Introduction
This workshop brought together developmental biologists and oncologists for three intense days of discussions about the recent progress that has been made on the roles of homeodomain proteins in development, haematopoiesis and leukaemogenesis. The homeo-domain is a 61-amino-acid DNA-binding motif with specific sequence and structural characteristics (Gehring & Hiromi, 1986). Homeodomain proteins were initially identified in Drosophila as being responsible, when mutated, for homeotic transformations—that is, the conversion of a part of the body into the likeness of another. Since their initial discovery, the number of homeodomain proteins has increased enormously, and they comprise several different families of transcription factors. The main stars of the meeting were members of the Hox family and of the three amino-acid loop extension (TALE) families, with some other homeodomain proteins making guest appearances. TALE proteins include the PBC family (Exd in Drosophila and Pbx proteins in vertebrates) and the Hth/Meis superfamily (Hth in Drosophila, and Meis and Prep proteins in vertebrates). The link between Hox and TALE proteins is their functional interactions, which were initially revealed in flies by genetic analyses and later supported by biochemical data (Mann & Affolter, 1998; Moens & Selleri, 2006). PBC proteins have been shown to act as Hox cofactors that mainly determine Hox DNA-binding specificity and selectivity. Hth/Meis proteins interact with PBC proteins, participating in ternary complexes with Hox proteins, or modulating PBC protein stability and subcellular localization.

This European Molecular Biology Organization/European School of Molecular Medicine workshop on Homeodomain Proteins, Hematopoietic Development and Leukemias took place in Riva del Garda, Italy, between 23 and 25 March 2006, and was organized by F. Blasi, M. Cleary, P.G. Pelicci and M. Torres.
Functional specificities and redundancies
A large part of the meeting was devoted to the presentation and discussion of the consequences of the complete or partial inactivation of different TALE products in several vertebrates. These studies arrived at two main conclusions: first, TALE proteins are essential for the development of most areas of the organism; and second, they have both individual functions and a large degree of redundancy. This was clearly shown in the Pbx gene family, for which an advanced phenotypic analysis of single and compound mutants was presented. In mice, the individual contribution of each member of the family seemed to be different because, whereas the Pbx1 single mutant had a wide range of malformations, only minor or no phenotypes were obvious in Pbx2 and Pbx3 mutants. However, when a reduced Pbx2 and/or Pbx3 dosage was added to the Pbx1−/− background, strong exacerbations of the malformations were detected, as reported by L. Selleri (New York, NY, USA) and M. Cleary (Stanford, CA, USA). In particular, the cardiovascular system, the axial skeleton, the limbs and the craniofacial area were shown to be strongly affected. A.J. Waskiewicz (Edmonton, Canada) also described redundancy among Pbx genes in zebrafish, in which the functions of the Pbx2 and Pbx4 gene products seem to overlap in a range of tissues, including the hindbrain and tectum, and during early haematopoiesis. Hox gene products are known to be largely redundant, especially within paralogous groups. In the case of the Meis and Prep gene families, pleiotropic functions in patterning and differentiation processes were also described (see below); however, genetic analyses designed to evaluate the possible extent of redundancy among the different members of these gene families are still at an early stage.
At the meeting, several tissues were reported to require the activity of homeodomain proteins for proper development; we highlight some of these data below.
Mechanisms of TALE protein function
TALE proteins are usually assumed to work as Hox partners. However, data presented during the workshop indicated that this might be just one of several possible mechanisms for the action of these proteins. For instance, E. Ferretti (New York, NY, USA) reported that compound Pbx1-mutant mice have craniofacial malformations that affect maxillary and frontonasal mass derivatives, in which the 39 murine Hox genes are not expressed (Krumlauf, 1993). Selleri reported that the limbs of the same embryos also show pronounced phenotypes with decreasing Pbx dosage on the Pbx1-mutant background. The similarity of the skeletal phenotype in the distal regions of the limb (zeugopod and autopod) to those recently reported in mice lacking the HoxA and HoxD complexes (Kmita et al, 2005) could indicate the requirement for Pbx proteins as Hox partners in limb development. However, analysis of the Pbx-mutant limb buds revealed that all Hox genes were downregulated, suggesting that they are involved in the Pbx-mutant phenotype as downstream targets rather than as essential cofactors. This was also apparent for some members of the Meis/Prep superfamily. For instance, F. Argenton (Padua, Italy) reported that inactivation of Prep1.1 in zebrafish produced a phenotype that affected, among other structures, the formation of cartilages from the facial and branchial region, apparently as a consequence of increased apoptosis. These cartilages were in both Hox-positive and Hox-negative areas of the embryo, suggesting a common Hox-independent mechanism. A similar finding was reported by M. Torres (Madrid, Spain) for haematopoiesis; inactivation of Meis1 in the mouse results in a severe impairment of the stem and progenitor cell compartments, and the absence of the megakaryocytic lineage, which leads to embryonic lethality. Importantly, Meis1 is strongly expressed in the aortic-gonad-mesonephros region, where haematopoietic stem cells are generated, but Hox transcription is undetectable in this area.
So, how do the Pbx/Meis–Prep genes work? Their classical activity as Hox cofactors can probably account for some of their functions (Fig 1, steps 6 and 7). They could also act as partners of other non-Hox homeodomain proteins. For instance, V. Zappavigna (Modena, Italy) reported that Emx2 and Pbx1 interact genetically in the patterning of the most proximal part of the mouse forelimb, which is an area that apparently does not depend on Hox gene activity (Kmita et al, 2005). Waskiewicz showed that Engrailed also functionally interacts with Pbx proteins to establish the midbrain–forebrain boundary in zebrafish embryos, which strengthened previous biochemical data (Peltenburg & Murre, 1996). Likewise, Waskiewicz suggested Cdx proteins as Pbx partners in the regulation of haematopoiesis in zebrafish, and Hox11 was shown to interact with Pbx1 in the murine splenic mesenchyme (A. Brendolan, New York, NY, USA). Most importantly, the relevance of the interaction of TALE proteins with non-homeodomain proteins, in both the presence and absence of Hox proteins (Fig 1, steps 2, 3, 6 and 7), became clear at this meeting. An example was provided by C. Sagerstrom (Worcester, MA, USA) in the patterning of the zebrafish nervous system. In particular, he showed that Pbx4 and Hoxb1b bind to the Hoxb1a promoter, and a histone deacetylase is recruited to the complex to inhibit transcription. However, Meis3 can displace the histone deacetylase from Pbx4 to promote—together with Pbx4 and Hoxb1b—the transcriptional activation of Hoxb1a. Furthermore, F. Blasi (Milan, Italy) showed that the p160Myb-binding protein can compete with Pbx1 for binding to Prep1 to regulate murine haematopoiesis. Finally, an even more divergent mechanism for Pbx functional activity was proposed by S. Tapscott (Seattle, WA, USA) in the muscle lineage. Pbx proteins, together with Meis1, could 'instruct' MyoD by labelling specific promoter target sites in the genome for MyoD-dependent activation. This interaction seems to be essential for transcriptional activation, as L. Mares (Seattle, WA, USA) reported that knocking down Pbx activity in zebrafish embryos resulted in downregulation of myogenin without any apparent effects on MyoD expression. Therefore, Pbx proteins can act as cofactors of other non-homeodomain-containing developmental regulators.
Figure 1.
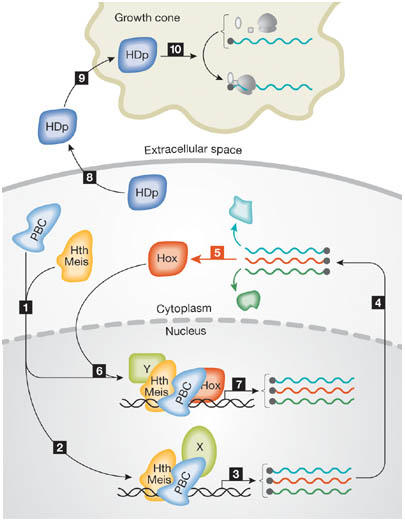
Molecular activities of homeodomain proteins. In step 1, homeodomain proteins of the Hth/Meis family relocate PBC family proteins to the nucleus. In steps 2–5, PBC and Hth/Meis proteins, together with other factors (X), activate transcription of a range of genes, including Hox genes (shown in red), which are translated into proteins. In steps 6 and 7, Hox proteins regulate gene transcription in association with proteins that include those of the PBC and Hth/Meis families, and a range of other factors (Y). In steps 8–10, in addition to transcriptional regulators, homeodomain proteins (HDp) are also secreted and taken up by other cells (such as neuronal growth cones) to regulate translation of specific messenger RNAs.
Interestingly, MyoD was shown to follow a feed-forward mechanism in the activation of its targets, which might also apply to the Pbx family. Tapscott showed that the activation of different MyoD targets in murine cells follows different dynamics, and that the products of some primary MyoD targets are themselves transcriptional activators that function, together with MyoD, in the activation of secondary targets. These could, in turn, have a similar effect on tertiary targets. An intriguing hypothesis, based on data presented during this workshop, is that Pbx could act through a similar feed-forward mechanism (Fig 1, steps 2–7). Findings supporting this view include the evidence that Hox genes are downstream targets of Pbx or Meis proteins in the following model systems: mouse limb buds (as previously mentioned); the haematopoietic tissue of zebrafish embryos, in which Hoxb6, Hoxb7 and Hoxa9 are downregulated when Pbx2/4 activity is removed, as reported by Waskiewicz; the hindbrain of Xenopus, in which Hoxd1 is regulated by Meis3 as shown by D. Frank (Haifa, Israel); and the hindbrain of zebrafish, where Argenton reported that some Hox genes are downregulated when Prep1.2 is inactivated. Hox genes could act as primary targets that would cooperate with Pbx to activate secondary targets. The evaluation of this possibility requires the design of specific experimental strategies, including the use of conditional mutants, as the strong phenotype of Pbx mutants during early developmental phases hinders the analysis of possible later functions. Actually, increasing evidence indicates that homeodomain proteins have roles at both early and late stages of development or differentiation. Hox genes, which are required early in central nervous system development for positional and patterning information, are also involved at later stages in the wiring of somatosensory circuits, as shown by F. Rijli (Strasbourg, France) using conventional and conditional inactivation of Hoxa2 in the mouse. Similarly, H. Simon (Heidelberg, Germany) reported that En1 and En2 are essential not only for early patterning processes, but also for survival of mesencephalic dopaminergic neurons, even in the adult mouse.
The existence of Hox-related and Hox-unrelated functions for the TALE proteins is not a vertebrate-specific phenomenon, as it has also been reported in Drosophila (Casares & Mann, 1998). Interestingly, work from Drosophila presented at the workshop provided some clues about the molecular mechanisms of this process. According to R. Mann (New York, NY, USA), the Hth gene has, in addition to the classical transcript, two further splice variants, both of which resulted in the formation of a truncated protein that does not contain a homeodomain. These alternative transcripts and their protein products are distributed in the embryo with a pattern indistinguishable from that of the full-length version of the messenger RNA (mRNA) and protein. However, they are functionally specific. Surprisingly, the homeodomain-containing protein is responsible for the Hox-independent functions of Hth and, for many of the Hox cofactor functions, it is the homeodomain-less isoform that has the dominant role. Apparently, similar homeodomain-less versions also exist for the Hth mammalian homologues Meis1 and Prep2. It will be important to test whether they also share similar functional mechanisms with their Drosophila counterparts.
Downstream from the homeodomain
In addition to the molecular mechanisms of homeodomain protein function, downstream analysis of these proteins was also addressed in various model systems. Expression of several genes was reported to be regulated by homeodomain proteins. One group of targets comprises other transcription factors that are also involved in patterning processes. Among these, the above-mentioned Hox genes and the Pax genes deserve special mention. In particular, the expression of Pax3 in the cardiogenic neural crest was reported by Cleary to be under the control of the Pbx gene family in the mouse, as revealed by its downregulation in Pbx1-null mutant embryos. This regulation might be direct, as a Hox/Pbx-binding site was found in the Pax3 promoter. Similarly, B. Peers (Liége, Belgium) reported that Pax6b was under the control of a Pbx–Prep–Pdx ternary complex in the dorsal pancreas primordium of the zebrafish endoderm.
Another salient group of molecules identified as downstream targets of homeodomain proteins encompass components of several signalling pathways. In some cases, homeodomain proteins have been shown to modulate these signals. For instance, Sagerstrom reported that Sonic hedgehog (Shh) seems to be downregulated by Meis3 in zebrafish endoderm, which is an effect that could be important for pancreatic development. Conversely, Selleri reported that Pbx activity is required for Shh expression in murine limb buds, although it is possible that this activity is mediated by Hox genes. Indeed, Zappavigna showed that Hoxa13 and Hoxd13 could activate a regulatory element of the mouse Shh gene located ∼1 Mbp away from its transcriptional start site. A. Care (Rome, Italy) reported that basic fibroblast growth factor (bFGF) was also a direct target of Hoxb7 in melanomas and breast carcinomas. In other cases, homeodomain proteins seem to modulate the ability of cells to respond to signals. For instance, M. Mallo (Oeiras, Portugal) reported that Hoxb4 modulates the response of murine embryonic stem cells to several signals, thereby promoting expansion of haematopoietic progenitors, probably through the Hoxb4-dependent production of specific modifications in the sugar moieties of heparan sulphate proteoglycans. M. Kamps (La Jolla, CA, USA) reported that an increase in responsiveness to cytokines was also produced by Hoxa9 and Meis1 overexpression during leukaemogenesis, which was mediated, at least in part, by the upregulation of Flt3, CD34, IL7R, Erg and GPR56.
The regulation of gene expression by homeodomain proteins might differ from conventional models in additional unexpected ways. As the homeodomain is known to be a DNA-binding motif, it is generally assumed that proteins containing this domain act at the transcriptional level. However, this is not always the case. A. Trembleau (Paris, France) showed that the homeodomain proteins Engrailed 2 (En2) and Emx2 can also act as translational regulators in developing sensory systems, such as the visual and olfactory systems. Even more surprising is the fact that En2 is secreted, and forms an extracellular gradient in the tectum that can repel axonal growth cones originating in the temporal retina of Xenopus and, conversely, can attract axons originating in the nasal retina. En2 is internalized into growth cones, where it modulates the translational machinery (Fig 1, steps 8–10). Consistent with this, its effect requires protein synthesis, but not transcription. In addition, another homeodomain protein, Emx2, was shown to interact with the translational initiation factor eIF4E in the axon of olfactory sensory neurons, indicating that it might regulate the translation of unknown mRNAs. Apparently, other homeodomain proteins (such as Bicoid and Hoxa9) can interact with the translational machinery (Topisirovic & Borden, 2005), and many homeodomain proteins can reportedly also be secreted (Prochiantz & Joliot, 2003), suggesting that the mechanism of action described for En2 and Emx2 might also apply to other homeodomain-containing proteins in different systems.
Homeodomain proteins in haematopoiesis
The contribution of homeodomain proteins to haematopoiesis and leukaemogenesis was another major focus of the workshop. The strong role of TALE proteins in haematopoiesis contrasts with the relatively weak haematopoietic phenotypes observed in a wide range of Hox mutants. In general, most TALE proteins have more than one role during normal haematopoiesis, and these functions are specific for each gene product. For instance, in the mouse, Meis1, Prep1 and Pbx1 were all reported to be essential for haematopoietic stem cell homeostasis. However, at later stages of development and differentiation, Meis1 is required for specification of the megakaryocytic lineage (as reported by Torres), Prep1 is required for proliferation of erythroid progenitors and for T-cell differentiation (as shown by Blasi), and Pbx1 is required for the expansion of the common lymphoid progenitor and to a lesser extent the common myeloid progenitor (as reported by F. Ficara, Stanford, CA, USA). In addition, M.C. Magli (Pisa, Italy) reported that the non-Hox homeodomain protein Otx1 is essential for the control of both the erythroid and the myelo-monocytic lineages. All these late functions of homeodomain proteins seem to be linked to specific pathways of haematopoietic differentiation, or to modulation of alternative cell-fate decisions. The involvement of these genes in these processes was clearly shown and characterized in appropriate mutants, and some information about their mechanism of action was also reported. Interestingly, analysis of Prep1 hypomorphic-mutant mice revealed a dosage-dependent effect of the protein on blood-cell development, and its function was dependent on its subcellular localization. Furthermore, Blasi suggested that Prep1 might be an oncosuppressor gene, as its downregulation in vivo is associated with the development of lymphoid malignancies.
Much more information was provided about the involvement of homeodomain proteins in leukaemogenesis. Several Hox genes (such as A9, A7 and B8) have been associated with a number of leukaemic processes, mostly acute myeloid leukaemia (AML). However, upregulated Hox gene expression alone (or even that of some activating fusion proteins) is not enough to induce leukaemia. Kamps reported that Hox proteins produced the expansion of late myeloid progenitors, mostly controlling the proliferation/differentiation switch at specific points of haematopoietic differentiation. For the leukaemogenic process to occur, misregulated activity of other genes is also required. Several studies attempting to identify such genes were reported, and Meis1 turned out to be one of the most common counterparts of Hox genes in malignant transformation. For this, the carboxy-terminal domain of Meis1 was reported to be essential. Kamps reported that this domain seems to confer transactivation activity, as it can be replaced by the VP16 activation domain. However, M. Featherstone (Montreal, Canada) showed that this carboxy-terminal domain does not seem to have such an activity by itself; rather, it recruits protein kinase A-dependent transcriptional-activation complexes. In particular, transducers of regulated CREB activity (TORC) coactivators, which are known to form nuclear complexes with the cAMP-response-element-binding protein (CREB) in response to protein kinase A (Cheng & Saltiel 2006), activate transcription through Meis1 on binding to its carboxyl terminus. T. Nakamura (Tokyo, Japan) reported that other factors were also shown to cooperate with Hox/Meis in the induction of myeloid leukaemias, using an in vivo retroviral screen. Among the most frequently found factors were Trib1 and Evi1. Trib1 enhances the activity of the Flt3/mitogen-activated protein kinase (MAPK) pathway by a direct interaction with MAPK/ERK kinase (MEK). This makes it particularly interesting in the context of Meis1-induced AML, because, in these cells, Flt3 is itself upregulated in a Meis1-dependent fashion.
The involvement of Hox proteins in leukaemogenic processes often results from the genesis of fusion proteins derived from translocations. The contribution of these fusion proteins to malignancy could derive not only from their increased transcriptional activity, but also from increased accumulation resulting from their insensitivity to degradation. In particular, P. Zhou (New York, NY, USA) reported that the NUP98/Hoxa9 fusion protein, which is often associated with leukaemias, was insensitive to degradation mediated by the E3 ubiquitin enzyme Cul-4A, which normally mediates Hoxa9 turnover.
The final day of the workshop was mainly dedicated to studies focused on normal and leukaemic haematopoietic stem cells. P.G. Pelicci (Milan, Italy) presented data supporting the view that the hierarchical organization of haematopoietic cells is maintained after malignant transformation, and that a leukaemic stem-cell population is responsible for the maintenance and growth of the disease. In addition, he showed that the induction of leukaemia by several fusion proteins that have been associated with AML requires the cell-cycle inhibitor p21, which is essential for the maintenance of quiescent stem cells. These findings highlight the importance of a resting pool of self-maintaining cells, which are crucial for leukaemia development and are refractory to chemotherapy.
Several studies were presented in which microarray chip analysis was used to identify the molecular signature of normal and leukaemic stem cells. M. Goodell (Houston, TX, USA) reported on a comparison of gene-expression profiles between haematopoietic stem cells and populations of mature cells of different haematopoietic lineages, which allowed the identification of unique transcriptional fingerprints for each specific cell type. The 350 genes unique to haematopoietic stem cells included genes encoding transcription factors, some of them also of the Hox family. In addition, M. Alcalay (Milan, Italy) reported that transcriptional activity of members of the Hox and TALE families was detected in a gene-expression analysis of AML cells, suggesting that they might reflect the molecular status of leukaemic stem cells.
Conclusions
This workshop highlighted the pivotal role of members of the Hox and TALE families in the development/differentiation of most prenatal and/or postnatal tissues, as well as illustrating their high degree of redundancy. Furthermore, increasing evidence indicates that these genes are involved in normal and malignant haematopoiesis, although the latter requires dysregulation of more than one of them. An emerging view at the meeting was that the function of TALE proteins is not restricted to Hox cofactors (Fig 1); they also act upstream from Hox genes, and as partners of non-Hox proteins, both with and without a homeodomain. In addition, it should be stressed that homeodomain proteins might not only be transcription factors, but could also be regulators of translation.

Moisés Mallo

Maria Cristina Magli
Acknowledgments
We acknowledge the support of Ministerio dell'Università e della Ricerca-Programma Nazionale di Ricerca (MIUR-PNR) grant RBNE01R4MJ_002 (M.C.M.), Progetto Nazionale Cellule Staminali grant CS 73 (M.C.M.), Fundação para a Ciência e a Tecnologia (FCT) grant POCI/SAU-MMO/60419/2004 (M.M.) and the Centro de Biologia do Desenvolvimento (M.M.). We thank R. Cassada for reading the manuscript. We apologize to those speakers whose work we could not discuss owing to space constraints.
References
- Casares F, Mann RS (1998) Control of antennal versus leg development in Drosophila. Nature 392: 723–726 [DOI] [PubMed] [Google Scholar]
- Cheng A, Saltiel AR (2006) More TORC for the gluconeogenic engine. Bioessays 28: 231–234 [DOI] [PubMed] [Google Scholar]
- Gehring W, Hiromi Y (1986) Homeotic genes and the homeobox. Annu Rev Genet 20: 147–173 [DOI] [PubMed] [Google Scholar]
- Kmita M, Tarchini B, Zàkàny J, Logan M, Tabin CJ, Duboule D (2005) Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature 435: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Krumlauf R (1993) Hox genes and pattern formation in the branchial region of the vertebrate head. Trends Genet 9: 106–112 [DOI] [PubMed] [Google Scholar]
- Mann R, Affolter M (1998) Hox proteins meet more partners. Curr Opin Genet Dev 8: 423–429 [DOI] [PubMed] [Google Scholar]
- Moens CB, Selleri L (2006) Hox cofactors in vertebrate development. Dev Biol 291: 193–206 [DOI] [PubMed] [Google Scholar]
- Peltenburg LT, Murre C (1996) Engrailed and Hox homeodomain proteins contain a related Pbx interaction motif that recognizes a common structure present in Pbx. EMBO J 15: 3385–3393 [PMC free article] [PubMed] [Google Scholar]
- Prochiantz A, Joliot A (2003) Can transcription factors function as cell–cell signalling molecules? Nat Rev Mol Cell Biol 4: 814–819 [DOI] [PubMed] [Google Scholar]
- Topisirovic I, Borden KL (2005) Homeodomain proteins and eukaryotic translation initiation factor 4E (eIF4E): an unexpected relationship. Histol Histopathol 20: 1275–1284 [DOI] [PubMed] [Google Scholar]


