Diseases date back to the dawn of humankind. As humans have evolved, so too have their diseases: some that were once rare have become common, others have disappeared and new varieties have emerged. Many of these changes have taken place in the wake of important transformations in human civilizations and ecology. It is therefore feasible to propose that diseases succeed and fail in response to humanity's advances. My hypothesis to explain the appearance and disappearance of some of these diseases—both infectious and chronic—is that changes in human ecology result in changes in the microbes that populate our bodies. This, in turn, affects our physiology and ultimately our health.
…diseases succeed and fail in response to humanity's advances
The invention of agriculture about 10,000 years ago led to a massive increase in the human population, but at a price: by living in closer proximity to domesticated animals and those that parasitized their food supply (such as rats), humans became more susceptible to measles, plague, tuberculosis and other zoonoses. Urbanization during the Middle Ages was one of the main factors facilitating the devastating spread of Yersinia pestis through the crowded cities of Europe—the Black Death subsequently killed about 30% of the continent's population. Improved hygiene in the nineteenth century eventually reduced the prevalence of plague, along with cholera, dysentery, materno-fetal mortality, childhood infections and other ancient foes. The agricultural revolution after the Second World War relegated famine and its associated diseases—such as pellagra, which is a deficiency of niacin—to history, at least in the industrialized world.
In the nineteenth and twentieth centuries, industrialization—and the scientific revolution that preceded and facilitated it—also gave humans new tools for controlling infectious diseases. Due to improvements in sanitation, antibiotics and vaccines, smallpox has now been eradicated, polio is on the brink of extinction and many other infectious diseases are on the decline. However, industrialization also made possible the rise of new diseases (Table 1). Chronic ailments, such as allergies, asthma, coronary heart disease and cancer, are taking a major toll in developed countries. Man-made toxic compounds affect our immune, neurological and reproductive systems. Predictably, as humans move through a ‘post-modern' era, different diseases are becoming prominent. For some, such as HIV/AIDS, the cause is obvious: in this case, the introduction of a new microbial agent into a population that has no immunity against it, and its dissemination through both ancient and modern routes of transmission, namely sexual activity and needle sharing.
Predictably, as humans move through a ‘post-modern' era, different diseases are becoming prominent
Table 1.
Examples of emerging diseases
|
Early-modern and modern diseases |
| Peptic ulcer disease |
| Appendicitis |
| Lung cancer |
| Asbestosis |
| Hypertension |
| Atherosclerosis |
| Rheumatic fever |
| Tuberculosis |
|
Post-modern diseases |
| Gastroesophageal reflux disease, adenocarcinoma of the oesophagus |
| Asthma |
| Atopic dermatitis |
| Type II diabetes |
| Metabolic syndrome |
| Obesity |
| Acquired immunodeficiency syndrome |
For many other post-modern diseases (Table 1), the aetiology is more obscure; however, I postulate that changes in the ecology of our indigenous microbiota have important roles in their emergence. This view parallels the well-known hygiene hypothesis, which posits that diminished exposure to parasites and pathogens early in life—through improved hygiene, vaccines and antibiotics—might be partly responsible for an increased prevalence of allergic and autoimmune disorders in later years (Strachan, 1989; Cookson & Moffatt, 1997). The lack of pathogenic and invading organisms might cause the immune system to shift its immunological response away from a balance between type 1 and type 2 T-helper cells to one skewed towards the latter (Prescott et al, 1999).
However, my view differs in the following respects: what constitutes ‘hygiene'; the mechanisms that cause disease, which might be metabolic, physiological and immunological; and the emphasis on understanding the problem in the light of natural selection. My hypothesis to explain these diseases is centred on indigenous, or autochthonous, microbes. Although my arguments apply to a variety of biological systems, for relevance and ease of understanding, here I focus on humans.
Most cells in our body are not human but microbial: the ratio of ‘them' to ‘us' is about 10:1 (Savage, 1977). In addition to being numerous, our microbes also are enormously varied—more than 1,000 bacterial species abound in a variety of niches in our bodies. This immediately raises the question of who we are. Newborn babies are essentially sterile at birth; however, from the moment they pass through the birth canal and draw their first breath, they are colonized by successions of microbes that form the communities found in adults (Savage, 1977). The most numerous bacterial populations are in the gastrointestinal tract, on the skin, in the upper respiratory tract and inside the vagina. Although these populations are highly stable, they are still prone to perturbations by environmental insults (Sullivan et al, 2001), with important consequences for nearly all of our bodily functions and, consequently, our health. My theory rests on the six simple and mostly self-evident tenets described below.
Most cells in our body are not human but microbial: the ratio of ‘them' to ‘us' is about 10:1
First, the existence of an indigenous—or residential—microbiota is ancient. Such microbes are found in all animals that are at least as complex as annelids (earthworms); this provides evidence for the co-evolution of animals and bacteria for more than 800 million years (Schramm et al, 2003; Savage, 1977). The microbiota at specific sites in our bodies and those of our mammalian relatives have many similarities in composition and organization, which is consistent with their common ancestry and/or convergent evolution. Similarly, the structures of human populations (as affected by migration and mating over the millennia) and populations of indigenous microbes show overlapping phylogenies, which provides evidence of shared inheritance for more than 50,000 years (Wirth et al, 2005).
Second, since the beginning of clinical microbiology it has been known that conserved microbes or microbial families colonize specific niches in the body; for example, staphylococci colonize the skin, Escherichia coli colonizes the colon and lactobacilli colonize the vagina. Molecular and genomic techniques in taxonomy have further confirmed and extended this idea, and indicate that enormous variations within phyla, genera and species are found at every site studied (Kroes et al, 1999; Sghir et al, 2000; Zoetendal et al, 2006). Although there is substantial, but poorly defined, variation between individuals, there is also a conserved site-specific microbiome—that is, all the genomes of the microbiota—in each definable ecological niche in our bodies (Eckberg et al, 2005; Gill et al, 2006).
Third, this conservation is not accidental, but rather is the result of ongoing selection for co-evolving organisms. As most of the species found in our microbiota appear to be obligate host-specific microbes, the death of their host means their own demise unless they can colonize a new one. This stringent requirement selects either for virulence with a short period of intense transmission—as in the case of pathogens—or for organisms that can have persistent transmission opportunities. In short, any organism that impairs or kills its host needs to find a new one quickly to ensure its own reproductive success; by contrast, quiescent organisms have developed less intrusive, although also less efficient, methods to transmit to a new host. In the latter case, selection favours microbes that, at the least, have negligible cost to their host, or, at the most, increase the host's own reproductive fitness and are therefore symbiotic. Optimal host–microbial interaction leads to improved host fitness during the reproductive years, and contributes to host demise in the senescent years; this maximizes the allocation and use of limited resources for the overall host population and their guests (M.J.B. & G.F. Webb, unpublished data).
Optimal host–microbial interaction… maximizes the allocation and use of limited resources for the overall host population and their guests
Fourth, microbial selection is based on the fitness of hosts and on host populations. When humans lived in small isolated groups—as was the case for most of our history—the extinction of a group also caused the loss of all its microbial genes. Consequently, evolution has selected for those microbial populations that maintain and increase the fitness of both individual hosts and the group as a whole. This concept of group selection—in which the main target is the gene, the individual or the group, depending on the context—is an extension of Darwinian principles. The evidence for such co-evolution is abundant: microbes produce essential metabolites, such as menaquinone (vitamin K2); they aid organ development and metabolism, for instance by affecting fat storage (Backhed et al, 2004); and they protect against intrusions by exogenous pathogens (Bohnhoff & Miller, 1962). Overall, their beneficial activities affect many metabolic, physiological and immunological functions in our bodies.
Fifth, as human ecology changes, so does our microbiota. In the past century, the human condition, especially in developed countries, has undergone dramatic changes that affect the transmission and maintenance of the indigenous microbiota. Caesarean sections limit the perinatal transfer of the maternal flora in the vagina to newborns. Replacing breast milk with formula changes the selection for gastric lactobacilli. Cleaner water diminishes our exposure to faecal organisms, albeit with many beneficial effects. Demographic developments in whole human populations—including the trend towards smaller families with fewer children—lessen the intra-familial transmission and selection of microbes that are pre-adapted to the particular phenotypes in a family. Most importantly, we are more than 60 years into the antibiotic era. Data from 1992 indicated that by the age of 15 years, US children had received, on average, more than four courses of antibiotics to treat a single disorder, otitis media, a common ear infection (McCaig & Hughes, 1995). There is substantial evidence that such short-term exposures to antibiotics select for resistant organisms; this trend has long-term effects on the composition and stability of endogenous microbial populations, as has been shown for intestinal enterococci (Sjolund et al, 2003) and cutaneous staphylococci (Sjolund et al, 2005).
Sixth, such changing selection for our endogenous microbial populations is responsible for some of the emerging patterns in human health and disease. If our microbiota affects human physiology, and its composition has been changing, it should be no surprise that this might alter host homeostasis and, therefore, disease risk. This is my main proposition, which I further explain in the following two examples.
Shortly after antibiotics were first used to treat infectious diseases in humans, agricultural scientists found that feeding farm animals subtherapeutic doses of antibiotics caused them to gain weight (Jukes, 1955; Feighner & Dashkevicz, 1987). This phenomenon was observed for several classes of antibiotics, and reflects their antibacterial activities. Antifungal agents do not have the same effects. The economics of this improved feed efficiency—the ratio of feed consumed to meat produced—means that the subtherapeutic use of antibiotics to promote growth in farm animals now accounts for more than 50% of all antibiotics sold in the USA (McEwen & Fedorka-Cray, 2002). In Europe, their use is much more tightly regulated owing to fears that resistance genes could transfer from animal microbes to human pathogens. Nevertheless, research in the USA, Europe and elsewhere continues to develop antibiotics for growth-promoting activities in livestock that are used for food production (Butaye et al, 2003).
The fact that these agents clearly cause animals to gain weight raises an important question: by what mechanisms do antibiotics exert this effect? If one assumes that natural selection has favoured indigenous microbes that help to maintain the body weight of vertebrates, their suppression or even elimination with antibiotics would disturb the system and could tip it towards weight gain. In such a model, indigenous bacteria maintain energy homeostasis by influencing metabolic processes (Fig 1). The host population and its density affect the microbiota and its transmission between individuals. At the same time, variation in the bacterial composition, owing to changing host-population dynamics and/or antibiotic usage, affect the homeostasis in individual hosts. Furthermore, selective pressures on the community (such as famine) favour bacteria that transduce population signals (for example, to keep food resources optimally allocated), and thereby influence the structure of the whole population. This model explains how the use of antibiotics leads not only to increased weight in livestock but also to other metabolic developments in humans.
Figure 1.
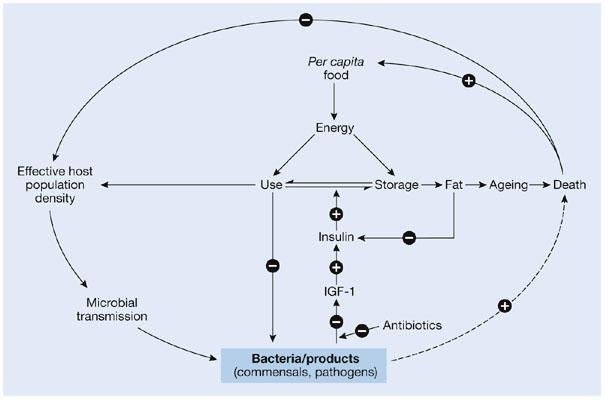
Relationships between indigenous bacteria and host energy homeostasis, longevity and population structure. Indigenous bacteria and their products—as well as pathogens, albeit for shorter periods—are involved in energy homeostasis, through intermediary host metabolic processes, which are modelled here with insulin-like growth factor-1 (IGF-1) and insulin. In a closed system, host population number and density affect microbial transmission and concentrations, and vice versa, through endocrine effects on fertility. Diminished levels of particular interactive bacteria, which can occur, for example, after antibiotic treatment, can lead to increased energy storage, leading to fat accumulation and accelerated ageing.
It was previously postulated that changes in microbial ecology are influencing human height, with diminished microbial transmission early in life contributing to the overall height increases that took place in all developed nations during the twentieth century (Beard & Blaser, 2002). By extension, I now postulate that a similar effect will be seen, or is already being seen, on human weight.
The second example is the interaction of the gastric bacterium Helicobacter pylori with humans, and the consequences of ending this intricate and long-standing relationship. In fact, H. pylori can be regarded as a paradigm of how changes in indigenous microbiota affect human health (Fig 2). Studies of the gastric helicobacters in other mammals, and of human migration patterns over time (Ghose et al, 2002; Falush et al, 2003), suggest that the forebears of modern H. pylori have been present in our ancestors since well before we became humans. The bacterium is also of interest because diagnostic tools are now able to determine its presence or absence in the human stomach, and epidemiological methods can study its effects on human health.
Figure 2.
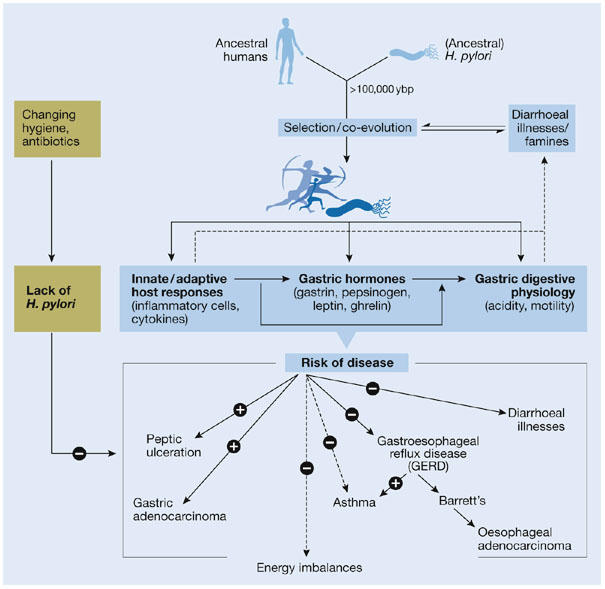
Hypothetical relationship of Helicobacter pylori with human health and disease. There is now considerable evidence that H. pylori has colonized the stomach of humans for more than 100,000 years. Its persistence and ubiquity before modern times suggest selection for its presence, possibly based on enhanced protection against diarrhoeal diseases and improved energy homeostasis (for example, leptin/ghrelin-mediated; in parallel with Fig 1). H. pylori colonization of the stomach induces inflammation, and affects gastric hormones and digestive physiology. In turn, the interaction of particular constellations of H. pylori strains with certain hosts affects the risk of specific diseases. Epidemiological and pathophysiological data indicate that the presence of H. pylori increases lifetime risk for peptic ulcer disease (PUD) and gastric cancer (GC), but protects against gastroesophageal reflux disease (GERD) and its consequences, including oesophageal adenocarcinoma. In response to changing hygiene demographics and antibiotic usage, H. pylori is disappearing, and reduced incidences of PUD and GC, along with increasing incidences of GERD, have been observed (Peek & Blaser, 2002).
Although H. pylori was once present in almost every adult human, the bacterium is now rapidly disappearing from human populations owing to changes in sanitation, demographics and antibiotic usage. Today, fewer than 10% of children in the USA harbour this bacterium in their stomach. When present, H. pylori is the single dominant species in the stomach (Bik et al, 2006), so its disappearance is potentially significant.
Through its pro-inflammatory effects, H. pylori modulates immunological, endocrine and physiological functions in the stomach (Blaser & Atherton, 2004), with both local and systemic manifestations (Fig 2). The biological costs of carrying H. pylori include peptic ulcers and adenocarcinoma of the distal stomach. The (cag+) strains that interact to the greatest extent chemically with their hosts convey the highest risks. Conversely, these strains also protect against gastroesophageal reflux disease (GERD) and its consequences, including oesophageal adenocarcinoma, owing in part to their effects on gastric-acid secretion (Peek & Blaser, 2002). These observations are consistent with the rise of these diseases wherever H. pylori is disappearing, and therefore provide the first definitive example of a group of chronic diseases caused by changes in the human microbiota. GERD has been linked to asthma, and preliminary results now support a link between the disappearance of H. pylori and the increase in asthma cases. In addition, H. pylori affects gastric hormones that have a role in energy homeostasis, such as leptin and ghrelin; a link between its disappearance and the increasing prevalence of metabolic syndrome, type II diabetes and obesity has also been postulated (Blaser & Atherton, 2004).
Because of the great clinical interest in H. pylori and its dominance in the stomach, diagnostic tests are now available to detect it. By contrast, if a bacterial species disappeared from the human colon, mouth, skin or genital tract, it would be difficult or impossible to tell using current knowledge and technologies. H. pylori might therefore be considered an ‘indicator organism' for changing human microecology and disease risk. I predict that there will be others. Indigenous organisms have been evolutionarily selected as a result of their interactions with human tissues, so many of their adaptations involve cross-signalling with host cells; this explains how changes in their populations alter human physiology. In developed countries, new generations are growing up without our ancient companion, H. pylori, to orchestrate their gastric hormones. How will this dysregulation affect the formation of adipose tissue and energy homeostasis during crucial periods of development, at a time when other factors, including the availability of food, are also changing? A similar question can be asked about the effects of this ecological change on immunological balance.
In developed countries, new generations are growing up without our ancient companion, H. pylori. to orchestrate their gastric hormones
A stronger research focus on such changes in human microecology should provide important clues for the prevention, diagnosis and treatment of many post-modern human diseases. The first human genome project has been completed. It is now time to invest in a second human genome project: sequencing the cumulative genomes of our microbiota. This is likely to uncover more indicator organisms, the presence or absence of which indicates important disease trends. Furthermore, harnessing these sophisticated host-adapted organisms to do our bidding could uncover new medical treatments.

Acknowledgments
This work was supported by the Senior Scholar Award in Infectious Diseases from the Ellison Medical Foundation, National Institutes of Health grant RO1 GM63270, and by the Diane Belfer Program for Human Microbial Ecology.
References
- Backhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI (2004) The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 101: 15718–15723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard AS, Blaser MJ (2002) The ecology of height: the effect of microbial transmission on human height. Perspect Biol Med 45: 475–498 [DOI] [PubMed] [Google Scholar]
- Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA (2006) Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci USA 103: 732–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser MJ, Atherton JC (2004) Helicobacter pylori persistence: biology and disease. J Clin Invest 113: 321–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnhoff M, Miller CP (1962) Enhanced susceptibility to Salmonella infection in streptomycin-treated mice. J Infect Dis 111: 117–127 [DOI] [PubMed] [Google Scholar]
- Butaye P, Devriese LA, Haesebrouck F (2003) Antimicrobial growth promoters used in animal feed: effects of less well known antibiotics on Gram-positive bacteria. Clin Microbiol Rev 16: 175–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson WO, Moffatt MF (1997) Asthma: an epidemic in the absence of infection? Science 275: 41–42 [DOI] [PubMed] [Google Scholar]
- Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA (2005) Diversity of the human intestinal microbial flora. Science 308: 1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D et al. (2003) Traces of human migrations in Helicobacter pylori populations. Science 299: 1582–1585 [DOI] [PubMed] [Google Scholar]
- Feighner SD, Dashkevicz MP (1987) Subtherapeutic levels of antibiotics in poultry feeds and their effects on weight gain, feed efficiency, and bacterial cholyltaurine hydrolase activity. Appl Environ Microbiol 53: 331–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghose C, Perez-Perez GI, Dominguez-Bello MG, Pride DT, Bravi CM, Blaser MJ (2002) East Asian genotypes of Helicobacter pylori strains in Amerindians provide evidence for its ancient human carriage. Proc Natl Acad Sci USA 99: 15107–15111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE (2006) Metagenomic analysis of the human distal gut microbiome. Science 312: 1355–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukes TH (1955) Antibiotics in Nutrition. New York, NY, USA: Medical Encyclopedia [Google Scholar]
- Kroes I, Lepp PW, Relman DA (1999) Bacterial diversity within the human subgingival crevice. Proc Natl Acad Sci USA 96: 14547–14552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaig LF, Hughes JM (1995) Trends in antimicrobial drug prescribing among office-based physicians in the United States. JAMA 273: 214–219 [PubMed] [Google Scholar]
- McEwen SA, Fedorka-Cray PJ (2002) Antimicrobial use and resistance in animals. Clin Infect Dis 34 (Suppl 3): S93–S106 [DOI] [PubMed] [Google Scholar]
- Peek RM Jr, Blaser MJ (2002) Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer 2: 28–37 [DOI] [PubMed] [Google Scholar]
- Prescott SL, Macaubas C, Smallacombe T, Holt BJ, Sly PD, Holt PG (1999) Development of allergen-specific T-cell memory in atopic and normal children. Lancet 353: 196–200 [DOI] [PubMed] [Google Scholar]
- Savage DC (1977) Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol 31: 107–133 [DOI] [PubMed] [Google Scholar]
- Schramm A, Davidson SK, Dodsworth JA, Drake HL, Stahl DA, Dubilier N (2003) Acidovorax-like symbionts in the nephridia of earthworms. Environ Microbiol 5: 804–809 [DOI] [PubMed] [Google Scholar]
- Sghir A, Gramet G, Suau A, Rochet V, Pochart P, Dore J (2000) Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization. Appl Environ Microbiol 66: 2263–2266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjolund M, Wreiber K, Andersson DI, Blaser MJ, Engstrand L (2003) Long-term persistence of resistant Enterococcus species after antibiotics to eradicate Helicobacter pylori. Ann Intern Med 139: 483–487 [DOI] [PubMed] [Google Scholar]
- Sjolund M, Tano E, Blaser MJ, Andersson DI, Engstrand L (2005) Persistence of resistant Staphylococcus epidermitis after single course of clarithromycin. Emerg Infect Dis 11: 1389–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strachan DP (1989) Hay fever, hygiene, and household size. BMJ 299: 1259–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan A, Edlund C, Nord CE (2001) Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect Dis 1: 101–114 [DOI] [PubMed] [Google Scholar]
- Wirth T, Meyer A, Achtman M (2005) Deciphering host migrations and origins by means of their microbes. Mol Ecol 14: 3289–3306 [DOI] [PubMed] [Google Scholar]
- Zoetendal EG, Vaughan EE, de Vos WM (2006) A microbial world within us. Mol Microbiol 59: 1639–1650 [DOI] [PubMed] [Google Scholar]


