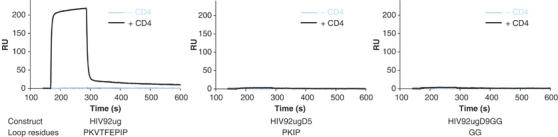
Binding of mAb 17b to HIV-1 gp120 core proteins in the absence or presence of CD4. mAb 17b was immobilized on a CM5 chip, and various gp120 proteins at 500 nM were passed over the chip surface with or without preincubation with equimolar amounts of two-domain soluble CD4. All injections were carried out in duplicate, which gave essentially identical results. The recorded sensorgrams for binding in the presence of CD4 are shown in black, and those for binding in the absence of CD4, in cyan (one of the duplicates). The residues in the β3–β5 loop are also shown for each protein at the bottom.
Several observations rule out the possibility that mAb 17b fails to bind the loop-deletion variants because the excised residues are part of its epitope. In the high-resolution structure of the ternary complex of gp120, CD4, and 17b, none of residues in the β3–β5 loop makes a direct contact with 17b (Kwong et al, 1998). The closest distance between an atom of a β3–β5 loop residue and an atom in 17b is greater than 8 Å. Moreover, HIV92ugD5, which has the same properties as the full deletant, has only five residues eliminated from the middle of the loop, and these residues are even more distant (greater than 15 Å) from the 17b binding site in the CD4-bound conformation. Finally, mutation of residues in the β3–β5 loop published by other groups does not significantly affect 17b binding (Rizzuto et al, 1998).
The β3–β5 loop helps anchor gp120 to gp41
Our model for the unliganded trimer suggests that the β3–β5 loop participates in the gp120/gp41 interaction (Figure 1C). To test this hypothesis, we systematically mutated the loop residues (220–228) in SIV Env and examined these mutants for induction of cell–cell fusion and spontaneous shedding of gp120. We chose the envelope protein from SIV, because SIV gp120 sheds from the Env trimer much less readily than does HIV-1 gp120 (Sattentau et al, 1993). Each residue in the loop was replaced with alanine by polymerase chain reaction (PCR)-based site-directed mutagenesis, and the mutations were confirmed by DNA sequencing. All mutant Envs were tested first for membrane fusion activity by a cell–cell fusion assay based on a reporter gene activation technique described previously (Ferrer et al, 1999). As shown in Figure 6A, all the mutant Envs support membrane fusion at a level similar to that of the wild type, regardless of the nature of the substitution. For example, D220A, K221A, D225A and R228A are all nonconservative substitutions that eliminate one charged side chain at a time, and Y223A and W224A replace bulky side chains with smaller ones. Mutants K221A and T226A had significantly higher levels of cell-associated gp120 than did wild type, probably due to more efficient processing of gp160 (Figure 6B). These two mutants may therefore be more strongly fusogenic than the wild type. Thus, a single change in this region does not seem to affect either folding or functionality of the protein. It is not clear why the lysine residue is so conserved; the effect of the alanine mutation could be masked by the high level of expression of Env in a cell–cell fusion assay.
Figure 6.
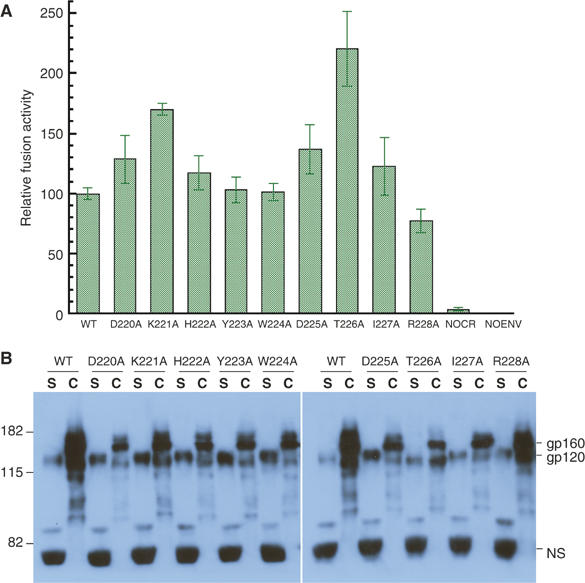
Fusion activity of mutant envelope glycoproteins in a cell–cell fusion assay and spontaneous dissociation of gp120 from the cell surface. (A) Cell–cell fusion assays were carried out in triplicate for each construct as described previously (Ferrer et al, 1999). The readouts of the assay were normalized based on the activity of the wild-type Env (100) to give the relative fusion activity of the mutant Envs. WT: wild-type Env transfected into effector cell; indicated mutant construct transfected into effector cell; NOCR: chemokine receptor, CCR5, omitted from target cell; NOENV: Env construct omitted from effector cell. NOCR and NOENV are negative controls. The standard deviations derived from the three measurements for each construct are indicated by the error bars. The experiment was repeated twice using independent transfections and gave similar results. (B) Wild-type and mutant Env expression constructs were transfected into 293T cells. Expression and distribution of gp120 were monitored by Western blot using mAbs KK19 and SIV-101, specific for SIV gp120. Mutations are indicated at the top; S, cell supernatants; C, cell-associated. Equal volumes for all supernatants or cell lysates were loaded in each lane. The equal intensity of a very abundant protein (NS) from the medium recognized nonspecifically by the antibodies indicates uniform sample loading. The experiment was repeated four times, using independent transfections, and gave the same result each time.
We then examined the stability of the association of gp120 and gp41, by monitoring the distribution of gp120 between cell and supernatant. All mutated Env constructs were transfected into 293T cells. Both supernatants and lysates were then harvested and analyzed by Western blot using a mixture of mAbs, KK19 and SIV-101 (Kent et al, 1991 and unpublished). Figure 6B shows that the antibodies detect dissociated gp120 in cell supernatants (S) and both gp120 and gp160 in cell pellets (C). In addition, they also detect a strong band of a very abundant protein migrating around 60 kDa from supernatants, presumably by nonspecific binding, as the same band was detected from a mock transfection (Supplementary Figure S1). The sample loading was normalized by volume and the nonspecific band showed the same intensity throughout, suggesting that the differences among dissociated gp120 mutant proteins in the supernatants are not due to variations in sample loading. All mutants except R228A showed a significant increase in the ratio between shed gp120 and cell-associated gp120. Mutation of residues between 220 and 225 increased the amount of free gp120, whereas the amount of cell-associated gp120 decreased. In contrast, the change R228A produced an Env very similar to the wild type in its gp120 dissociation properties. These results are consistent with our suggestion, that residues in the β3–β5 loop, disordered in the unliganded conformation, contribute to anchoring gp120 to gp41.
Discussion
CD4 binding to the HIV-1 envelope glycoprotein induces large structural rearrangements in gp120. These changes lead to formation of the coreceptor binding site and may prime the protein for further changes triggered by the coreceptor interaction. In the absence of CD4, various structural elements of gp120 may be relatively flexible, allowing it to sample a range of conformations. Thus, ‘conformational masking' of the CD4 binding site is believed to be among the viral strategies for immune evasion (Kwong et al, 2002). Monomeric gp120 has failed to elicit a protective immune response against HIV infection (Maek et al, 2003), but whether a gp120 immunogen fixed in a defined conformation can be more effective remains to be investigated. We have derived a simple strategy, based on our recent crystal structure of the unliganded SIV gp120 core, to lock gp120 in its unliganded (and presumably prefusion) conformation. Comparison of the unliganded and CD4-bound gp120 core structures shows that the β3–β5 loop connecting the V1V2 stem and the inner-domain β-sheet is disordered in the unliganded gp120 structure, but stretches into a well-ordered strand upon CD4 binding (Chen et al, 2005b). Shortening of this loop should prevent the structural rearrangements induced by CD4 binding. The data presented here are in excellent agreement with this prediction. Deletions in the β3–β5 loop prevent the docking (tight-binding) step of CD4 association, but do not prevent formation of a weaker encounter complex. Moreover, deletion of just five residues in the β3–β5 loop completely abolishes the formation of 17b binding site—and probably also the coreceptor binding site—even in the presence of CD4. Thus, deleting the β3–β5 loop in the inner domain represents a simple strategy for producing a gp120 immunogen restrained in the unliganded conformation.
Neutralizing antibodies must bind a relevant, functional conformation of Env. Many antibodies recognize epitopes near the receptor binding site (CD4 BS), but among well-studied CD4 BS antibodies, only b12 has potent, broadly neutralizing activity that inhibits viral infection by a variety of HIV-1 strains (Burton et al, 1994). Another CD4 BS antibody, b6, is non-neutralizing, but it has essentially the same affinity for gp120 as does b12 (Pantophlet et al, 2003). Thermodynamic studies show that binding of b6 to gp120 produces a large negative entropy change and therefore seems to induce major conformational changes (Kwong et al, 2002). The corresponding entropy change for b12 binding is relatively small, suggesting that b12 recognizes the unliganded conformation of gp120 (Kwong et al, 2002). The data in Table II are consistent with this conclusion, as b12 binding is unaffected by deletion of the β3–β5 loop. The b12 binding data also strengthen our earlier suggestion that the conformation of unliganded, free gp120, as seen in the SIV gp120 crystal structure, closely resembles its structure when part of trimeric gp120/gp41 on the surface of a virion (Chen et al, 2005b). The most compelling previous argument for this suggestion has been identification, in the unliganded gp120 structure, of a likely binding pocket for BMS-378806, a viral-entry inhibitor (Lin et al, 2003; Wang et al, 2003). As this small molecule is an active anti-viral, it clearly also interacts with gp120/gp41 trimers on virions. BMS-378806 appears to stabilize the unliganded gp120 conformation, thereby preventing the structural changes induced by CD4 binding (Ho et al, 2006). Residues that line the proposed BMS-378806 pocket, many of which are at positions of resistance mutations, come from elements of the inner domain, the outer domain, and part of the bridging sheet. They close up against each other in the liganded conformation, and CD4 binding would therefore require expulsion of the inhibitor.
Residues at the gp120/gp41 interface are likely to the coordinate structural changes in the two fragments. Previous mutagenesis studies have suggested that the N- and C-terminal segments of gp120 interact with gp41 (Helseth and Olshevsky, 1991; Wyatt et al, 1997). Moreover, the location of the so-called SOS disulfide mutation, which stabilizes gp120/gp41 trimers, indicates that at least one of the contacts of the gp120 C-terminal segment is the gp41 C–C loop (Binley et al, 2000). Likewise, in influenza virus HA, both ends of HA1 fold together with HA2 to form the stalk (Wilson et al, 1981; Skehel and Wiley, 2000). We have proposed that in addition to the N- and C-terminal segments, the spatially adjacent β3–β5 loop of gp120 might also interact directly with gp41 (Figure 1C). Our data (Figure 6) support this proposal, as mutations in the β3–β5 loop weaken the gp120:gp41 interaction and enhance gp120 shedding. Comparison of the unliganded and liganded conformations of gp120 further suggests how CD4 (and coreceptor) binding could trigger fusion. In the CD4-bound conformation of gp120, the β3–β5 loop interacts with other parts of the gp120 inner domain. Thus, we expect that CD4 binding will peel the loop away from gp41 and release constraints that hold it in a prefusion configuration. We suggest that the interaction between the β3–β5 loop and gp41 detected by our experiments intervenes directly in the cascade of conformational changes that ultimately leads to membrane fusion and viral entry.
Eliciting broadly neutralizing antibodies against HIV-1 remains the most elusive goal for AIDS vaccine development as illustrated by the many failed strategies for envelope-based immunogen design (Burton et al, 2004). Most efforts have used recombinant, cell-associated, or virion-associated envelope glycoproteins, known to be structurally heterogeneous. For instance, recombinant, uncleaved, HIV-1 gp140 is often a mixture of monomer, dimer, trimer, and higher-order oligomers, the conformations of which are difficult to define (Jeffs et al, 2004). Using such preparations as immunogen may distract the immune system with irrelevant decoys, contributing to production of noneffective antibody responses. Lack of adequate high-resolution structural information on the envelope glycoprotein has impeded efforts to make Env immunogens with well-defined conformations. Thus, we do not know whether gp120 restrained in the unliganded conformation could induce more b12-like response than have previously studied immunogens. The loop deletion described here should permit us to answer this question.
Materials and methods
Expression and purification of gp120 core protein and its deletion variants
Expression constructs pHIV92ug, pHIV92ugD5 and pHIV92ugD9GG were generated by standard PCR techniques. For gp120 core, we deleted residues from the N- and C-terminal of gp120, and we substituted short linkers for the V1–V2 and V3 loops as described (Chen et al, 2005a). A His-tag was added to the N-terminus to facilitate purification. Two additional residues (His-Met) were introduced by the restriction site (NdeI) at the N-terminus. pHIV92ugD5 and pHIV92ugD9GG were derived from pHIV92ug with five residues deleted from the β3–β5 loop and a short linker, GG, replacing the entire nine-residue β3–β5 loop, respectively. Both restriction digestion and DNA sequencing verified the expression constructs. The proteins were expressed using the Bac-to-Bac system (Invitrogen, Carlsbad, CA) following a procedure described previously (Chen et al, 2005a), except that Sf9 insect cells were used for large-scale protein production and that the cell supernatant was harvested 84 h after infection. Gp120 core proteins were purified by metal chelate affinity chromatography with NTA-nickel resin (Qiagen, Hilden, Germany) as described (Chen et al, 2000). The protein was eluted with 300 mM imidazole, and the fractions containing gp120 core protein were pooled, concentrated, and further purified by gel filtration chromatography on Superdex 200 (GE Healthcare, Piscataway, NJ) with a buffer containing 25 mM Tris–HCl (pH 8.0) and 150 mM NaCl. The protein was concentrated and stored at −80°C. His-tagged two-domain soluble CD4 was also expressed in insect cells and purified by a similar protocol.
SPR binding assays
All experiments were performed in duplicate with a Biacore 3000 instrument (Biacore Inc., Piscataway NJ) at 20°C in HBS-EP running buffer (10 mM N-2-hydroxyl piperazine-N′-2-ehane sulfonic acid pH 7.4, 150 mM NaCl, 3 mM ethylene diaminetetra acetic acid, 0.005% surfactant P20). Immobilization of ligands, four-domain sCD4 (Protein Sciences, Meriden, CT), 2G12 (Polymun Scientifc Inc., Vienna, Austria), b12 (a gift from Dr Dennis Burton), 17b (hybridoma was provided by Dr James Robinson), to CM5 chips (Biacore) was performed following the standard amine coupling procedure. Briefly, carboxyl groups were activated by injection of 50 μl of EDC:NHS (200 mM 1-ethyl-3[3-dimethyl aminopropyl] carbodiimide hydrochloride, 50 mM N-hydroxysuccinimide) at a flow rate of 5 μl/min (10 min contact time). Ligand (20 μg/ml in 10 mM sodium acetate pH 5.0) was passed over the activated surface until the desired immobilization level was reached. Excess carboxyl groups were blocked with 1 M ethanolamine (35 μl at a flow rate of 5 μl/min). A reference surface was prepared by activating and blocking a flow cell in the absence of ligand. The final immobilization levels for CD4 was 800 response units (RUs), for 2G12 1200 RUs, for b12 950 RUs, and for 17b 900 RUs. For kinetic measurements of gp120 binding to immoblilized CD4, sensorgrams were obtained by passing various concentrations (10 nM–1.0 μM) of gp120 over the ligand surface at a flow rate of 50 μl/min using a 2-min association phase and 5 min dissociation phase. The sensor surface was regenerated between each experiment using a single injection (3 s) of 35 mM NaOH, 1.3 M NaCl at a flow rate of 100 μl/min. Identical injections over blank surfaces were subtracted from the data for kinetic analysis. All the injections were carried out in duplicate; the results of the duplicate measurements were essentially identical. Binding kinetics was evaluated using BiaEvaluation software (Biacore) using a two-state reaction model. Kinetic measurements of gp120 to IgGs were performed in an identical manner, with the exception that regeneration was achieved using a single injection (3 s) of 10 mM HCl and a flow rate of 100 μl/min. Binding kinetics were evaluated using BiaEvaluation software (Biacore) using 1:1 Langmuir binding model.
Cell–cell fusion assay
Each residue was replaced with alanine by PCR-based site-directed mutagenesis, and the mutations were confirmed by DNA sequencing. All mutant Envs were tested for membrane fusion activity by a cell–cell fusion assay based on a reporter gene activation technique described previously (Ferrer et al, 1999). Briefly, 293T cells were cotransfected by the calcium phosphate method with equal amounts of an SIV gp160 expression construct and a plasmid expressing T7 polymerase; these were designated effector cells. CD4-and CCR5-expressing cells with a luciferase reporter gene under the control of a T7 promotor were designated target cells. The two types of cells were resuspended and mixed 40 h post-transfection and then incubated at 37°C for another 8 h. Fusion activity was measured by a luciferase assay (Promega, Madison, WI) following protocols recommended by the manufacturer.
Gp120 shedding assay by Western blot
The stability of the association of gp120 and gp41 was examined by monitoring the distribution of gp120 between cell and supernatant. Transfected 293T cells were harvested 40 h post-transfection. After a clarifying spin, supernatants were separated from cell pellets. Cells were then resuspended in the same volume of phosphate-buffered saline and lysed by mixing with an equal volume of 2 × lysis buffer (100 mM Tris pH 7.5, 300 mM NaCl, 1% NP-40, 0.2% sodium dodecyl sulfate (SDS), 0.25 mM MgCl2, 0.02 mg/ml RNase A (Sigma, St Louis, MO) and 2000 U/ml DNase I (Sigma)). Both supernatants and lysates were then boiled with SDS-loading buffer and resolved in a 6% SDS polyacrylamide gel. Proteins were transferred to polyvinylidine diflouride membrane (Millipore, Billerica, MA) and detected by a mixture of mAbs, KK19 and SIV-101 (Kent et al, 1991; and unpublished) and an ECL Plus Western blotting detection kit (GE Healthcare).
Supplementary Material
Supplementary Figure S1
Acknowledgments
We thank Haiyun Gong, John-William Carroll and Hanqin Peng for technical assistance, members of Harrison/Wiley laboratory for discussion, Dr James Robinson of Tulane University, for hybridoma, Dr Dennis Burton of the Scripps Research Institute for antibodies, and Drs Dan Barouch and Ethan Settembre for comments. The work was supported by NIH Grants GM-75752 (to SCH) and by the Consortium for HIV/AIDS Viral Immunology (NIH Grant AI-67854, to Barton Haynes, Duke University). SCH is an Investigator of the Howard Hughes Medical Institute.
References
- Allan JS, Coligan JE, Barin F, McLane MF, Sodroski JG, Rosen CA, Haseltine WA, Lee TH, Essex M (1985) Major glycoprotein antigens that induce antibodies in AIDS patients are encoded by HTLV-III. Science 228: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP (2000) A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol 74: 627–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Desrosiers RC, Doms RW, Koff WC, Kwong PD, Moore JP, Nabel GJ, Sodroski J, Wilson IA, Wyatt RT (2004) HIV vaccine design and the neutralizing antibody problem. Nat Immunol 5: 233–236 [DOI] [PubMed] [Google Scholar]
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, Lamacchia M, Garratty E, Stiehm ER, Bryson YJ, Cao Y, Moore JP, Ho DD, Barbas CF (1994) Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266: 1024–1027 [DOI] [PubMed] [Google Scholar]
- Center RJ, Leapman RD, Lebowitz J, Arthur LO, Earl PL, Moss B (2002) Oligomeric structure of the human immunodeficiency virus type 1 envelope protein on the virion surface. J Virol 76: 7863–7867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center RJ, Schuck P, Leapman RD, Arthur LO, Earl PL, Moss B, Lebowitz J (2001) Oligomeric structure of virion-associated and soluble forms of the simian immunodeficiency virus envelope protein in the prefusion activated conformation. Proc Natl Acad Sci USA 98: 14877–14882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC, Fass D, Berger JM, Kim PS (1997) Core structure of gp41 from the HIV envelope glycoprotein. Cell 89: 263–273 [DOI] [PubMed] [Google Scholar]
- Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC (2005a) Determining the structure of an unliganded and fully glycosylated SIV gp120 envelope glycoprotein. Structure (Camb) 13: 197–211 [DOI] [PubMed] [Google Scholar]
- Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, Harrison SC (2005b) Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433: 834–841 [DOI] [PubMed] [Google Scholar]
- Chen B, Zhou G, Kim M, Chishti Y, Hussey RE, Ely B, Skehel JJ, Reinherz EL, Harrison SC, Wiley DC (2000) Expression, purification, and characterization of gp160e, the soluble, trimeric ectodomain of the simian immunodeficiency virus envelope glycoprotein, gp160. J Biol Chem 275: 34946–34953 [DOI] [PubMed] [Google Scholar]
- Douglas NW, Munro GH, Daniels RS (1997) HIV/SIV glycoproteins: structure–function relationships. J Mol Biol 273: 122–149 [DOI] [PubMed] [Google Scholar]
- Edinger AL, Ahuja M, Sung T, Baxter KC, Haggarty B, Doms RW, Hoxie JA (2000) Characterization and epitope mapping of neutralizing monoclonal antibodies produced by immunization with oligomeric simian immunodeficiency virus envelope protein. J Virol 74: 7922–7935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TG, Hoffman TL, Baribaud F, Wyss S, LaBranche CC, Romano J, Adkinson J, Sharron M, Hoxie JA, Doms RW (2001) Relationships between CD4 independence, neutralization sensitivity, and exposure of a CD4-induced epitope in a human immunodeficiency virus type 1 envelope protein. J Virol 75: 5230–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer M, Kapoor TM, Strassmaier T, Weissenhorn W, Skehel JJ, Oprian D, Schreiber SL, Wiley DC, Harrison SC (1999) Selection of gp41-mediated HIV-1 cell entry inhibitors from biased combinatorial libraries of non-natural binding elements. Nat Struct Biol 6: 953–960 [DOI] [PubMed] [Google Scholar]
- Furuta RA, Wild CT, Weng Y, Weiss CD (1998) Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol 5: 276–279 [DOI] [PubMed] [Google Scholar]
- Gallo SA, Clore GM, Louis JM, Bewley CA, Blumenthal R (2004) Temperature-dependent intermediates in HIV-1 envelope glycoprotein-mediated fusion revealed by inhibitors that target N- and C-terminal helical regions of HIV-1 gp41. Biochemistry 43: 8230–8233 [DOI] [PubMed] [Google Scholar]
- Harrison SC (2005) Mechanism of membrane fusion by viral envelope proteins. Adv Virus Res 64: 231–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helseth E, Olshevsky U (1991) Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J Virol 65: 2119–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho HT, Fan L, Nowicka-Sans B, McAuliffe B, Li CB, Yamanaka G, Zhou N, Fang H, Dicker I, Dalterio R, Gong YF, Wang T, Yin Z, Ueda Y, Matiskella J, Kadow J, Clapham P, Robinson J, Colonno R, Lin PF (2006) Envelope conformational changes induced by human immunodeficiency virus type 1 attachment inhibitors prevent CD4 binding and downstream entry events. J Virol 80: 4017–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs A, Sen J, Rong L, Caffrey M (2005) Alanine scanning mutants of the HIV gp41 loop. J Biol Chem 280: 27284–27288 [DOI] [PubMed] [Google Scholar]
- Jeffs SA, Goriup S, Kebble B, Crane D, Bolgiano B, Sattentau Q, Jones S, Holmes H (2004) Expression and characterisation of recombinant oligomeric envelope glycoproteins derived from primary isolates of HIV-1. Vaccine 22: 1032–1046 [DOI] [PubMed] [Google Scholar]
- Kent KA, Gritz L, Stallard G, Cranage MP, Collignon C, Thiriart C, Corcoran T, Silvera P, Stott EJ (1991) Production and characterisation of monoclonal antibodies to simian immunodeficiency virus envelope glycoproteins. Aids 5: 829–836 [DOI] [PubMed] [Google Scholar]
- Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J (2002) HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420: 678–682 [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA (2000) Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Struct Fold Des 8: 1329–1339 [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA (1998) Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393: 648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner TT, Hahn B, Marx P, McCutchan F, Mellors J, Wolinsky S (eds) (2003) HIV Sequence Compendium 2003. NM: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, LA-UR 04-7420 [Google Scholar]
- Lin PF, Blair W, Wang T, Spicer T, Guo Q, Zhou N, Gong YF, Wang HG, Rose R, Yamanaka G, Robinson B, Li CB, Fridell R, Deminie C, Demers G, Yang Z, Zadjura L, Meanwell N, Colonno R (2003) A small molecule HIV-1 inhibitor that targets the HIV-1 envelope and inhibits CD4 receptor binding. Proc Natl Acad Sci USA 100: 11013–11018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maek ANW, Pitisuttithum P, Phonrat B, Bussaratid V, Naksrisook S, Peonim W, Thantamnu N, Muanaum R (2003) VaxGen vaccine trial fails the test but may offer insights. AIDS Alert 18: 43–45 [PubMed] [Google Scholar]
- Maerz AL, Drummer HE, Wilson KA, Poumbourios P (2001) Functional analysis of the disulfide-bonded loop/chain reversal region of human immunodeficiency virus type 1 gp41 reveals a critical role in gp120-gp41 association. J Virol 75: 6635–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, McKeating JA, Huang YX, Ashkenazi A, Ho DD (1992) Virions of primary human immunodeficiency virus type 1 isolates resistant to soluble CD4 (sCD4) neutralization differ in sCD4 binding and glycoprotein gp120 retention from sCD4-sensitive isolates. J Virol 66: 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, McKeating JA, Weiss RA, Sattentau QJ (1990) Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 250: 1139–1142 [DOI] [PubMed] [Google Scholar]
- Myszka DG, Sweet RW, Hensley P, Brigham-Burke M, Kwong PD, Hendrickson WA, Wyatt R, Sodroski J, Doyle ML (2000) Energetics of the HIV gp120-CD4 binding reaction. Proc Natl Acad Sci USA 97: 9026–9031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantophlet R, Ollmann Saphire E, Poignard P, Parren PW, Wilson IA, Burton DR (2003) Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J Virol 77: 642–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto C, Sodroski J (2000) Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res Hum Retroviruses 16: 741–749 [DOI] [PubMed] [Google Scholar]
- Rizzuto CD, Wyatt R, Hernández-Ramos N, Sun Y, Kwong PD, Hendrickson WA, Sodroski J (1998) A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280: 1949–1953 [DOI] [PubMed] [Google Scholar]
- Sattentau QJ, Moore JP (1991) Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med 174: 407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattentau QJ, Moore JP, Vignaux F, Traincard F, Poignard P (1993) Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J Virol 67: 7383–7393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel JJ, Wiley DC (2000) Receptor binding and membrane fusion in virus entry: the influenza haemagglutinin. Ann Rev Biochem 69: 531–569 [DOI] [PubMed] [Google Scholar]
- Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J, Sodroski J (1998) CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J Virol 72: 4694–4703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J (1993) Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol 67: 3978–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP (1996a) CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384: 184–187 [DOI] [PubMed] [Google Scholar]
- Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H (1996b) Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol 70: 1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese FD, DeVico AL, Copeland TD, Oroszlan S, Gallo RC, Sarngadharan MG (1985) Characterization of gp41 as the transmembrane protein coded by the HTLV-III/LAV envelope gene. Science 229: 1402–1405 [DOI] [PubMed] [Google Scholar]
- Wang T, Zhang Z, Wallace OB, Deshpande M, Fang H, Yang Z, Zadjura LM, Tweedie DL, Huang S, Zhao F, Ranadive S, Robinson BS, Gong YF, Ricarrdi K, Spicer TP, Deminie C, Rose R, Wang HG, Blair WS, Shi PY, Lin PF, Colonno RJ, Meanwell NA (2003) Discovery of 4-benzoyl-1-[(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)oxoacetyl]-2-(R)-methylpiperazine (BMS-378806): a novel HIV-1 attachment inhibitor that interferes with CD4-gp120 interactions. J Med Chem 46: 4236–4239 [DOI] [PubMed] [Google Scholar]
- Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC (1997) Atomic structure of the ectodomain from HIV-1 gp41. Nature 387: 426–430 [DOI] [PubMed] [Google Scholar]
- Wilson IA, Skehel JJ, Wiley DC (1981) Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289: 366–373 [DOI] [PubMed] [Google Scholar]
- Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, Sodroski J (1996) CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384: 179–183 [DOI] [PubMed] [Google Scholar]
- Wyatt R, Desjardin E, Olshevsky U, Nixon C, Binley J, Olshevsky V, Sodroski J (1997) Analysis of the interaction of the human immunodeficiency virus type 1 gp120 envelope glycoprotein with the gp41 transmembrane glycoprotein. J Virol 71: 9722–9731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J (1995) Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol 69: 5723–5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Sodroski J (1998) The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280: 1884–1888 [DOI] [PubMed] [Google Scholar]
- York J, Nunberg JH (2004) Role of hydrophobic residues in the central ectodomain of gp41 in maintaining the association between human immunodeficiency virus type 1 envelope glycoprotein subunits gp120 and gp41. J Virol 78: 4921–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Godillot AP, Wyatt R, Sodroski J, Chaiken I (2001) Antibody 17b binding at the coreceptor site weakens the kinetics of the interaction of envelope glycoprotein gp120 with CD4. Biochemistry 40: 1662–1670 [DOI] [PubMed] [Google Scholar]
- Zhu P, Chertova E, Bess J Jr, Lifson JD, Arthur LO, Liu J, Taylor KA, Roux KH (2003) Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc Natl Acad Sci USA 100: 15812–15817 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1


