Abstract
A suitable system for expression of the rhodopsin kinase (RK) gene and its mutants is needed for structure–function studies of RK. Previously, investigation of the baculovirus system showed satisfactory production of RK, but posttranslational isoprenylation was deficient. We now report on a comparative study of expression of the RK gene in yeast (Pichia pastoris), COS-1 cells and in an HEK293 stable cell line. Expression in COS-1 cells, by using pCMV5 vector, is the most satisfactory. A two-step procedure for purification of the expressed enzyme with an N-terminal histidine tag has been developed. The purified enzyme has correct posttranslational modifications and shows a somewhat broader pH vs. catalytic activity profile than the wild-type enzyme.
Keywords: G-protein-coupled receptor kinases, rod outer segment, autophosphorylation, isoprenylation
Rhodopsin kinase (RK; EC 2.7.1.125) specific to the mammalian retina, is the best characterized member of a family of G-protein coupled receptor kinases (GRKs) that bind to activated G-protein coupled receptors and initiate desensitization of the receptors by phosphorylation. GRKs contain three domains, the N-terminal domain, believed to be involved in receptor recognition (1, 2), the central highly conserved catalytic domain (2, 3), and the carboxy terminus domain; the latter contains autophosphorylation sites (4), the consensus isoprenylation site, and additional regulatory elements (5). Activation of RK on binding to illuminated rhodopsin is presumed to result from a conformational change in the enzyme. The nature of this structural change is unknown. Nor is there any detailed information on the interactions between the light-activated rhodopsin and RK, although recent studies indicate that the binding of the enzyme involves the cytoplasmic loops in rhodopsin (6–8). Study of these questions by the mutagenesis approach requires a suitable system for expression and purification of RK mutants. Although expression in baculovirus enables the preparation of RK in reasonable amounts, isoprenylation is incomplete and heterogeneous (9). This posttranslational modification is important for studies of at least some aspects of RK function (5). Here, we report on a comparative study of the expression of the RK gene in yeast (Pichia pastoris), COS 1 cells and in an HEK293 stable cell line.§ We find that expression in COS cells is the most satisfactory, and we describe a two-step procedure for purification and characterization of the expressed enzyme containing a histidine tag at the amino terminus.
Materials and Methods
Materials.
Frozen bovine retinae were from J. A. Lawson Corporation (Lincoln, NE). [γ-32P]-ATP was from Dupont/NEN. Dodecyl-β-d-maltoside (DM) was from Anatrace (Maumee, OH). Nitrocellulose filters were from Intermountain Scientific (Kaysville, UT). Heparin–sepharose and cobalt–sepharose were from Pharmacia and CLONTECH, respectively. The enhanced chemiluminescence detection kit and the restriction enzymes were from New England Biolabs. DNA purification columns were from Qiagen (Chatsworth, CA). The yeast P. pastoris strains and expression vectors were from Invitrogen. The mammalian expression vector pCMV5 containing a cDNA fragment encoding the bovine RK gene was provided by Inglese (Pharmacopeia, Princeton, NJ). Geneticin (G418) was from GIBCO/BRL. Trypsin-EDTA, penicillin, streptomycin, l-glutamine, DMEM, and DMEM/F12 were from Irvine Scientific. FBS was from Sigma. Cell growth media were: medium 1, DMEM supplemented with 4.5 g/liter glucose/292 μg/ml l-glutamine/100 units/ml penicillin G/100 μg/ml streptomycin sulfate. Medium 2, medium 1 supplemented with 10% heat-treated FBS. The protease inhibitor mixture used was 0.5 mM [4-(2-aminoethyl)-benzenesulfonyl]/20 μg/ml leupeptin/1 mM benzamidine-hydrochloride.
Buffers used were: buffer A, 20 mM 1,3-bis[tris(hydroxymethyl)methylamino]propane (BTP) (pH 7.5)/250 mM KCl/0.4% Tween 80; buffer B, 20 mM BTP (pH 7.5)/protease inhibitor mixture; buffer C, 20 mM BTP (pH 7.5)/250 mM KCl/1 mM EDTA/0.125% DM/protease inhibitor mixture; buffer D, 10 mM BTP (pH 7.5)/0.05% DM/100 mM KCl; buffer E, 10 mM BTP (pH 7.5)/0.05% DM/130 mM KCl/1 mM MgCl2/0.2 mM ATP/protease inhibitor mixture; buffer F, 10 mM BTP (pH 7.5)/0.05% DM/300 mM KCl; buffer G, 10 mM BTP (pH 7.5)/0.05% DM; buffer H, 50 mM Tris⋅HCl (pH 7.4)/5 mM EDTA; buffer I, 20 mM BTP (pH 7.5)/2 mM MgCl2/1 mM DTT; buffer J, 0.05% Tween 20 in PBS.
Materials and Methods
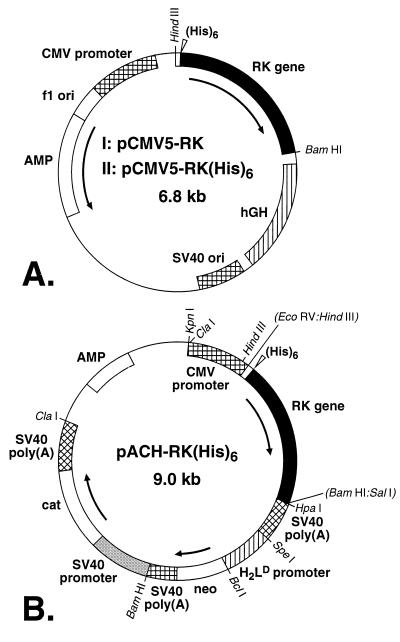
pCMV5-RK and pCMV5-RK(His)6 Plasmids (Fig. 1A).
Figure 1.
Vectors for expression of the RK gene and mammalian cells. (A) pCMV5-RK; (I) pCMV5-RK(His)6; (II) vectors (Materials and Methods) used for the transient transfection of COS-1 mammalian cells. (B) pACH-RK(His)6 vector for the preparation of stable HEK293S mammalian cell lines (Materials and Methods).
The noncoding DNA sequences at 5′- and 3′-ends of the RK gene in the vector pCMV5 were removed by using the PCR. The 5′ end of the gene was further modified to introduce an optimally positioned Kozak sequence. Six histidines were inserted at the N terminus between aspartic residue at position two and phenylalanine at position three. The RK(His)6 gene was also constructed by PCR.
Plasmid pACHRK(His)6 and Construction of Stable Cell Lines (Fig. 1B).
The HindIII–BamHI DNA fragment containing the RK(His)6 gene was excised from pCMV5-RK(His)6 (Fig. 1AII). The single-stranded ends of the RK(His)6 DNA HindIII–BamHI fragment were filled by using the Klenow fragment of DNA polymerase I and dNTPs. This blunt-ended fragment was transferred to pACHEnc, and stable cell lines were constructed as described previously (11).
Construction of P. pastoris Yeast Transformants Carrying the RK Gene.
The DNA fragment containing the RK and His-6 genes was purified from the pPIC3-RK plasmid after digestion with BglII. The fragment was introduced into yeast strain GS115 by electroporation, and transformants prototrophic for histidine and exhibiting a slow utilization of methanol were selected. Confirmation of insertion into the AOX1 locus was confirmed by the PCR by using yeast genomic DNA as a template.
Preparation of Yeast Protein Extract.
Yeast cells grown to saturation in 50 ml of minimal medium containing 3% glycerol were collected by centrifugation and used to inoculate 100 ml of minimal medium containing 0.5% methanol in 1-liter baffled flasks. The culture was agitated for 48 h at 28°C, 0.5% methanol being added after 24 h. After centrifugation, the cells were washed with cold water and benzamidine, 62 μl, 0.1 M, 20 μl of leupeptin 2 mg/ml, 25 μl of 4-(2-aminoethyl)benzenesulfonyl fluoride 0.1 M, and 12 μl of EDTA 0.5 M were added per gram of cells in chilled buffer A to a final volume of 40 ml in two tubes. Ten milliliters of acid-washed glass beads (0.45 μm) were added to each tube, and the samples were mixed by vortexing (×10 for 30 s each time) with 1-min intervals on ice. The extract was collected by filtration, clarified by centrifugation, and the supernate diluted 2-fold in chilled buffer B. After clarification by centrifugation, the extract was used for purification of RK by FPLC on heparin-Sepharose. The procedure used was as described below except that DM was replaced by 0.2% Tween 80.
Transient Transfection of COS-1 Cells.
COS-1 cells were transfected by using pCMV5-RK or pCMV5-RK(His)6 as described previously (12).
Purification of RK and RK(His)6 from Transfected COS-1 Cells.
Transfected COS-1 cells (5–6 × 108) were suspended in buffer C (10 ml), and the protein extract was prepared as described previously (9). RK was purified as previously described (9), whereas RK(His)6 was purified as follows.
Step 1: FPLC on Heparin-Sepharose Column.
(i) In the absence of ATP: The protein extract from above (20 ml) was applied to a heparin–sepharose column (5 ml, 1.6 cm diameter) preequilibrated with buffer D. After a wash with the same buffer (10 bed volumes), elution was performed at 4°C by using a linear KCl gradient of 100–500 mM in 80 ml of the same buffer. Two-milliliter fractions were collected at a flow rate of 0.5 ml/min. A280 and RK activity were monitored in the fractions. (ii) In the presence of ATP: The extract was applied to the heparin–sepharose column and the column washed as above. A linear gradient of KCl (100–130 mM in 6 ml) was first used, and subsequently the salt concentration was kept constant for 40 ml. The column was then washed with buffer E (20 ml) at flow rate 0.02 ml/min. The flow rate was then increased to 0.5 ml/min, and the column was washed with 30 ml of buffer D containing 130 mM KCl. A linear gradient of KCl (130–500 mM) in 60 ml was finally applied. Four-milliliter fractions were collected, and those containing RK activity were combined.
Step 2: FPLC on Cobalt–Sepharose Column.
The combined fractions from Step 1 (i or ii) were applied at a flow rate of 0.02 ml/min to a cobalt–sepharose column (1 ml, 8 mm diameter) preequilibrated with buffer F. The column was washed with the same buffer at a flow rate of 0.5 ml/min. Elution was carried out by using a two-phase imidazole gradient (0.7–50 mM) in buffer G at a flow rate of 0.1 ml/min. Two-milliliter fractions were collected, and both A280 and RK activity were monitored.
Urea-Stripped Rod Outer Segments (ROS).
ROS were prepared from frozen retinae (13) and washed in the dark at 4°C for 15 min with 5 M urea in buffer H (14).
Assay of RK Activity.
Aliquots from fractions (2 μl, 0–0.03 units) were dispensed in a 96-well microplate, and the plate was kept on ice (see ref. 9). A reaction mixture (8 μl) containing 40 μM [γ-32P]ATP (200–2,000 cpm/pmol) plus 20 μM urea-stripped ROS in buffer H was added to each well. Phosphorylation initiated by illumination was allowed to proceed for 5–7 min at 30°C. As controls, parallel reactions were carried out (i) in the dark; (ii) in the absence of ROS; and (iii) in the absence of an enzyme fraction. The reactions were terminated by the addition of 40 μl of a solution containing 20 mM ATP plus 20 mM EDTA. The reaction mixture from each well was transferred to a nitrocellulose filter by using a microdot blot system. The filters were washed in 20 mM EDTA three times for 5 min each time, and the radioactivity in the dots was measured by Cerenkov counting. In time course experiments, the reaction mixtures were 60 μl, and 10-μl aliquots were taken and treated with 40 μl of the stop solution.
3H Labeling in Vivo of the Isoprenyl Group in RK.
COS-1 cells (15 × 15 cm dia dishes) were transiently transfected with pCMV5-RK (12). Twenty-four hours after addition of the DNA mixture, the growth medium was removed and replaced with fresh medium (15 ml) containing [3H]-DL-mevalonolactone (1,000 μCi; 1 μCi = 2.2 × 106 DPM). The cells were harvested 60 h posttransfection.
Characterization of Isoprenoid Groups.
RK was purified from COS-1 cells transiently expressing the RK gene in the presence of [3H]-mevalonolactone, by heparin–sepharose chromatography followed by immunoaffinity chromatography as described (15). Isoprenoid characterization was as described (9).
Other Methods.
Immunoblotting: Proteins were separated by SDS/PAGE (16) and electroblotted onto a nitrocellulose filter. The filter was incubated for 5 min in a 2% dry skim milk solution, washed twice for 5 min each in buffer J, and then incubated for 2 h in 20 ml of buffer J containing 5 μg of anti-RK monoclonal antibody 6D8 or 1C3 (15). The filter was washed four times in buffer J and incubated for 1 h in 20 ml of the same buffer containing 3.5 μl of the secondary antibody (anti-mouse IgG horseradish peroxidase conjugate, New England Biolabs). The filter was washed four times in buffer J before performing the chemiluminescent reaction.
Protein concentrations were determined by the method of Bradford (16).
Results
Attempted Purification of Recombinant RK from Yeast Cells.
Yeast (P. pastoris) transformants containing the RK gene were grown after induction with methanol. Production of RK peaked at 48 h. A cell extract from 200 ml of culture was applied to a heparin–sepharose column (see Materials and Methods). RK activity was detected in fractions eluted at 250 mM KCl concentration. Combined fractions were applied to a Mono-Q Sepharose column. Although RK was detected in fractions eluted at about 150 mM KCl by immunoblotting, no significant enzymatic activity could be recovered.
Expression of the Recombinant RK Gene in COS-1 Cells by Transient Transfection.
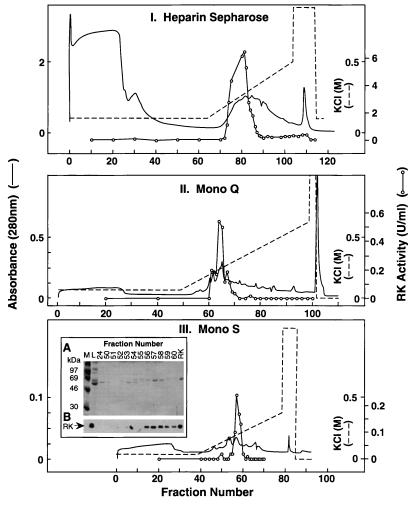
COS-1 cell extracts prepared after transient transfection with the vector pCMV5-RK (Fig. 1AI) indicated about 15 μg of RK from 107 transfected cells by SDS/PAGE and immunodetection (15). A three-step purification procedure as described previously (9) was used to purify RK from 5 × 108 transfected cells (Fig. 2). The total protein extract was separated by chromatography on heparin–sepharose (Fig. 2I); RK bound to the column, whereas the bulk of the proteins did not. The bound proteins were eluted by using a linear KCl gradient; RK eluted as a single peak centered at about 280 mM KCl. The combined RK fractions from this step were chromatographed on Mono Q Sepharose (Fig. 2II). The bound proteins were eluted by using the same gradient as above and RK eluted at about 180 mM KCl. Fractions with RK activity from Step 2 were combined and chromatographed on Mono S Sepharose (Fig. 2III). By using a 50–300 mM KCl gradient, RK eluted at about 120 mM KCl. RK (100 μg) was obtained with a purity of about 80%, representing a yield of 13% with specific activity, 40 units/mg. Thus, RK constituted 0.3% to 0.5% of the total protein in the transfected COS-1 cell extract.
Figure 2.
Purification of RK from mammalian cells. (I) The protein extract from 5 × 108 transfected COS-1 cells was chromatographed on a heparin–sepharose column as in Materials and Methods. (II) Fractions 78–84 (14 ml) from I were diluted and applied to a Mono Q column. (III) Fractions 63–66 (8 ml) from II were diluted and applied to a Mono S column. (Inset) SDS/PAGE of selected fractions followed by protein detection by using Coomassie blue staining (A) or immunodetection (B) by using an anti-RK monoclonal antibody. L, preloading sample; M, molecular weight marker.
Purification of RK(His)6 from Transiently Transfected COS-1 Cells.
COS-1 cells were transiently transfected with the vector pCMV5-RK(His)6 (Fig. 1AII) as described (12), and RK(His)6 was purified from the extract in the absence or in the presence of ATP.
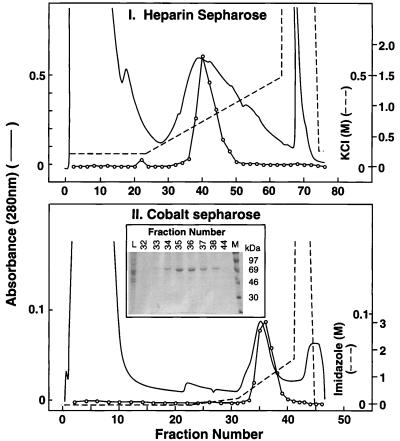
Purification in the Absence of ATP.
Step 1: RK from the protein extract from 6 × 108 cells was purified on a heparin–sepharose column as in Materials and Methods. RK(His)6 eluted as a single peak at about 280 mM KCl (Fig. 3I). The recovery from this step was about 80% (Table 1). Fractions (38–44) with RK activity were combined. Step 2: The combined fractions were applied to a cobalt–sepharose column preequilibrated with 300 mM KCl and elution performed as in Materials and Methods. Of the bulk of proteins, some showed affinity for cobalt and eluted at about 10 mM imidazole concentration (Fig. 3II). RK(His)6 eluted at about 25 mM imidazole concentration. The recovery at this step was about 70%. The purity of RK in fractions 32–38 was determined by SDS/PAGE (Fig. 3II Inset) followed by Coomassie blue staining and densitometry. RK(His)6 formed the major protein band in these fractions. RK(His)6 was most concentrated in fractions 35 and 36, and the protein was the purest in fraction 36. A total of 400 μg (60% recovery) of RK(His)6 was obtained from this experiment with a purity of about 80% and a specific activity of 40 units/mg (Table 1).
Figure 3.
Purification of RK(His)6 from mammalian cells. (I) Chromatography on heparin–sepharose of RK(His)6 as in Materials and Methods. (II) Chromatography on cobalt–sepharose. Fractions 38–44 from I were pooled and chromatographed on a cobalt–sepharose column (Materials and Methods). (Inset) SDS/PAGE followed by Coomassie blue detection of fractions 32–44. L, preloading sample; M, molecular weight marker.
Table 1.
Purification of RK(His)6 produced in transiently transfected COS-1 cells
| RK(His)6 | Vol, ml | Total Protein, mg | Specific Activity, nmol/min/mg | Purification, fold | Yield, % |
|---|---|---|---|---|---|
| In the absence of ATP | |||||
| Crude | 20 | 120 | 0.18 | 1 | 100 |
| Heparin-sepharose | 14 | 14 | 1.27 | 7 | 82 |
| Cobalt-sepharose | 10 | 0.4 | 40 | 222 | 74 |
| In the presence of ATP | |||||
| Crude | 20 | 102 | 0.19 | 1 | |
| Heparin-sepharose | 20 | 8 | 4.9 | 26 | 48 |
| Cobalt-sepharose | 8 | 0.2 | 40 | 210 | 43 |
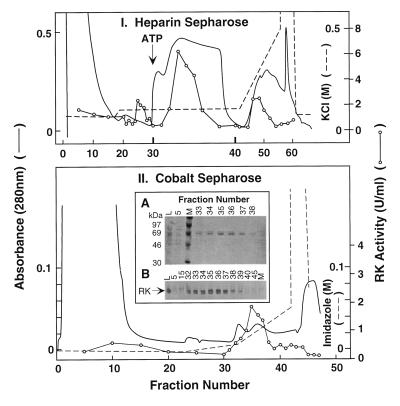
Purification in the Presence of ATP.
Elution of nonphosphorylated RK from heparin–sepharose chromatography requires higher salt concentration than phosphorylated RK (9, 17). Thus, treatment of RK while bound to a heparin–sepharose column with a buffer containing ATP and MgCl2 results in RK autophosphorylation and expedites elution from the column. This rationale was used for the purification of RK from ROS (17) and in the procedure described below (Fig. 4I). The protein extract from 6 × 108 cells was applied to the heparin–sepharose column and the column first washed with 130 mM KCl (Fig. 4I). A small amount (17%) of RK(His)6 eluted from the column during this wash. Maintaining the salt concentration at 130 mM, the column was treated with ATP + MgCl2 (Materials and Methods). About 48% of RK(His)6 eluted from the column in the second bed volume after ATP addition. Removal of the ATP buffer and further wash with 130 mM KCl did not elute RK(His)6. When the salt concentration was subsequently raised to elute the remaining proteins, 35% of kinase activity was present in the leading fractions. Fractions 32–35 were pooled and applied to the cobalt–sepharose column (Fig. 4II). As observed above in the experiment of Fig. 3II, the bulk of the proteins did not bind to the column, but some proteins eluted from the column at 8–10 mM imidazole concentration. Phosphorylated RK(His)6 eluted at about 25 mM, similar to the nonphosphorylated counterpart. However, the peak of phosphorylated RK(His)6 was broader than that of nonphosphorylated RK(His)6. As shown in Fig. 4II (Inset), SDS/PAGE showed the presence of RK(His)6 in fractions 33–38. Phosphorylated RK(His)6 in fractions 35–38 was homogeneous, and it was most concentrated in fractions 35 and 36. A total of 200 μg of RK(His)6 was recovered (43%) with a purity above 90% with a specific activity of 40 units/mg (Table 1).
Figure 4.
Purification of RK(His)6 from mammalian cells by using ATP elution. (I) Chromatography on heparin–sepharose. RK(His)6 was purified as in Fig. 3, except that the elution was performed by using a MgCl2/ATP solution as described in the text. Bolder lines at fractions 30 and 40 indicate that the flow rate for elution was slower between these fractions. The volume in each fraction (4 ml) was constant. (II) Chromatography on cobalt–sepharose. Fractions 32–35 from I were chromatographed on cobalt–sepharose as in Fig. 3. (Inset) Selected fractions were examined by SDS/PAGE followed by (A) Coomassie blue staining or (B) immunodetection. L, preloading sample; M, molecular weight marker.
Expression and Attempted Purification of RK(His)6 in HEK293S Stable Cell Lines.
HEK293S stable cell lines were prepared by using the vector pACHRK(His)6 (Fig. 1B), as described previously (11). Three cell lines derived from single colonies tested positive for RK expression by immunodetection (13). The strength of the signal indicated that the expression level was in the order of 4- to 5-fold lower than that observed in transiently transfected COS-1 cells. HEK293S cells expressing the highest levels of RK were grown and harvested, and attempts were made to purify RK(His)6 by the two-step procedure described in Fig. 3. Analysis showed only a purity of about 50% for the product. Thus, the stable cell line approach did not produce practically useful amounts of RK(His)6.
Isoprenylation of RK Produced in Transfected COS-1 Cells.
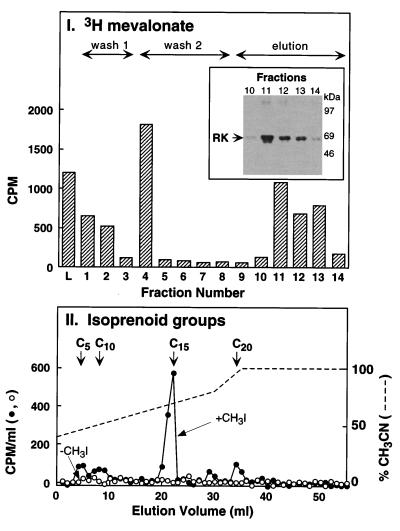
[3H]-RK prepared from COS-1 cells in the presence of [3H]-mevalonolactone (Materials and Methods) was purified in two steps, including the immunoaffinity procedure (15). Briefly, 3H-labeled RK was first chromatographed on heparin–sepharose. Next, [3]H-RK was bound to the monoclonal antibody column and eluted as in Materials and Methods (15). Radioactivity was monitored in fractionation by immunoaffinity chromatography (Fig. 5I). Washes with high and low salt concentrations, respectively, removed the contaminating 3H-labeled proteins. After addition of the elution peptide, a radioactive peak corresponding to RK appeared as determined by immunoblotting analysis (Fig. 5I Inset). No RK was detected by immunoblotting during the washes. After cleavage and extraction of the isoprenoid groups from the purified RK (Materials and Methods), the extract was applied to a C18 reverse-phase HPLC column and elution performed by using a two-phase acetonitrile gradient (Materials and Methods). As seen in Fig. 5II, elution of the major radioactivity peak (65%) superimposed with that of the C15 standard. Three smaller peaks (11%, 11%, and 13%) were detected, the positions of which corresponded to the C5, C10, and C20 standards, respectively. Thus, RK expressed in COS-1 cells was mainly farnesylated.
Figure 5.
Analysis of RK isoprenylation expressed in COS-1 cells. (I) Transfected COS-1 cells were grown in the presence of [3H]-mevalonolactone, and RK was purified by immunoaffinity chromatography as described in Materials and Methods. The radioactivity present in the loading (L), washing (no. 1, high salt; no. 2, low salt), and elution fractions was quantitated by fluorography. The presence of RK in the elution fractions was verified by immunoblotting (Inset). (II) HPLC analysis of the isoprenoid groups. The isoprenoid groups from the purified RK molecules were cleaved, extracted, and analyzed as described in Materials and Methods. Elution from the HPLC column was performed by using a gradient of acetonitrile (solid line), and the radioactivity present in the eluted fractions was counted (dotted line). A control sample, not cleaved, was analyzed in parallel (triangles). The position of C5 (mevalonic acid), C10 (geranol), C15 (trans-farnesol and nerolidol), and C20 (geranylgeranol) standards is indicated.
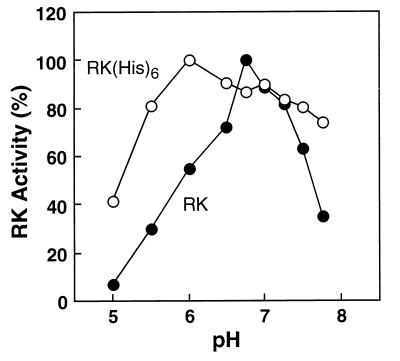
Activity-pH Profile of RK(His)6.
Previously, RK purified from baculovirus showed the same pH-activity profile as RK purified from ROS (9). Here, RK and RK(His)6 purified from transiently transfected COS-1 cells (Figs. 2 and 3, respectively) were tested for activity in rhodopsin phosphorylation (Fig. 6). Maximum activity for RK was at pH 6.75, 60% of the activity being lost with one pH unit change. However, maximum activity for RK(His)6 was shifted at pH 6.0, and the activity profile was broader as a function of pH (Fig. 6).
Figure 6.
Effect of pH on RK(His)6 activity. RK (filled circles) and RK(His)6 (open circles) were purified as described in the text. The activity of RK(His)6 was measured under various pH conditions and compared with that of RK. The reaction conditions were as described in Materials and Methods, except that 50 mM BTP buffer was used instead of 20 mM
Discussion
Baculovirus-infected insect cells are frequently used for expression of proteins. This system was previously investigated for expression of RK, and a procedure was developed for purification of the expressed protein to near homogeneity (9). However, isoprenylation, a biologically important posttranslational modification, was incomplete and heterogeneous in the above product. Although the enzyme thus prepared is satisfactory for a number of studies of RK, correct isoprenlyation is desirable for studies such as functional translocation of the enzyme (5). We have now investigated the main expression systems that are in current use for preparation of RK with desired posttranslational modifications. The production of RK in the yeast P. pastoris was so low that the expressed protein was not amenable to purification. Similarly, expression of the RK gene in the yeast Saccharomyces cerevisiae in a galactose inducible manner was not satisfactory (data not shown). In contrast, the production of RK after transient transfection of COS-1 cells by using the modified pCMV5 expression vector was satisfactory. This encouraged us to explore the construction of HEK293S cell lines that would stably express RK. Stable mammalian cell lines have proved to be useful for the large-scale production of rhodopsin and its mutants (10, 11). However, in the present work, the stable cell lines harboring the RK gene that we obtained produced only low levels of RK, and purification of the protein was not feasible. The failure to obtain high-level expression is very likely because of the toxicity of high levels of RK to the cells.
Although the COS-1 cell expression system afforded a reasonable level of RK, purification by about 300-fold was necessary to obtain homogeneous RK from these cells. The use of immunoaffinity chromatography was investigated in attempts to develop a rapid purification procedure (15). In the present work, satisfactory purification of RK was achieved by introducing a hexahistidine tag at the N terminus of RK. The two-step purification procedure developed used cobalt affinity chromatography as a second step, after heparin–sepharose chromatography. About 0.4 mg of purified active RK from 6 × 108 transfected cells can thus be prepared. It is noteworthy that when the sequence of the two purification steps was reversed, inactivation of the enzyme was observed during heparin chromatography, which then was the second step. The use of Mono Q or Mono S columns instead of heparin also caused inactivation. The inactivation does not seem to be caused by the lability of the enzyme accompanying increase in purity because control samples remained active.
The purity of RK preparation was ascertained by using relatively large amounts of material in the analytical method used (densitometry of Coomassie blue stained gels), such that the contaminating proteins, each less than 4% of the total proteins, could be detected. The purity of the purified RK preparations thus estimated was greater than 80%. Purification of above 90% could be achieved by triggering autophosphorylation of RK by ATP treatment on the heparin column. The specific activity of the present preparations of RK(His)6 is about 60% of that of RK purified from ROS or baculovirus-infected insect cells. The lower activity is not caused by the variation in pH/activity profile observed for RK(His)6 (Fig. 6). It is possible that the presence of the histidine tag at the N terminus of RK affects in some way the function of the enzyme. On the other hand, it is possible that because of the inherent instability of the purified enzyme, the relatively long cobalt–sepharose chromatography results in loss of activity. The presence of glycerol during the purification procedure afforded no protection against inactivation (data not shown).
RK contains three autophosphorylation sites (4), the role of which in the function or regulation of RK remains unclear. When purified from ROS, RK was obtained mainly as a mixture of mono- and diphosphorylated forms (17). RK expressed in insect cells was also a mixture of mono- and diphosphorylated species (9). RK as now obtained from COS-1 cells is mainly diphosphorylated. Furthermore, the bulk (65%) of the produced RK has been characterized as correctly farnesylated. This result is in agreement with that obtained previously by Inglese et al. (18).
Acknowledgments
We are grateful to Professor U. L. RajBhandary for reading the manuscript and making helpful suggestions. Research reported here was supported by National Institutes of Health Grants GM28289 and EY117165 (H.G.K.) C.B. was the recipient of a Human Frontier Science Program Postdoctoral Fellowship (LT-449/96), whereas E.G. was the recipient of an Association for Research in Vision and Ophthalmology/Japan National Society for the Prevention of Blindness Research Fellowship. We gratefully acknowledge Ms. Judy Carlin's assistance in the preparation of the manuscript.
Abbreviations
- RK
rhodopsin kinase
- ROS
rod outer segments
- BTP
1,3-bis[tris(hydroxymethyl)methylamino]propane
- DM
dodecyl-β-d-maltoside
Footnotes
This is paper 39 in the series “Structure and Function in Rhodopsin.” Paper 38 is ref. 10.
References
- 1.Pitcher J A, Freedman N J, Lefkowitz R J. Annu Rev Biochem. 1998;67:653–692. doi: 10.1146/annurev.biochem.67.1.653. [DOI] [PubMed] [Google Scholar]
- 2.Inglese J, Freedman N J, Koch W J, Lefkowitz R J. J Biol Chem. 1993;268:23735–23738. [PubMed] [Google Scholar]
- 3.Hanks S K, Quinn A M. Methods Enzymol. 1991;200:38–62. doi: 10.1016/0076-6879(91)00126-h. [DOI] [PubMed] [Google Scholar]
- 4.Palczewski K, Ohguro H, Premont R T, Inglese J. J Biol Chem. 1995;270:15294–15298. doi: 10.1074/jbc.270.25.15294. [DOI] [PubMed] [Google Scholar]
- 5.Inglese J, Koch W J, Caron M G, Lefkowitz R J. Nature (London) 1992;359:147–150. doi: 10.1038/359147a0. [DOI] [PubMed] [Google Scholar]
- 6.Thurmond R L, Creuzenet C, Reeves P J, Khorana H G. Proc Natl Acad Sci USA. 1997;94:1715–1720. doi: 10.1073/pnas.94.5.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi W, Osawa S, Dickerson C D, Weiss E R. J Biol Chem. 1995;270:2112–2119. doi: 10.1074/jbc.270.5.2112. [DOI] [PubMed] [Google Scholar]
- 8.Palczewski K, Buczylko J, Kaplan M W, Polans A S, Crabb J W. J Biol Chem. 1991;266:12949–12955. [PubMed] [Google Scholar]
- 9.Cha K, Bruel C, Inglese J, Khorana H G. Proc Natl Acad Sci USA. 1997;94:10557–10582. doi: 10.1073/pnas.94.20.10577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein-Seetharaman J, Getmanova E, Loewen M, Reeves P J, Khorana H G. Proc Natl Acad Sci USA. 1999;96:13744–13749. doi: 10.1073/pnas.96.24.13744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reeves P J, Thurmond R L, Khorana H G. Proc Natl Acad Sci USA. 1996;93:11487–11492. doi: 10.1073/pnas.93.21.11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oprian D, D, Molday R S, Kaufman R J, Khorana H G. Proc Natl Acad Sci USA. 1987;84:8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papermaster D S. Methods Enzymol. 1982;81:240–246. doi: 10.1016/s0076-6879(82)81037-9. [DOI] [PubMed] [Google Scholar]
- 14.Shichi H, Somers R L. J Biol Chem. 1978;253:7040–7046. [PubMed] [Google Scholar]
- 15.Bruel C, Cha K, Niu L, Reeves P J, Khorana H G. Proc Natl Acad Sci USA. 2000;97:3010–3015. doi: 10.1073/pnas.97.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bradford M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 17.Palczewski K. Methods Neurosci. 1993;15:217–225. [Google Scholar]
- 18.Inglese J, Glickman J F, Lorenz W, Caron M G, Lefkowitz R J. J Biol Chem. 1992;267:1422–1425. [PubMed] [Google Scholar]