Abstract
CPEB is a sequence-specific RNA-binding protein that promotes polyadenylation-induced translation in oocytes and neurons. Vertebrates contain three additional genes that encode CPEB-like proteins, all of which are expressed in the brain. Here, we use SELEX, RNA structure probing, and RNA footprinting to show that CPEB and the CPEB-like proteins interact with different RNA sequences and thus constitute different classes of RNA-binding proteins. In transfected neurons, CPEB3 represses the translation of a reporter RNA in tethered function assays; in response to NMDA receptor activation, translation is stimulated. In contrast to CPEB, CPEB3-mediated translation is unlikely to involve cytoplasmic polyadenylation, as it requires neither the cis-acting AAUAAA nor the trans-acting cleavage and polyadenylation specificity factor, both of which are necessary for CPEB-induced polyadenylation. One target of CPEB3-mediated translation is GluR2 mRNA; not only does CPEB3 bind this RNA in vitro and in vivo, but an RNAi knockdown of CPEB3 in neurons results in elevated levels of GluR2 protein. These results indicate that CPEB3 is a sequence-specific translational regulatory protein.
Keywords: CPEB, CPEB3, CPEB4, GluR2, translation
Introduction
One widely used mechanism to activate the translation of dormant mRNAs is cytoplasmic polyadenylation. Although this process was first described in invertebrates, it is also important for vertebrate oocyte development (Hake and Richter, 1994; Sheets et al, 1995; Stebbins-Boaz et al, 1996; Tay and Richter, 2001), cell cycle progression (Groisman et al, 2002), and neuronal synaptic plasticity (Alarcon et al, 2004). CPEB is the key protein that controls this process; it binds the 3′ UTR cytoplasmic polyadenylation element (CPE; consensus UUUUUAU) of target mRNAs (Hake and Richter, 1994). CPEB also interacts with a number of proteins that are important for polyadenylation and include (i) cleavage and polyadenylation specificity factor (CPSF), which binds the hexanucleotide AAUAAA, another cis element in the RNA essential for polyadenylation, (ii) symplekin, a scaffold protein that helps link CPEB to CPSF, and (iii) Gld-2, a cytoplasmic poly(A) polymerase (Barnard et al, 2004). CPEB also binds a guanine nucleotide exchange factor (Reverte et al, 2003), an RNA helicase (Minshall and Standart, 2004), and amyloid precursor proteins (Cao et al, 2005), all of which influence CPEB-dependent polyadenylation. Polyadenylation is initiated when CPEB is phosphorylated by Aurora A, which results in an enhanced interaction between CPEB and CPSF and between CPEB and Gld-2 (Mendez et al, 2000a, 2000b; Barnard et al, 2004). These events induce Gld-2 to extend the poly(A) tail. Translation of CPE-containing RNAs is most proximally controlled by Maskin, which simultaneously binds CPEB and the cap-binding factor eIF4E. The association of Maskin with eIF4E inhibits assembly of the eIF4F (eIF4E, eIF4G, eIF4A) initiation complex (Stebbins-Boaz et al, 1999; Richter and Sonenberg, 2005). Phosphorylation (Barnard et al, 2005) and polyadenylation and poly(A)-binding protein (PABP) help Maskin dissociate from eIF4E, thereby allowing translation initiation to proceed (Cao and Richter, 2002; Barnard et al, 2005).
In neurons, CPEB promotes the dendritic transport (Huang et al, 2003) and polyadenylation-induced translation of CPE-containing mRNAs following synaptic stimulation (Wu et al, 1998; Huang et al, 2002; Du and Richter, 2005). Because local mRNA translation modulates synaptic efficacy (Steward and Schuman, 2003; Bailey et al, 2004), it is not surprising that CPEB knockout (KO) mice display defects in synaptic plasticity (Alarcon et al, 2004), as do Aplysia neurons treated with an antisense oligonucleotide against CPEB mRNA (Si et al, 2003). However, three additional genes encode CPEB-like proteins in vertebrates (Mendez et al, 2002) that, at least at the RNA level, are expressed in the brain (Theis et al, 2003; Human Unidentified Genome Encoded Large Protein Database). The possibility that these proteins might partially compensate for the loss of CPEB caused us to investigate not only their RNA-binding specificities, but also their involvement in translational control in neurons.
All CPEB-like proteins in both vertebrates and invertebrates have a similar structure in which most of the carboxy-terminal region is composed of two RNA recognition motifs (RRMs) and two zinc-fingers. At least for CPEB, all of these domains are important for binding to the CPE with high affinity (Hake et al, 1998). In spite of these structural similarities, however, a sequence comparison of the RNA-binding regions indicated that CPEB is distinct from CPEB2–4 (Mendez and Richter, 2001). Indeed, mouse CPEB is more similar to Drosophila CPEB (also known as Orb) than it is to mouse CPEB2–4. This observation suggests that CPEB2–4 might bind a sequence other than the CPE. Using the RNA-binding region of CPEB4, we now report that SELEX (systematic evolution of ligands by exponential enrichment) analysis has identified a new binding sequence for these proteins. RNA gel shifts using this sequence as well as the CPE demonstrate that although CPEB binds the CPE and CPEB2–4 bind the SELEX sequence with high affinity (Kd of 100–160 nM), CPEB does not bind the SELEX sequence nor do CPEB3–4 bind the CPE. Although CPEB recognition of the CPE does not appear to involve RNA secondary structure, such structure is important for CPEB3–4 interaction with the SELEX sequence. CPEB3–4 are expressed in partially overlapping regions in the brain and are found in dendrites; CPEB3 colocalizes with a synaptic marker whereas CPEB4 does not. Experiments employing reporter RNAs transfected into neurons demonstrate that CPEB3 represses and then stimulates translation in response to NMDA treatment. CPEB3 neither interacts with CPSF nor requires the AAUAAA hexanucleotide for translational activation, implying that, in contrast to CPEB, it regulates translation in a polyadenylation-independent manner. The AMPA receptor GluR2 mRNA is a target of CPEB3 regulation; not only does CPEB3 bind this RNA in vivo, but an RNAi knockdown of CPEB3 in neurons results in elevated translation of GluR2 mRNA. Thus, based on RNA-binding specificity and functional regulation of translation, CPEB2–4 form a class of proteins distinct from CPEB.
Results
All CPEB-like proteins have a carboxy-terminal RNA-binding domain (RBD), which is comprised of two RRMs and two zinc-fingers, and an amino-terminal domain, which, in the case of CPEB, stimulates polyadenylation-induced translation once it is phosphorylated on T171 (in the mouse protein, S174 in Xenopus) by Aurora A. Although there is no significant identity among the amino-terminal domains of the CPEB proteins within a species (e.g., the mouse) or between species (e.g., mouse and fly), there is strong identity among the RBDs. For example, mouse CPEB and mouse CPEB2 are 45% identical in this region. However, mouse CPEB2, CPEB3, and CPEB4 are >95% identical. Interestingly, mouse CPEB has a higher identity to fly CPEB (also known as Orb) than to mouse CPEB2–4. Moreover, fly CPEB2 is more similar to mouse CPEB2–4 than it is to fly CPEB (Figure 1A). These comparisons imply that CPEB2–4 might interact with a sequence different from that of CPEB. Such a possibility was further suggested by experiments in injected Xenopus oocytes. Although mRNA encoding CPEB or CPEB3 had no effect on progesterone-induced oocyte maturation, mRNA encoding a chimeric protein composed of the regulatory domain of CPEB3 fused to the RBD of CPEB inhibited maturation (Figure 1B, left, the right shows Western blots of the resulting proteins). We infer that the chimeric protein acted as a repressor of translation of CPE-containing mRNAs required for maturation because it could not respond to progesterone stimulation; CPEB3 did not repress translation because it could not bind these mRNAs (see below).
Figure 1.
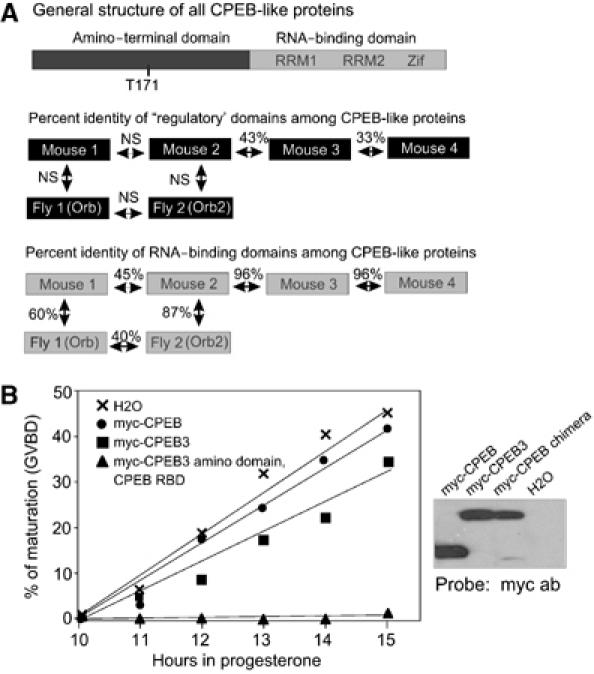
Structural features and comparison of CPEB-like proteins. (A) All CPEB-like proteins have an amino-terminal region and a carboxy-terminal region containing two RRMs and two zinc-fingers (Zif). CPEB T171, which is not conserved in CPEB2–4, must be phosphorylated for polyadenylation to occur. Among the amino-terminal regions of the CPEB proteins, there is little identity (NS, not significant). Among the RBDs, there is considerable identity. However, mouse CPEB (designated CPEB1 for convenience) is closer to Drosophila CPEB (Orb) than it is to mouse CPEB2; in addition, Drosophila CPEB2 (Orb2) is more similar to mouse CPEB2 than it is to Orb. Mouse CPEB2–4 are nearly identical in the RBDs. (B) Xenopus oocytes were injected with water or RNA encoding myc-CPEB or CPEB3, or myc fused with the amino domain of CPEB3 and the RBD of CPEB. The oocytes were incubated with progesterone and scored for oocyte maturation as assessed by germinal vesicle breakdown (GVBD). A Western blot probed with myc antibody shows the level of the myc fusion proteins in oocytes.
SELEX identifies CPEB2–4 binding sequences
To determine whether the RBDs of CPEB2–4 indeed interact with sequences other than the CPE, a SELEX experiment was performed. The RBD of CPEB4 was mixed with in vitro-synthesized RNA derived from an oligonucleotide library composed of a randomized 25-mer flanked by constant regions for PCR; the mixture was subjected to eight rounds of binding and elution. After the final elution, the RNA was cloned and the sequences of 50 plasmid inserts were determined, some of which are shown in Figure 2A. In vitro gel shifts confirmed that CPEB4 RBD bound all cloned RNAs tested (Figure 2B). Two point mutations that disrupted the CPEB4 zinc-fingers abrogated binding to a selected RNA, 1904 (Figure 2C), indicating the importance of this domain for RNA interaction, which is consistent with a previous finding that the zinc-fingers are important for CPEB to bind the CPE (UUUUAU derived from Xenopus mos; Hake et al, 1998; de Moor and Richter, 1999). Further analysis showed that while CPEB3 and CPEB4 bound the 1904 sequence, CPEB did not. Moreover, CPEB did bind CPE-containing RNA, as expected, but CPEB3 and CPEB4 did not (Figure 2D). When analyzed kinetically, the binding constant (Kd) of CPEB for the CPE was 130 nM (Hake et al, 1998); CPEB did not interact with 1904. In contrast, while the CPEB3 and CPEB4 RBDs (>95% identical; Figure 1) did not bind the CPE, the Kd values for the 1904 sequence were 166 and 100 nM, respectively. Thus, CPEB and CPEB2–4 have different RNA-binding specificities.
Figure 2.
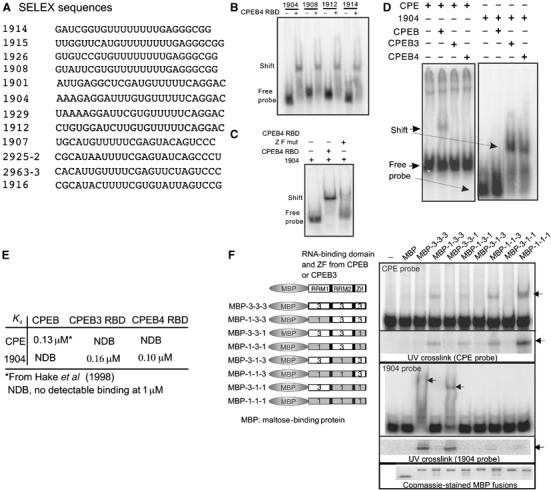
Analysis of CPEB and CPEB4 interaction with RNA. (A) The CPEB4 RBD was mixed with RNA synthesized in vitro from oligonucleotides containing a randomized 25-mer central domain flanked by constant regions used for transcription and PCR. The RNA–protein complexes were collected on filters, eluted, and the cycle repeated for eight rounds before cloning. Representatives of 50 clones are shown. (B) Several SELEX RNAs were synthesized in vitro with 32P-UTP and used for gel shifts with 500 nM CPEB4RBD. (C) The CPEB4 RBD, containing or lacking the zinc-finger (ZF), was used in gel shifts with SELEX sequence 1904. (D) RNA gel shifts were performed with CPEB, CPEB3, or CPEB4 with CPE-containing RNA or the 1904 sequence. (E) A kinetic analysis of CPEB3 and CPEB4 RBD interactions with the 1904 sequence or the CPE was used to calculate equilibrium dissociation constants (Kd). Similar experiments were performed with CPEB binding to the 1904; the binding of CPEB to the CPE was taken from Hake et al (1998). (F) Various regions of the RBDs of CPEB and CPEB3 were fused to MBP, expressed in bacteria, and used for RNA gel shifts and UV crosslinks with the CPE or the 1904 sequence. The bottom panels show that equal amounts of the MBP fusion proteins were used on all experiments.
To identify the origin of these different RNA-binding specificities, chimeric molecules composed of RRM1, RRM2, and the zinc-fingers (Zif) from CPEB and CPEB3 were fused to maltose-binding protein (MBP) and subjected to RNA gel shifts and UV crosslinking analysis with the CPE and the 1904 SELEX sequence (Figure 2F). Although the entire CPEB RBD was the most efficient at binding the CPE, RRM1 was essential for this binding (gel shift and UV crosslink). Similarly, the entire CPEB3 RBD was the most efficient at binding the 1904 sequence (compare gel shift and UV crosslink), but in this case, both RRMs were important for binding. Exchange of the zinc-fingers had no effect on RNA binding. Thus, while certain domains within CPEB (RRM1) and CPEB3 (RRMs 1 and 2) are important for binding specificity, the zinc-fingers of both proteins (see also Hake et al, 1998), while important for RNA interaction, do not confer specificity.
CPEB3–4 RBD recognizes RNA secondary structure
One of the goals of the SELEX experiments was to identify endogenous mRNAs that are bound by the CPEB3–4 RBD. To make a search more specific, we delineated the nucleotides necessary for binding. Using 1904 as a substrate, mutations in several regions of the 25-mer destroyed or reduced RNA binding (Figure 3A). Surprisingly, deletion of the 3′ but not 5′ constant region destroyed binding; the 25-mer alone was not bound by the RBD (Figure 3A). Using the mfold program (Zuker, 2003), the 5′ constant region deleted in RNA M14 is predicted to fold into a secondary structure with poly(U) in the loop; similar structures were predicted for most of the other selected RNAs (Figure 3B and data not shown). To assess whether such a possible structure (ΔG of −21.3 kcal/mol) could be important for CPEB4 binding, three bases were mutated in the bottom stem (CAC for GUG), which completely abrogated CPEB4 binding (Figure 3B). Compensatory changes in the complementary sequence (denoted REV for reverse, GUG for CAU) restored binding by CPEB4; however, substitution of the GUG for CAU alone did not restore binding. These results suggest that RNA secondary structure could be necessary for CPEB4 binding.
Figure 3.
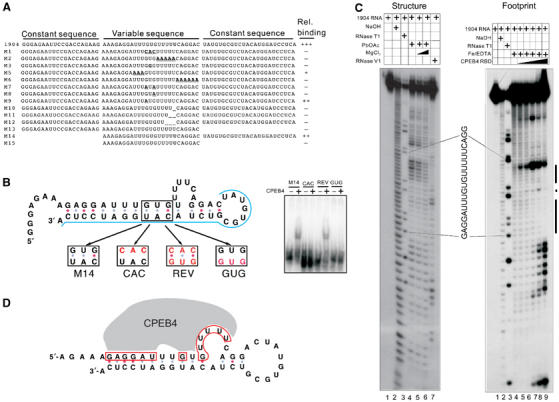
The CPEB4 RBD recognizes RNA secondary structure. (A) Nucleotide changes were introduced into the 1904 sequence and the resulting RNAs were used for gel shifts with the CPEB4 RBD; the relative amount of binding is indicated. (B) Compensatory mutagenesis of M14 RNA. The mfold program generated a predicted secondary structure of M14 RNA. The blue line denotes the nucleotides derived from the constant sequence, the black box indicates the part of the stem used for the mutagenesis, and the red nucleotides indicate the mutated sequences. Gel shifts of these RNAs using the CPEB4 RBD is shown at right. (C) Structure mapping and RNA footprinting. 5′ end-labeled RNA was untreated (lane 1), or subjected to partial alkaline hydrolysis (lane 2), digested with RNase T1 (lane 3), subjected to lead acetate cleavage in the absence or presence of MgCl2 (lanes 4–6, left panel), or digested with the double-strand-specific nuclease RNase V1 (lane 7, left panel). For RNA footprinting, RNA samples either lacked added protein (lane 4, right), or were 0.25, 0.5, 1, 2, or 4 μM CPEB4 RBD and were treated with the hydroxyl radical (lanes 5–9, right panel). The products were then analyzed on a sequencing gel. The protected region is denoted by a black bar. (D) Predicted RNA secondary structure and nucleotides protected from hydroxyl radical cleavage by CPEB4 RBD; the red box indicates the protected nucleotides. (For colour figure see online version.)
To further assess this hypothesis, 5′ end-labeled RNA was cleaved with the single-strand-specific lead acetate (Darnell et al, 2005), and RNase V1, which cleaves double-stranded regions (Lockard and Kumar, 1981). These samples were resolved on a sequencing gel and compared to parallel lanes containing untreated RNA, RNA partially hydrolyzed with NaOH, and RNA cleaved with RNase T1 to locate the guanosines (Figure 3C). Lane 4 shows that the uridine residues predicted to reside in a loop structure (panel B) were susceptible to lead cleavage, which was reduced by inclusion of MgCl2, a competitor of the lead ion (lanes 5 and 6). These uridine regions were also resistant to RNase V1 cleavage, further demonstrating that they are not base-paired with other residues. To determine the region of the RNA bound by the CPEB4 RBD, RNA footprinting was performed. An increasing amount of CPEB4 RBD was mixed with 5′ end-labeled RNA; the RNA was then cleaved by the hydroxyl radical generated from a mixture of Fe/EDTA (Wang and Padgett, 1989). Compared to a sample digested with hydroxyl radical only (no protein) (lane 4), CPEB4 protected two regions of the RNA, one was the single-stranded uridines together with the 5′ proximal stem and the other was an adjacent double-stranded region (lane 9). The binding of CPEB4 to the 5′ proximal stem is consistent with the compensatory mutagenesis result and further suggests the significance of RNA structure for CPEB4 binding. Two other SELEX clones gave similar RNA footprinting patterns (data not shown). Figure 3D depicts a revised secondary structure of the minimal RNA required for CPEB4 RBD binding based on the data in panels B and C. The residues protected by the CPEB4 RBD are also indicated.
CPEB3 and CPEB4 in the brain
Western blotting shows that CPEB3 and CPEB4 are present in many tissues including the brain (Figure 4A). Immunohistochemistry demonstrates that although both proteins were evident in the hippocampus and granule cells of the cerebellum, only CPEB4 was detected in Purkinje cells of the cerebellum. In contrast, only CPEB3 was detected in mitral cells of the olfactory bulb and interneurons of the cerebellum (Figure 4B). In hippocampal neurons cultured in vitro and stained for Map2 to identify dendrites (not shown), CPEB3 often colocalized with synaptophysin, a synaptic marker (Figure 5A). Although CPEB4 appeared to be adjacent to synaptophysin immunoreactivity, both proteins were strongly detected in the post-synaptic density (PSD) fraction (Figure 5B). CPEB4, the only one tested, colocalized with RNA as assessed by Syto-RNAselect staining (Figure 5A). Finally, both CPEB3 and CPEB4, when fused to GFP, were detected in dendrites, often as puncta (Figure 5C). Taken together, these data show that CPEB3 and CPEB4 are expressed in only partially overlapping regions of the brain; within hippocampal neurons, where both are expressed, they appear to be synaptic.
Figure 4.
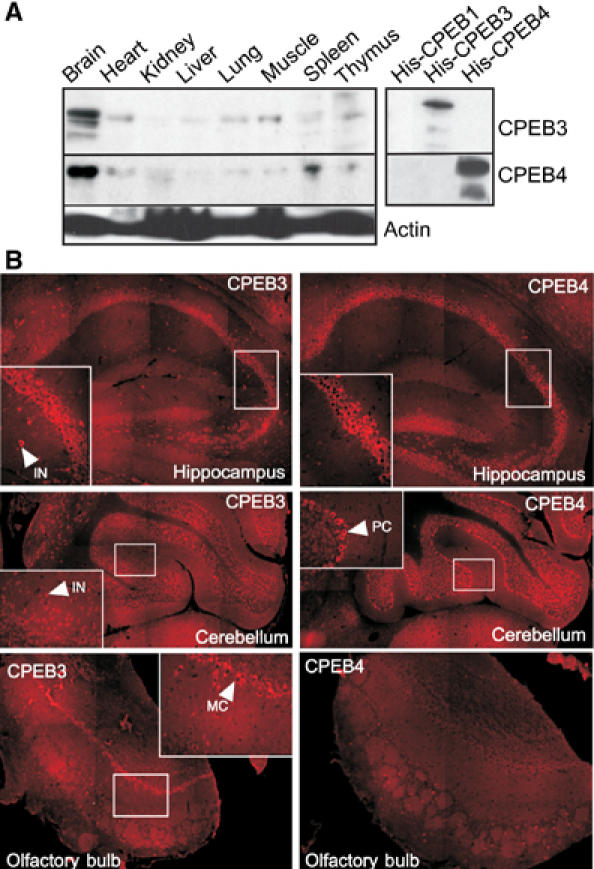
CPEB3 and CPEB4 in the brain. (A) Western blots of several mouse tissues probed for CPEB3 and CPEB4. The blot also shows that the antibodies for CPEB3 and CPEB4 do not crossreact and do not recognize CPEB. The calculated molecular sizes of CPEB3 and CPEB4 are ∼74 and 78 kDa, respectively. However, on SDS gels, their apparent molecular sizes are ∼100 and ∼98 kDa, respectively. (B) Immunocytochemistry for CPEB3 and CPEB4 in rat hippocampus, cerebellum, and olfactory bulb. The arrows point to specific regions of immunoreactivity in interneurons (IN) and Purkinje cells (PC) of the cerebellum and Mitral cells (MC) of the olfactory bulb.
Figure 5.
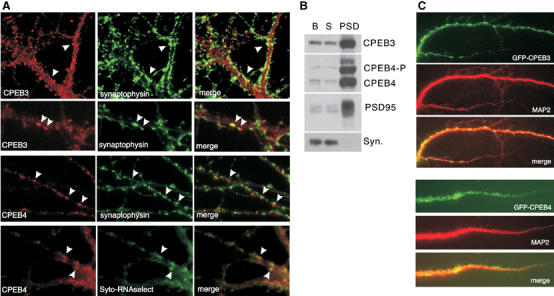
Localization of CPEB3 and CPEB4 in neurons. (A) Co-staining of CPEB3 or CPEB4 with synaptophysin in 21-day-old cultures of hippocampal neurons. CPEB4 is also co-stained with the RNA marker Syto-RNAselect after UV crosslinking and fixation. (B) Detection of CPEB3 and CPEB4 in the PSD fraction. P-CPEB4 refers to putative phosphorylated CPEB4, B refers to the brain extract, S refers to the synaptoneurosome fraction, and syn refers to synaptophysin. (C) Detection of transfected GFP-CPEB3 and GFP-CPEB4 proteins in dendrites of hippocampal neurons co-stained with MAP2.
CPEB-like proteins and translation
To investigate whether the CPEB3 could be involved in translational control, we employed a tethered function assay in hippocampal neurons that were transfected with several sets of reporter RNAs (Figure 6A). They encoded the dimeric MS2 coat protein (MS2CP) fused to CPEB3 or mutant CPEB3 proteins that lacked the amino- or carboxy-terminal regions, or as a control, MS2CP fused to GFP. These RNAs were mixed with RNA encoding firefly luciferase that contained or lacked the stem–loops recognized by MS2CP. The mix also contained RNA encoding Renilla luciferase, which served as an internal control. In transfected neurons, MS2CP-CPEB3 and MS2CP-GFP were synthesized (Figure 6B, left) and gel shifted a probe containing the MS2CP stem–loops (Figure 6B, right). Figure 6C shows that relative to MS2CP-GFP, MS2CP-CPEB3 and MS2CP-CPEB3 amino terminus reduced firefly luciferase expression by ∼30–40%, which was statistically significant (P<0.01). This reduction was not due to a general repression, as the substitution of the MS2CP moiety with myc had no effect on translation. Moreover, removal of the MS2 stem–loop from firefly luciferase abrogated the NMDA-induced translation by MS2CP-CPEB3 (Figure 6D). Thus, CPEB3 can reduce translation when tethered to a specific mRNA.
Figure 6.
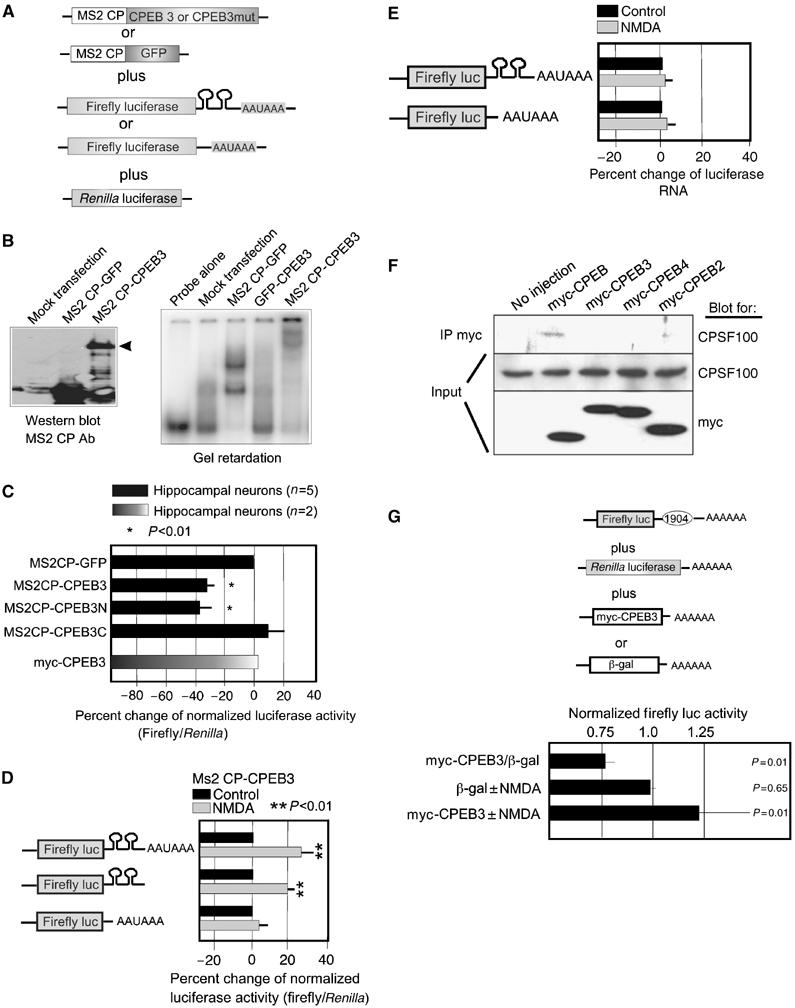
Translational repression in neurons. (A) Hippocampal neurons were transfected with mRNAs encoding (1) dimeric MS2CP fused to CPEB3 or amino- or carboxy-terminal truncations of this protein, or MS2CP fused to GFP, and (2) firefly luciferase whose 3′ UTR contained or lacked the MS2 stem–loop, and (3) Renilla luciferase. (B) Western blot showing the expression of the fusion proteins in transfected neurons. The right panel shows an RNA gel shift of transfected proteins binding to RNA containing the MS2 stem–loops. (C) The ratio of firefly/Renilla luciferase activity in neurons (normalized to the MS2CP-GFP control) transfected with the RNAs noted in panel A. Neurons were transfected with mRNA encoding myc-CPEB3 in place of the MS2CP fusions. All the firefly luciferase RNAs contained the MS2 stem–loop in the 3′ UTR.(D) Luciferase values calculated as those in panel C from neurons transfected with RNA encoding firefly luciferase that contained or lacked the MS2 stem–loop and that was treated with NMDA. (E) Semiquantitative RT–PCR of luciferase RNA containing or lacking the MS2 stem–loops following RNA transfection into neurons; some of the neurons were treated with NMDA. (F) Xenopus oocytes were injected with mRNAs encoding fusions between myc and CPEB or CPEB2–4. The CPEB proteins were then immunoprecipitated with myc antibody and probed for myc, to note the level of expression, and the 100 kDa subunit of CPSF. (G) Hippocampal neurons were transfected with firefly luciferase mRNA appended with a 3′ UTR containing the 1904 SELEX sequence, Renilla luciferase mRNA, and either myc-CPEB3 or β-galactosidase. Some of the neurons were also treated with NMDA. Firefly luciferase, normalized to Renilla luciferase, was then determined. Probability (p) values were derived from the Student's t-test.
To assess whether CPEB3 can stimulate translation and if so, whether it requires AAUAAA, a firefly luciferase containing or lacking this sequence was transfected into neurons together with MS2CP-CPEB3 as before and then stimulated with NMDA. MS2CP-CPEB3 enhanced translation by 20–30% irrespective of whether the AAUAAA was present (Figure 6D). The MS2 stem–loops were required for MS2CP-CPEB3 translation. This change in translation occurred even though the luciferase RNA levels were unchanged (panel E). In injected Xenopus oocytes, CPEB was co-immunoprecipitated with CPSF, as shown previously (Mendez et al, 2000b), but CPEB2–4 were not (panel F). Finally, luciferase mRNA was appended with a 3′ UTR containing the 1904 SELEX sequence and analyzed for translation when cotransfected with myc-CPEB3 or, as a control, β-galactosidase. CPEB3 reduced translation of the reporter RNA by ∼25%. Although NMDA had no effect on luciferase activity when β-galactosidase was cotransfected, it stimulated luciferase activity by nearly 25% when CPEB3 was cotransfected (panel G). These data, as well as those in Figure 1B, suggest that although CPEB3 repressed and then activated translation in response to NMDA, it probably does not do so by changing poly(A) tail length because it does not require the cis-acting AAUAAA and does not bind CPSF.
CPEB3 controls GluR2 mRNA translation
Because RNA secondary structure appears to be important for binding by CPEB3, BLAST searches for endogenous RNAs based on SELEX sequence alone would not be fruitful. Consequently, we considered several neuronal RNAs whose translation might be regulated and whose 3′ UTR could contain a stem–loop structure similar to that shown in Figure 3B. The mRNA encoding the AMPA receptor GluR2 is dendritically localized and may be subject to translational control (Kacharmina et al, 2000); the mfold program also predicts several stem–loop structures that resemble those predicted to form in the 1904 SELEX sequence (data not shown). To assess whether CPEB3 might bind this mRNA, the 3′ UTRs of GluR2 and Arc (a control) mRNAs were subjected to UV crosslinking in vitro with the RBDs of CPEB and CPEB3 fused to MBP. Figure 7A shows that the RBD of CPEB3, but not of CPEB, strongly crosslinked to the 3′ UTR of GluR2 but not of Arc, suggesting that GluR2 RNA could be a direct binding substrate of CPEB3. To identify the region of GluR2 3′ UTR bound by CPEB3, a deletion series was constructed and used for in vitro crosslinking to CPEB3 RBD (Figure 7B). With the exception of nucleotides 1–474, CPEB3 RBD bound to multiple regions throughout the length of the 3′ UTR. Additional deletions were constructed that were used for RNA gel shifts (Figure 7C). Again, CPEB3 bound to multiple regions of the 3′ UTR. The binding to one of the regions that was chosen, L4, was specific, as the shift was competed away when the 1904 SELEX sequence was added to the mix but not when 1904-M1 was added. Moreover, a 202-base fragment (S4) derived from the L4 RNA was bound by CPEB3, which again was competed by the 1904 sequence but not by the 1904-M1 sequence (Figure 7D).
Figure 7.
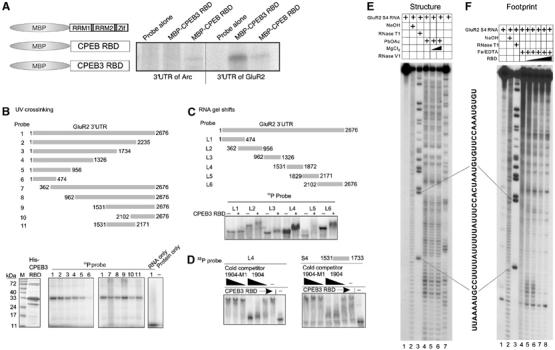
CPEB binds the GluR2 3′ UTR. (A) Maltose-binding fusion proteins containing the RBD of CPEB or CPEB3 were expressed in bacteria and used for in vitro UV crosslinking with 32P-labeled 3′ UTRs of Arc and GluR2 mRNAs. (B) Deletions of the GluR2 3′ UTR were used for UV crosslinking with His-tagged CPEB3 RBD. (C) Additional deletions of GluR2 3′ UTR were used for RNA gel shift reactions with His-tagged CPEB3 RBD. (D) Some of the gel shift mixtures also contained unlabeled 1904 or 1904-M1 RNAs as competitors for CPEB3 RBD binding. (E) Structure mapping of GluR2 3′ UTR fragment, S4. 5′ end-labeled S4 RNA was either untreated (lane 1), subjected to partial alkaline hydrolysis (lane 2), RNase T1 digested (lane 3), treated with 100 mM lead acetate in the absence or presence of 10 mM or 100 mM MgCl2 (lanes 4–6, left panel), or digested with nuclease V1 (lane 7, left panel) and analyzed on a sequencing gel. The sequence corresponding to the uridine-rich region is shown. (F) RNA footprinting of S4. 5′ end-labeled S4 RNA was cleaved by hydroxyl radical in the absence or presence of 0.25, 0.5, 1.0, 2.0, and 4.0 μM CPEB3 RBD (lanes 4–8). Lanes 1–3 is the same as in panel E.
The S4 RNA was used for structure mapping as in Figure 3. Multiple regions were cleaved by lead acetate, including one containing multiple uridine residues (Figure 7E, lane 4). This region was not cleaved by the double-strand-specific RNase V1 (lane 7). Sequences including these single-stranded uridines were protected by CPEB3 RBD from cleavage by the hydroxyl radical (Figure 7F, lanes 4–8). Thus, the CPEB3 RBD may interact with a sequence and structure in GluR2 similar to the sequence and structure identified by SELEX. We also note that S4 contains a CPE-like sequence, which interacts with CPEB in vitro (data not shown).
To determine whether CPEB3 binds GluR2 mRNA in vivo, living cultures of hippocampal neurons were irradiated with UV light, which was followed by cell homogenization in detergent-containing buffer to reduce nonspecific adsorption followed by immunoprecipitation with CPEB3 antibody or IgG. The precipitates were then deproteinized and subjected to RT–PCR for Arc, Map2, neurofilament (NF), or GluR2 RNAs. Although all the RNAs were clearly amplified from input material, only GluR2 RNA was amplified from the CPEB3 co-immunoprecipitate. No RNAs were amplified from the control IgG immunoprecipitate (Figure 8A). Thus, GluR2 mRNA is an in vivo substrate of CPEB3.
Figure 8.
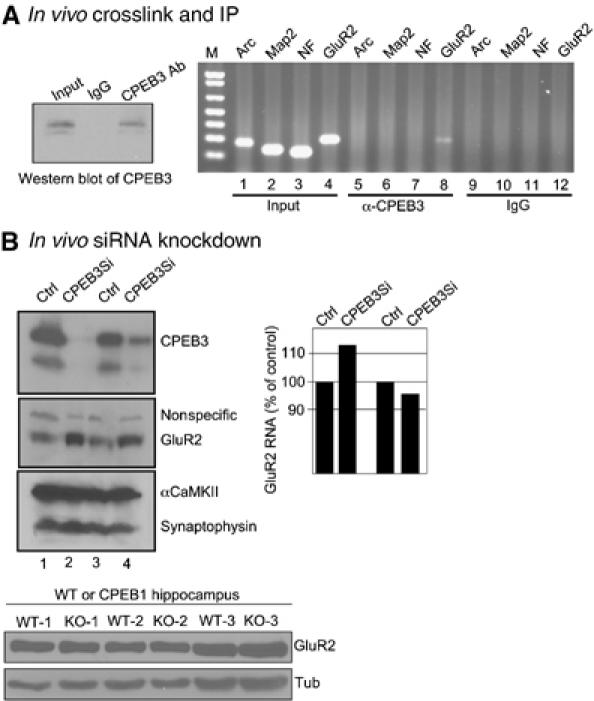
CPEB3 controls GluR2 mRNA translation. (A) Living cultures of hippocampal neurons were irradiated with UV light, homogenized, and subjected to immunoprecipitation with CPEB3 antibody or IgG in the presence of SDS-containing buffer. The precipitated RNA was extracted after proteinase K digestion and subjected to RT–PCR for Arc, Map2, NF, or GluR2 mRNAs. (B) Endogenous CPEB3 knockdown by RNAi. Cultured hippocampal neurons were infected with a control lentivirus or one expressing a short hairpin RNA against CPEB3 under the control of the U6 promoter. Extracts were then prepared from the cells and analyzed for levels of CPEB3, GluR2, αCaMKII, and synaptophysin. From other cultures, the RNA was extracted and analyzed for GluR2 mRNA by real-time PCR. The bottom panel shows the level of GluR2 in wild-type and CPEB KO hippocampus. Tubulin served as a loading control.
We next performed RNAi knockdown experiments to examine whether CPEB3 regulates GluR2 mRNA translation in neurons. Hippocampal neurons cultured for 4 days were infected with lentivirus containing or lacking a short hairpin sequence for CPEB3 under the control of the U6 promoter. After a further 4 days of culture, the cells were harvested and the extracted protein was probed on Western blots. In two independent experiments, CPEB3 protein was reduced by 80–99% (Figure 8B). In contrast, GluR2 levels increased by three-fold whereas αCaMKII and synaptophysin were unaffected. Because the CPEB3 knockdown had little effect on the level of GluR2 RNA level (Figure 8B), we infer that the translation of GluR2 mRNA is under negative regulation by CPEB3. It is possible that GluR2 mRNA localization could also be affected. Finally, we note that GluR2 levels were identical in wild-type and CPEB KO hippocampus, indicating that the expression of GluR2 RNA is controlled by CPEB3 and not CPEB.
Discussion
In this study, we demonstrate that in contrast to CPEB, CPEB proteins 2–4 do not avidly bind the CPE, but instead strongly interact with a U-rich loop within a stem–loop structure. While the zinc-fingers are necessary for RNA binding, it is RRMs 1 and 2 that confer the binding specificity. These results imply that CPEB2–4 probably do not functionally substitute for CPEB, as they interact with RNAs with different binding specificities. There could, however, be RNAs that are bound by CPEB and CPEB2–4. CPEB3 represses translation of a reporter RNA in transfected neurons and stimulates translation in response to NMDA. Although the mechanism of translational control by CPEB3 is not yet known, it does not bind CPSF nor does it require an AAUAAA cis element, implying that unlike CPEB, it does not promote cytoplasmic polyadenylation. Most importantly, CPEB3 interacts with GluR2 mRNA in vivo and a knockdown of CPEB3 in neurons stimulates the translation of this mRNA. Thus, CPEB3 is a sequence-specific translational repressor that governs the synthesis of the AMPA receptor GluR2.
Unlike CPEB, the CPEB3–4 RBD recognizes a secondary structure and interacts with uridines that are single-stranded as well as double-stranded stems. Although the zinc-fingers of the RBD are necessary for stable RNA binding, they do not confer binding specificity. In other proteins, zinc-finger domains have been shown to bind double-stranded RNA (Yang et al, 1999; Mendez-Vidal et al, 2002), and we would hypothesize that this could also be the case with the CPEB2–4 RBD.
CPEB2–4 are functionally distinct from CPEB and may also be distinct from one another. Although they bind the same cis element, the fact that CPEB3 and CPEB4 reside in only partially overlapping regions (i.e., both are in the hippocampus, but only CPEB4 is in Purkinje cells of the cerebellum and only CPEB3 is in mitral cells of the olfactory bulb) indicates that they may interact with at least some unique RNAs in vivo. In addition, the large amino-terminal regions of CPEB2–4 proteins are only 33–43% identical. This relatively low identity suggests that these proteins could respond to different signaling pathways and/or interact with different sets of proteins to modify their activities such as translation repression, stimulation, or RNA transport.
Translational control of GluR2 mRNA
The use of SELEX to identify CPEB2–4 binding sites in RNA was not particularly useful for recognizing endogenous RNA targets, as secondary structure was important for RNA–protein interactions. Consequently, we examined a number of neuronal mRNAs that might form similar secondary structures as determined by the mfold program; the mRNA encoding the AMPA receptor GluR2 was able to do so and was immunoprecipitated with CPEB3 following UV irradiation of living neurons. An RNAi knockdown of CPEB3 stimulated GluR2 levels while having little effect on GluR2 mRNA, suggesting that CPEB3 is a specific translational repressor protein. We do note the caveat that our attempts examine the translation of a reporter RNA appended with the GluR2 3′ UTR in transfected hippocampal neurons was variable (data not shown), perhaps because the RNA was not properly localized or because it exceeded the amount of the endogenous CPEB3. Nonetheless, the cumulative data in this report do indicate that CPEB3 is at least one factor that mediates GluR2 mRNA expression.
The molecular mechanism by which CPEB3 modulates translation is unknown; perhaps it interacts with an eIF4E-binding protein such as Maskin, competes with eIF4E for binding to the cap (Cho et al, 2005), or modulates ribosomal subunit joining (Ostareck et al, 2001). Irrespective of how CPEB3 controls translation, the observation that GluR2 is an endogenous target has important implications for AMPA receptor regulation. For example, GluR1 mRNA is present in dendrites (Miyashiro et al, 1994) and is regulated at least in part at the translational level (Kacharmina et al, 2000; Ju et al, 2004). AMPA receptors are also controlled at the protein localization level, as they are trafficked to the membrane in response to activity (Malinow and Malenka, 2002). Thus, both cell soma and local synthesis of GluR1 and GluR2 could contribute to the formation of functional AMPA receptors.
Translational control and synaptic plasticity
Intense interest has focused on local (dendritic) mRNA translation, as it was shown nearly a decade ago to be important for maintaining long-term changes in synaptic strength (Kang and Schuman, 1996). Although many studies have confirmed and extended these results (reviewed by Klann and Dever, 2004; Sutton and Schuman, 2005), in some ways local translation remains enigmatic. For example, although it has become almost axiomatic that activity-induced synthesis of new proteins helps distinguish experienced from naïve synapses (Steward and Schuman, 2001), a demonstration that specific proteins involved has not emerged. The synthesis of several proteins increases upon synaptic stimulation, but only ∼2–4-fold (e.g., Kelleher et al, 2004; Schratt et al, 2004). Such increases could certainly be physiologically significant, especially if they are concentrated at particular synapses. Moreover, relatively modest changes in many proteins could be essential for plasticity. Alternatively, perhaps the synthesis of less abundant proteins, while substantially stimulated by synaptic activity, is obscured by the general but low-level increase. Such a possibility is particularly intriguing, as it is known to occur in other cells. In Xenopus oocytes, progesterone stimulation of M-phase progression is accompanied by an ∼2-fold increase in general protein synthesis. In contrast, proteins such as Mos and cyclin B1, which are necessary for M-phase progression, increase from nearly undetectable levels to easily observed amounts when assayed by, for example, Western blots. However, because these proteins are relatively rare compared to the bulk of the newly made proteins, they are not readily detected by metabolic labeling unless they are specifically immunoprecipitated. Thus, critically important proteins could be synthesized at synapses, but because they are not abundant, they are difficult to detect. In contrast to specific protein synthesis, the productive capture of certain newly made proteins by stimulated synapses may be responsible for regulating synaptic efficacy (Frey and Morris, 1997; Kelleher et al, 2004).
Although one way to investigate the relationship between protein synthesis and plasticity is obviously to identify mRNAs that are translated in response to activity, an alternative approach is to first identify translational control proteins in the brain and then determine which mRNAs are bound and/or regulated by them. For example, a number of CPE-containing RNAs have now been identified that undergo activity-dependent polyadenylation (Wu et al, 1998; Du and Richter, 2005), presumably because they are bound by CPEB. In this study, we have shown that CPEB3 and CPEB4 are components of the PSD and have identified one mRNA that is bound by and under the translation control of CPEB3. By defining the precise binding site in GluR2 mRNA, we may be able to deduce additional RNAs that are regulated by this and the other CPEB-like proteins.
Materials and methods
Plasmids and protein expression
Escherichia coli strain BL21(DE3)pLysS (Novagen) was transformed with expression plasmids (pET28a) encoding CPEB and the RBDs of CPEB3–4. The cells were cultured to OD600 0.3–0.6 before the addition of 1 mM IPTG for 30 min. His-tagged proteins were purified using Ni-NTA agarose resin (Qiagen) and dialyzed against 1X GR buffer (10 mM HEPES, pH 7.6, 50 mM KCl, 1 mM MgCl2, 0.1 mM ZnCl2, 10% glycerol, 1 mM DTT) for 2 h. His-CPEB was denatured with 6 M urea and then renatured by stepwise lowering of the urea concentration in the wash buffer to 2 M urea before elution and dialysis against 1X GR with 2 M urea. The CPEB4RBD used for SELEX was further purified by FPLC (AKTA, Amersham Pharmacia Biotech) in a HiLoad 16/60 Superdex 200 column in 1X GR buffer; protein concentrations were determined by BCA protein assay reagent (Pierce).
For other experiments, DH5α cells transformed with plasmids encoding MBP fused to CPEB proteins were grown to OD600∼0.6 and then induced with 1 mM IPTG for 3 h. The bacterial pellet was resuspended in buffer A (20 mM HEPES, pH 7.6, 500 mM NaCl, 1 mM MgCl2, 0.5 mM DTT, 0.1% Triton X-100, 10% glycerol, 1 mM PMSF) and incubated for 30–40 min on ice with lysozyme (1 mg/ml). The cells were sonicated to loss of viscosity and clarified by centrifugation at 12 000 g for 15 min. The resulting supernatant was incubated with amylose resin (NEB) and washed with 100 × volume of buffer A. The protein was eluted with buffer A (100 mM NaCl, no Triton X-100) with 10 mM maltose. Gel shift assays typically employed 100 ng of fusion protein.
SELEX
For SELEX, three oligonucleotide primers were synthesized: PO-67, GGGAGAATTCCGACCAGAAGN25TATGTGCGTCTACATGGATCCTCA; PO-69, TGAGGATCCATGTAGACGCA; PO-71, TAATACGACTCACTATAGGTGGGAGAATTCCGAC CAGAAG. To generate templates for a SELEX library, these three primers were used in a PCR reaction at a ratio of PO-67:PO-69:PO-71=1:1:3. The PCR products were then used for in vitro transcription with T7 RNA polymerase. Free nucleotides from the in vitro transcription reaction were removed by FPLC using HiPrep 26/10 desalting column before further purification by denaturing TBE PAGE. For the SELEX, FPLC-purified CPEB4RBD was mixed with heat-denatured RNA library in 1 ml of 1X GR buffer containing 160 μg tRNA and 5 mg heparin. RNA–protein mixtures were kept on ice for 10 min and at room temperature for 10 min before being filtered through nitrocellulose membranes on a porous plate by gentle suction. The membranes were washed with 5 ml of 1X GR with tRNA (0.5 mg/ml). The membranes were cut into small pieces and mixed with Trizol for RNA extraction as described (Invitrogen). The extracted RNA was used for reverse transcription to generate cDNA by Superscript Reverse Transcriptase II (Invitrogen) and PCR amplification using PO-69 and PO-71. The amplified PCR products were used for transcribing RNA for the next round of SELEX. To increase the binding specificity of selected RNA to CPEB4RBD, the amount of this protein used in each cycle was reduced by half from 2 μM for the first cycle to 25 nM for the seventh and eighth cycles. The amount of purified RNA library used for each SELEX cycle was 20 μg except the first cycle, which was 60 μg.
Electrophoresis mobility shift assay
RNA probes used for mobility shift assays were labeled by in vitro transcription with α32P-UTP. Mutants of 1904 RNA were transcribed from PCR products using oligonucleotides with the T7 promoter. For gel shifts, 20 μl reactions in 1X GR buffer included probe RNA (various concentrations, ∼105 c.p.m.), protein (e.g., CPEB4 RBD), 1 μg tRNA, 50 μg heparin, and 12 U RNasin. The reactions were was kept on ice for 10 min and then at room temperature for 10 min, before being resolved by TBE PAGE.
Immunohistochemistry and RNA transfection in neurons
Two-month-old male mice were anesthetized and perfused with 4% formaldehyde. The fixed brains were embedded in paraffin, sectioned at 10 μm thickness, and treated with antigen retrieval procedure (Tay et al, 2003) before incubation with the affinity-purified CPEB3 and CPEB4 antibodies. Hippocampal neurons were cultured and immunostained as described (Huang et al, 2002). Other neuronal cultures were UV irradiated in a Stratagene 1800 Crosslinker, fixed, and stained with Syto-RNAselect (Molecular Probes).
Hippocampal neurons cultured for 9–10 days in Neurobasal medium with B27 supplement (Invitrogen) at a cell density of 30 000–40 000/cm2 were cotransfected (TransMessenger Transfection reagent, Qiagen) for 3 h with ∼12 pmol of Ms2CP-CPEB RNA, 1.7 pmol of firefly luciferase RNA appended with various 3′ UTRs, and 1 pmol of Renilla luciferase RNA. The transfected neurons were stimulated with 50 μM NMDA for 3 h before lysis in 100 μl of buffer for dual luciferase assay (Promega). To quantify the amount of firefly and Renilla luciferase RNAs, total RNA was extracted from transfected neurons, reverse transcribed, and subjected to real-time PCR amplification (Huang et al, 2003). The specific primers used were as follows: firefly sense 5′-GAGATGTATTACGCAAAGTAC and antisense 5′-CCAGTATGACCTTTATTGAGC; Renilla sense 5′-GTTGTGTTCAAGCAGCCTGG and antisense 5′-CCAGTGAGTAAAGGTGACAG.
Lentivirus infection of cultured neurons
To knock down rat CPEB3 (rCPEB3), the coding region of rCPEB3 was RT–PCR amplified from total RNA isolated from rat hippocampal neurons and cloned to pcDNA3.1+ plasmid. Five shRNA sequences designed against the mRNA were cloned into the lentiviral vector PLL3.7-Syn (gift of M Sheng); one that was particularly efficacious when tested in transfected 293T-17 cells corresponded to nucleotides 2320–2337 (CCGTACGTGCTGGATGAT) of rCPEB3. This particular construct was used to produce lentivirus using the viralpower packaging system (Invitrogen) according to the manufacturer's protocol. Generally, hippocampal neurons (4 days in vitro (DIV)) were infected with the virus (1–2 MOI) for 24 h. The infected neurons were cultured for another 3–4 days before RNA isolation or protein extraction.
Oocyte injection and immunoprecipitation
A 25 ng portion of RNA encoding myc-tagged CPEB, CPEB3, or chimeric CPEB was injected to Xenopus oocytes that were cultured for 14 h before stimulation with progesterone. For immunoprecipitation, 40 injected oocytes were homogenized in 200 μl of immunoprecipitation buffer (20 mM HEPES, pH 7.6, 150 mM NaCl, 1 mM MgCl2, 0.5 mM DTT, 0.1% Triton X-100, 100 μg/ml RNaseA) and centrifuged at 12 000 g for 5 min at 4°C. The supernatant was incubated with myc antibody for 1 h and then immunoprecipitated with Dynabeads conjugated with antibody raised against mouse IgG. After several washes, the co-immunoprecipitated proteins were eluted and analyzed on Western blots. For UV crosslinking and immunoprecipitation, three plates of hippocampal neurons (∼6–7 million cells, 21 DIV) were each covered with 200 μl of immunoprecipitation buffer and UV irradiated on ice for 30 min. The cells were collected and centrifuged at 1000 g for 5 min to remove nuclei. One-twentieth of the resulting supernatant was saved for total RNA isolation. The remaining solution was equally divided for IgG and CPEB3 antibody immunoprecipitation using Dynabeads conjugated with antibody against rabbit IgG. After 2 h incubation, the beads were washed 4 × with RIPA buffer and 1 × with genomic DNA lysis buffer (50 mM Tris, pH 7.4, 10 mM EDTA, 500 mM NaCl, 2.5 mM DTT, 0.5 mM spermidine, 1% Triton X-100). Approximately 300 μl of proteinase K solution (1 mg/ml in genomic DNA lysis buffer and 0.4 U/μl RNase inhibitor) was added to the total lysate and beads and incubated at 37°C for 30 min. The digested mixtures were used for RNA isolation and subsequent RT–PCR. The primer sequences are as follows: Map2 sense 5′-GACAATTGGGTACCTTGCAAC and antisense 5′-GGAGAAGGCCAGCTGTAG; NF sense 5′-GAGATGTATTACGCAAAGTACC and antisense 5′-CCAGTATGACCTTTATTGAGC; GluR2 sense 5′-CAGAGCTCAGTCTTAGGCAG and antisense 5′-GTTTGTCTCCTTGGAGTACG.
RNA structure probing and footprinting
Methods for RNA alkaline hydrolysis, RNase T1 digestion, and lead acetate-mediated RNA cleavage have been described (Darnell et al, 2005). 5′ end-labeled RNA was suspended in HEPES-SBB buffer (25 mM HEPES, pH 7.6, 200 mM KOAc, 5 mM Mg(OAc)2), heat denatured and cooled on ice, and digested in the presence of 20 μg tRNA with 0.035 U RNase V1 at 37°C for 5 min. The methods for RNA footprinting have been described (Hartmuth et al, 1999; Lee et al, 2003). The entire rat GluR2 3′ UTR can be found in accession number NM_017261.
Acknowledgments
We thank Biliang Zhang for providing T7 RNA polymerase and technical advice with the SELEX experiments, Morgan Sheng for the lentivirus vector, David Peabody for the MS2CP antibody, Allan Jacobson for MS2-containing clones, Oswald Steward for the Arc clone, Aldo Rossini for viral packaging plasmids, and Stephanie Nottrott for advice on structure mapping and RNA footprinting. We also thank Mario Stevenson, Maria Zapp, and Darcey Halley for use of their culture facilities and for discussions regarding lentivirus production. Y-SH was supported by a fellowship from the Charles A King Trust. This work was supported by grants from the NIH. Core support from the Diabetes Endocrinology Research Center Program Project (DK32520) is also gratefully acknowledged.
References
- Alarcon J, Hodgman R, Theis M, Kandel ER, Richter JD (2004) Deficit in some forms of Schaffer collateral-CA1 LTP in mice with a disrupted CPEB gene. Learn Mem 11: 318–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CH, Kandel ER, Si K (2004) The persistence of long-term memory: a molecular approach to self-sustaining changes in learning-induced synaptic growth. Neuron 44: 49–57 [DOI] [PubMed] [Google Scholar]
- Barnard C, Cao Q, Richter JD (2005) Differential phosphorylation controls Maskin association with eIF4E and localization on the mitotic apparatus. Mol Cell Biol 25: 7605–7615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard DC, Ryan K, Manley JL, Richter JD (2004) Symplekin and xGld2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119: 641–651 [DOI] [PubMed] [Google Scholar]
- Cao Q, Huang YS, Kan MC, Richter JD (2005) Amyloid precursor proteins anchor CPEB to membranes and promote polyadenylation-induced translation. Mol Cell Biol 25: 10930–10939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Richter JD (2002) Dissolution of the maskin-eIF4E complex by cytoplasmic polyadenylation and poly(A) binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J 21: 3852–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho PF, Poulin F, Cho-Park YA, Cho-Park IB, Chicoine JD, Lasko P, Sonenberg N (2005) A new paradigm for translational control: inhibition via 5′–3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell 121: 411–423 [DOI] [PubMed] [Google Scholar]
- Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, Eddy SR, Darnell RB (2005) Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev 19: 903–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Moor CH, Richter JD (1999) Cytoplasmic polyadenylation elements mediate masking and unmasking of cyclin B1 mRNA. EMBO J 18: 2294–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Richter JD (2005) Acivity-dependent polyadenylation in neurons. RNA 11: 1340–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey U, Morris RG (1997) Synaptic tagging and long-term potentiation. Nature 385: 533–536 [DOI] [PubMed] [Google Scholar]
- Groisman I, Jung M-Y, Sarkissian M, Richter JD (2002) Translational control of the embryonic cell cycle. Cell 109: 473–483 [DOI] [PubMed] [Google Scholar]
- Hake LE, Mendez R, Richter JD (1998) Specificity of RNA binding by CPEB: requirement for RNA recognition motifs and a novel zinc finger. Mol Cell Biol 18: 685–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake LE, Richter JD (1994) CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell 79: 617–627 [DOI] [PubMed] [Google Scholar]
- Hartmuth K, Raker VA, Huber J, Branlant C, Luhrmann R (1999) An unusual chemical reactivity of Sm site adenosines strongly correlates with proper assembly of core U snRNP particles. J Mol Biol 285: 133–147 [DOI] [PubMed] [Google Scholar]
- Huang Y-S, Carson JH, Barbarese E, Richter JD (2003) Facilitation of dendritic mRNA transport by CPEB. Genes Dev 17: 638–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y-S, Jung M-Y, Sarkissian M, Richter JD (2002) N-methyl-D-aspartate receptor signaling results in aurora kinase-catalyzed CPEB phosphorylation and αCaMKII mRNA polyadenylation at synapses. EMBO J 21: 2139–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju W, Morishita W, Tsui J, Gaietta G, Deerinck TJ, Adams SR, Garner CC, Tsien RY, Ellisman MH, Malenka RC (2004) Activity-dependent regulation of dendritic synthesis and trafficking of AMPA receptors. Nat Neurosci 7: 244–253 [DOI] [PubMed] [Google Scholar]
- Kacharmina JE, Job C, Crino P, Eberwine J (2000) Stimulation of glutamate receptor protein synthesis and membrane insertion within isolated neuronal dendrites. Proc Natl Acad Sci USA 97: 11545–11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Schuman EM (1996) A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science 273: 1402–1406 [DOI] [PubMed] [Google Scholar]
- Kelleher RJ III, Govindarajan A, Jung HY, Kang H, Tonegawa S (2004) Translational control by MAPK signaling in long-term synaptic plasticity and memory. Cell 116: 467–479 [DOI] [PubMed] [Google Scholar]
- Klann E, Dever TE (2004) Biochemical mechanisms for translational regulation in synaptic plasticity. Nat Rev Neurosci 5: 931–942 [DOI] [PubMed] [Google Scholar]
- Lee N, Gorelick RJ, Musier-Forsyth K (2003) Zinc finger-dependent HIV-1 nucleocapsid protein–TAR RNA interactions. Nucleic Acids Res 31: 4847–4855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockard RE, Kumar A (1981) Mapping tRNA structure using double-strand-specific ribonuclease V1 from cobra venom. Nucleic Acids Res 9: 5152–5167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinow R, Malenka RC (2002) AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci 25: 103–126 [DOI] [PubMed] [Google Scholar]
- Mendez R, Barnard D, Richter JD (2002) Differential mRNA translation and meiotic progression require Cdc2-mediated CPEB destruction. EMBO J 21: 1833–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Hake LE, Andresson T, Littlepage LE, Ruderman JV, Richter JD (2000a) Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature 404: 302–307 [DOI] [PubMed] [Google Scholar]
- Mendez R, Murthy KG, Ryan K, Manley JL, Richter JD (2000b) Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol Cell 6: 1253–1259 [DOI] [PubMed] [Google Scholar]
- Mendez R, Richter JD (2001) Translational control by CPEB: a means to the end. Nat Rev Mol Cell Biol 2: 521–529 [DOI] [PubMed] [Google Scholar]
- Mendez-Vidal C, Wilhelm MT, Hellborg F, Qian W, Wiman KG (2002) The p53-induced mouse zinc finger protein wig-1 binds double-stranded RNA with high affinity. Nucleic Acids Res 30: 1991–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall N, Standart N (2004) The active form of Xp54 RNA helicase in translational repression is an RNA-mediated oligomer. Nucleic Acids Res 32: 1325–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro K, Dichter M, Eberwine J (1994) On the nature and differential distribution of mRNAs in hippocampal neurites: implications for neuronal functioning. Proc Natl Acad Sci USA 91: 10800–10804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck DH, Ostareck-Lederer A, Shatsky IN, Hentze MW (2001) Lipoxygenase mRNA silencing in erythroid differentiation: The 3′UTR regulatory complex controls 60S ribosomal subunit joining. Cell 104: 281–290 [DOI] [PubMed] [Google Scholar]
- Reverte CG, Yuan L, Keady BT, Lacza C, Attfield KR, Mahon GM, Freeman B, Whitehead IP, Hake LE (2003) XGef is a CPEB-interacting protein involved in Xenopus oocyte maturation. Dev Biol 255: 383–398 [DOI] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N (2005) Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433: 477–480 [DOI] [PubMed] [Google Scholar]
- Schratt GM, Nigh EA, Chen WG, Hu L, Greenberg ME (2004) BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin–phosphatidylinositol 3-kinase-dependent pathway during neuronal development. J Neurosci 24: 7366–7377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets MD, Wu M, Wickens M (1995) Polyadenylation of c-mos mRNA as a control point in Xenopus meiotic maturation. Nature 374: 511–516 [DOI] [PubMed] [Google Scholar]
- Si K, Giustetto M, Etkin A, Hsu R, Janisiewicz AM, Miniaci MC, Kim JH, Zhu H, Kandel ER (2003) A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in Aplysia. Cell 115: 893–904 [DOI] [PubMed] [Google Scholar]
- Stebbins-Boaz B, Cao Q, de Moor CH, Mendez R, Richter JR (1999) Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol Cell 4: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Stebbins-Boaz B, Hake LE, Richter JD (1996) CPEB controls the cytoplasmic polyadenylation of cyclin, cdk2, and c-mos mRNAs and is necessary for oocyte maturation in Xenopus. EMBO J 15: 2582–2592 [PMC free article] [PubMed] [Google Scholar]
- Steward O, Schuman EM (2001) Protein synthesis at synaptic sites on dendrites. Annu Rev Neurosci 24: 299–325 [DOI] [PubMed] [Google Scholar]
- Steward O, Schuman EM (2003) Compartmentalized synthesis and degradation of proteins in neurons. Neuron 40: 347–359 [DOI] [PubMed] [Google Scholar]
- Sutton M, Schuman EM (2005) Local translational control in dendrites and its role in long-term synaptic plasticity. J Neurobiol 64: 116–131 [DOI] [PubMed] [Google Scholar]
- Tay J, Hodgman R, Sarkissian M, Richter JD (2003) Regulated CPEB phosphorylation during meiotic progression suggests a mechanism for temporal control of maternal mRNA translation. Genes Dev 17: 1457–1462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay J, Richter JD (2001) Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev Cell 1: 201–213 [DOI] [PubMed] [Google Scholar]
- Theis M, Si K, Kandel ER (2003) Two previously undescribed members of the mouse CPEB family of genes and their inducible expression in the principal cell layers of the hippocampus. Proc Natl Acad Sci USA 100: 9602–9607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Padgett RA (1989) Hydroxyl radical ‘footprinting' of RNA: application to pre-mRNA splicing complexes. Proc Natl Acad Sci USA 88: 7795–7799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Wells D, Tay J, Mendis D, Abbot M, Barnitt A, Quinlan E, Heynen A, Fallon J, Richter JD (1998) CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of α-CaMKII mRNA at synapses. Neuron 21: 1129–1139 [DOI] [PubMed] [Google Scholar]
- Yang M, May WS, Ito T (1999) JAZ required the double-stranded RNA-binding zinc finger motifs for nuclear localization. J Biol Chem 274: 27399–27406 [DOI] [PubMed] [Google Scholar]
- Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]


