Abstract
The lsa gene confers intrinsic resistance to lincosamides and streptogramins A in Enterococcus faecalis, probably by active efflux. The lsa-like genes of two clinical isolates of E. faecalis susceptible to lincosamides and dalfopristin contained mutations that produced premature termination codons. Revertant mutants were obtained by selection on agar plates containing clindamycin.
Resistance to lincosamides (lincomycin and clindamycin) and streptogramins A (dalfopristin, pristinamycin II, and virginiamycin M) that defines the LSA phenotype is intrinsic in Enterococcus faecalis. Resistance to factor A of streptogramins is responsible for the loss of synergism between factors A and B composing the streptogramin mixture (quinupristin-dalfopristin, pristinamycin, and virginiamycin) that explains the intrinsic resistance of E. faecalis to this class of antibiotics (3, 7). Recently, this resistance has been related to the expression of a chromosomal lsa gene in E. faecalis OG1RF, which appears to be species specific (11). Inactivation of the lsa gene resulted in susceptibility to clindamycin, dalfopristin, and quinupristin-dalfopristin, whereas complementation with a recombinant plasmid bearing an intact lsa gene restored resistance to clindamycin and dalfopristin (11). The Lsa protein shows similarities to members of a superfamily of transport-related proteins known as ABC transporters. ABC proteins are capable of transporting both small and large molecules in response to ATP hydrolysis. Several characterized or postulated ABC transporters have been reported in staphylococci and enterococci to export antimicrobials belonging to the macrolide-lincosamide-streptogramin family (1, 9, 10). The ABC transporter system requires two ATP-binding domains located in the cytoplasm that interact with two hydrophobic domains consisting generally in six transmembrane segments. The four core components of an ABC transporter can be synthesized as individual proteins or be fused into multifunctional polypeptides in a variety of combinations (6). Conserved motifs that are used to define ABC domains have been identified in Lsa, i.e., the Walker A and B motifs, which are involved in the binding and hydrolysis of ATP, and an ABC signature probably involved in energy transduction (6). Similar to the MsrA protein putatively responsible for macrolide efflux, the two ATP-binding regions of Lsa are fused into a single protein (9). However, no transmembrane partner has been found associated with the Lsa protein and the efflux mechanism has not been proven. The mechanism of resistance to lincosamides and streptogramins A therefore remains incompletely elucidated.
Clinical isolates of E. faecalis susceptible to lincosamides and streptogramins A.
Strains of E. faecalis UCN32 and UCN33 were isolated from urine samples. The isolates were identified as E. faecalis by API 20Strep (Biomérieux, Marcy l'Etoile, France) and conventional techniques (5). Antimicrobial susceptibility testing by the agar diffusion technique showed that the isolates were susceptible to ampicillin and low levels of gentamicin, and erythromycin and were surprisingly susceptible to clindamycin, lincomycin, and quinupristin-dalfopristin. Determination of MICs of clindamycin, lincomycin, and dalfopristin by the agar dilution technique (http://www.sfm.asso.fr) confirmed the susceptibility to these antimicrobials, which was unusual for E. faecalis isolates (Table 1). Since certain enterococcal species such as Enterococcus faecium, Enterococcus durans, and Enterococcus hirae do not display the intrinsic LSA resistance phenotype (2), identification of the isolates was questioned. However, identification of the isolates as E. faecalis was confirmed by amplification of a DNA fragment of the expected size, using primers specific for the ddl gene of E. faecalis (encoding a chromosomal d-alanyl-d-alanine ligase) (4).
TABLE 1.
MICs of erythromycin, lincosamides, and streptogramins for E. faecalis strains
| E. faecalis strain(s) | MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|
| ERYa | LIN | CLX | QUI | DAL | Q-D | |
| ATCC 29212 | 2 | 32 | 16 | 8 | >128 | 8 |
| UCN32 | 0.5 | 1 | 0.25 | 8 | 8 | 1 |
| UCN32-1, UCN32-2, and UCN32-3 | 0.5 | 32 | 32 | 8 | >128 | 8 |
| UCN33 | 0.5 | 2 | 0.5 | 8 | 8 | 0.5 |
| UCN33-1, UCN33-2, and UCN33-3 | 0.5 | 64 | 32 | 8 | >128 | 16 |
CLX, clindamycin; DAL, dalfopristin; ERY, erythromycin; LIN, lincomycin; QUI, quinupristin-dalfopristin; Q-D, quinupristin-dalfopristin.
Sequence of the lsa-like gene of isolates.
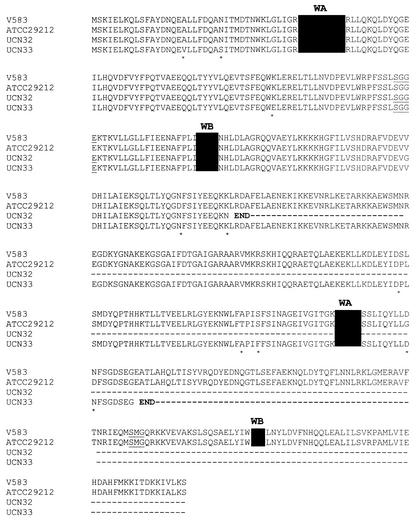
The analysis of the sequence of the lsa gene of E. faecalis V583, obtained from The Institute for Genomic Research website at http://www.tigr.org, allowed us to design four pairs of primers to amplify overlapping DNA fragments (Table 2). A DNA region of E. faecalis UCN32 and UCN33 and of the reference strain E. faecalis ATCC29212 (resistant to lincosamides and streptogramins A) that included the entire lsa-like gene and 436 nucleotides upstream was sequenced in both directions. The predicted Lsa sequences of E. faecalis ATCC 29212 (GenBank access no. AY225127) and E. faecalis V583 were nearly identical except for five substitutions (Fig. 1). In E. faecalis UCN32, in addition to a few substitutions, deletion of an adenine was detected at position 618 (base relative to ATG), which generated a stop codon four nucleotides downstream. In E. faecalis UCN33, a G-T mutation at position 987 produced a stop codon. For both strains, the mutations would lead to a premature termination of the LSA sequence before the second Walker A motif for E. faecalis UCN32 and before the second Walker B motif for E. faecalis UCN33 (Fig. 1). Our work suggested that the loss of only the second Walker B motif was deleterious to the function of LSA.
TABLE 2.
Oligonucleotides used for amplification of lsa-like genes
| Primer | Sequence (5′-3′)a | Positionb |
|---|---|---|
| LsaDIRA | +CGCTCCAGCTGTATGAGAACTGC | −459, −436 |
| LsaREVA | −TCAAGCGATTGACTTCTTTTTTG | +480, +460 |
| Lsa DIRB | +CAAGTGGCTGAATATTTGAAG | +657, +679 |
| Lsa REVB | −TCCCTTTACTTTTTCAAGATG | +1521, +1501 |
+, sense primer; −, antisense primer.
Base relative to ATG.
FIG. 1.
Multiple-sequence alignment of deduced amino acid sequences of the lsa-like genes from E. faecalis V583, ATCC 29212, UCN32, and UCN33. ClustalW at the Infobiogen website (http://www.infobiogen.fr) was used. The two ATP-binding domains consisting of Walker A (WA) and Walker B (WB) motifs (marked in solid black) and of a signature (SGG) sequence (underlined) are shown. Asterisks indicate differences in the amino acids.
Selection of revertant mutants.
Approximately 108 cells of E. faecalis UCN32 and E. faecalis UCN33 were spread onto brain heart infusion agar plates containing 20 μg of clindamycin/ml and were incubated for 24 h at 37°C. Resistant mutants were obtained at frequencies approximately equal to 10−6. Three mutants derived from each parental strain were selected and were further studied. All displayed an LSA phenotype (coresistance to clindamycin, lincomycin, and dalfopristin) (Table 1). lsa sequence analysis in the three mutants, UCN32-1, UCN32-2, and UCN32-3, derived from E. faecalis UCN32 showed an insertion of an adenine in place of the deletion in the parent strain, which restored a sequence identical to that of E. faecalis V583. In the mutants from E. faecalis UCN33, the stop codon (TAA) was changed to a glutamic acid codon (GAA) in mutant UCN33-1 or to a glutamine codon (CAA) in mutants UCN33-2 and UCN33-3. There were no differences between the MICs of lincosamides and dalfopristin for mutants (Table 1).
This study confirms that the lsa gene was responsible for intrinsic resistance to lincosamides and streptogramins A in E. faecalis. It also shows that clindamycin resistance is not always reliable as an identification marker of E. faecalis. Since revertant mutants resistant to clindamycin were readily obtained in the presence of this antimicrobial, the use of clindamycin should be discouraged in the case of infection by clindamycin-susceptible E. faecalis.
REFERENCES
- 1.Allignet, J., V. Loncle, and N. El Sohl. 1992. Sequence of a staphylococcal plasmid gene, vga, encoding a putative ATP-binding protein involved in resistance to virginiamycin A-like antibiotics. Gene 117:45-51. [DOI] [PubMed] [Google Scholar]
- 2.Bozdogan, B., and R. Leclercq. 1999. Effects of genes encoding resistance to streptogramins A and B on the activity of quinupristin-dalfopristin against Enterococcus faecium. Antimicrob. Agents Chemother. 43:2720-2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duh, R. W., K. V. Singh, K. Malathum, and B. E. Murray. 2001. In vitro activity of 19 antimicrobial agents against enterococci from healthy subjects and hospitalized patients and use of an ace gene probe from Enterococcus faecalis for species identification. Microb. Drug Resist. 7:39-46. [DOI] [PubMed] [Google Scholar]
- 4.Dutka-Malen, S., S. Evers, and P. Courvalin. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:24-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Facklam, R. R., and M. D. Collins. 1989. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J. Clin. Microbiol. 27:731-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linton, K. J., and C. F. Higgins. 1998. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol. 28:5-13. [DOI] [PubMed] [Google Scholar]
- 7.Maskell, J. P., A. M. Sefton, J. Yong, S. J. Chi, and J. D. Williams. 1988. Comparative in-vitro activity of erythromycin, vancomycin and pristinamycin. Infection 16:365-370. [DOI] [PubMed] [Google Scholar]
- 8.Ross, J. I., E. A. Eady, J. H. Cove, and S. Baumberg. 1996. Minimal functional system required for expression of erythromycin resistance by msrA in Staphylococcus aureus RN4220. Gene 183:143-148. [DOI] [PubMed] [Google Scholar]
- 9.Ross, J. I., E. A. Eady, J. H. Cove, W. J. Cunliffe, S. Baumberg, and J. C. Wootton. 1990. Inducible erythromycin resistance in staphylococci is encoded by a member of the ATP-binding transport super-gene family. Mol. Microbiol. 4:1207-1214. [DOI] [PubMed] [Google Scholar]
- 10.Singh, K. V., K. Malathum, and B. E. Murray. 2001. Disruption of an Enterococcus faecium species-specific gene, a homologue of acquired macrolide resistance genes of staphylococci, is associated with an increase in macrolide susceptibility. Antimicrob. Agents Chemother. 45:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh, K. V., G. M. Weinstock, and B. E. Murray. 2002. An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob. Agents Chemother. 46:1845-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]