Abstract
Hygromycin A, an antibiotic produced by Streptomyces hygroscopicus, is an inhibitor of bacterial ribosomal peptidyl transferase. The antibiotic binds to the ribosome in a distinct but overlapping manner with other antibiotics and offers a different template for generation of new agents effective against multidrug-resistant pathogens. Reported herein are the results from a series of stable-isotope-incorporation studies demonstrating the biosynthetic origins of the three distinct structural moieties which comprise hygromycin A. Incorporation of [1-13C]mannose and intact incorporation of d-[1,2-13C2]glucose into the 6-deoxy-5-keto-d-arabino-hexofuranose moiety are consistent with a pathway in which mannose is converted to an activated l-fucose, via a 4-keto-6-deoxy-d-mannose intermediate, with a subsequent unusual mutation of the pyranose to the corresponding furanose. The aminocyclitol moiety was labeled by d-[1,2-13C2]glucose in a manner consistent with formation of myo-inositol and a subsequent unprecedented oxidation and transamination of the C-2 hydroxyl group to generate neo-inosamine-2. Incorporation of [carboxy-13C]-4-hydroxybenzoic acid and intact incorporation of [2,3-13C2]propionate are consistent with a polyketide synthase-type decarboxylation condensation to generate the 3,4-dihydroxy-α-methylcinnamic acid moiety of hygromycin A. No labeling of hygromycin A was observed when [3-13C]tyrosine, [3-13C]phenylalanine, or [carboxy-13C]benzoic acid was used, suggesting that the 4-hydroxybenzoic acid is derived directly from chorismic acid. Consistent with this hypothesis was the observation that hygromycin A titers could be reduced by addition of N-(phosphonomethyl)-glycine (an inhibitor of chorismic acid biosynthesis) and restored by coaddition of 4-hydroxybenzoic acid. The convergent biosynthetic pathway established for hygromycin A offers significant versatility for applying the techniques of combinatorial and directed biosynthesis to production of new antibiotics which target the ribosomal peptidyl transferase activity.
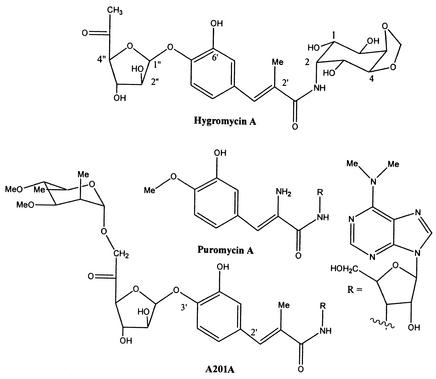
The isolation of hygromycin A (Fig. 1) (14, 19) from several strains of Streptomyces hygroscopicus was reported in 1953. A structurally unrelated antibiotic, the aminoglycoside hygromycin B, was later isolated from the same S. hygroscopicus strains (13). Initial studies demonstrated that hygromycin A had a relatively broad spectrum of modest activity against gram-positive and gram-negative bacteria (14, 19). Hygromycin A has also been shown recently to have immunosuppressant activity in the mixed poor lymphocyte reaction but does not work via suppression of interleukin 2 production (29).
FIG. 1.
Structures of hygromycin A, antibiotic A201A, and puromycin.
More than 2 decades after its initial isolation the mode of antibacterial action of hygromycin A was demonstrated to be inhibition of the ribosomal peptidyl transferase activity (7). Hygromycin B also disrupts protein synthesis, but by a different mechanism, that of inhibition of translocation and misreading of mRNA (4, 6, 12). Initial studies demonstrated that hygromycin A blocks binding of either radiolabeled chloramphenicol or lincomycin to the ribosome, suggesting that the binding sites of all three antibiotics are closely related (7). These analyses also demonstrated that hygromycin A is more potent than chloramphenicol and binds strongly to ribosomes (7). More recent competitive footprinting experiments have shown that the binding of hygromycin A is affected only by macrolides containing a mycarose moiety (22). Recent crystallographic analyses have clearly demonstrated that for the 16-membered macrolides tylosin and carbomycin, the C-5 disaccharide group containing this mycarose extends from the polypeptide exit channel of the large ribosomal subunit into the peptidyl transferase center (8). Taken together, the data clearly imply that hygromycin A binds to the ribosome in this region in a distinct but overlapping manner with other antibiotics. The decrease in clinical utility of macrolides due to the appearance of resistant pathogens has prompted a need to identify agents which bind differently to the ribosome. Analogs and derivatives of hygromycin A may offer important possibilities in the development of such compounds.
Renewed interest in hygromycin A has also been a result of the observation that the antibiotic is effective in the treatment of induced dysentery in pigs at levels of 5 to 20 g/ton of feed (15). Swine dysentery is a severe mucohemorrhagic diarrheal disease thought to be caused by Serpulina (Treponema) hyodysenteriae (16), an anaerobic spirochete against which hygromycin A has been shown to have good in vitro potency. These observations have led to the preparation of over 100 analogs and determination of their activity, in terms both of MICs for S. hyodysenteriae and of their ability to inhibit protein synthesis in an Escherichia coli cell-free system (9). This work has been used to construct a structure-activity relationship for hygromycin A, in which the aminocyclitol moiety is essential for activity. 6-Deoxy-5-keto-d-arabino-hexofuranose is not essential for antibacterial activity and can be replaced by a hydrophobic allyl group. This replacement decreases the intrinsic protein synthesis inhibition activity but may allow more efficient transport into the bacterial cell. The methyl group in the central 3,4-dihydroxy-α-methylcinnamic acid moiety is sensitive to change, and replacement with propyl, allyl, or hydrogen substituents leads to a reduction in antibacterial activity (9, 10). This work has also shown that while hygromycin A is a potent inhibitor of protein synthesis, its ineffectiveness against gram-negative enteric bacteria such as E. coli is mainly due to the efflux mechanism (AcrAB) (9).
The biological activity of hygromycin A and its analogs, coupled with the observation that it offers ribosomal binding that is distinct from other antibiotics, prompted us to investigate the biosynthesis of this antibiotic. This work was undertaken with a long-term view of generating potentially useful new antibiotic lead compounds through techniques such as combinatorial biosynthesis and mutasynthesis. In this paper we detail results of stable-isotope-incorporation studies which establish the biosynthetic origins of the three moieties of hygromycin A. The results indicate a convergent biosynthetic pathway, which suggests significant versatility for engineering production of new antibiotics which target the bacterial ribosomal peptidyl transferase activity.
MATERIALS AND METHODS
Cultural conditions for hygromycin A production.
Seed cultures of S. hygroscopicus NRRL 2388 were prepared by inoculating spores into a 500-ml baffled flask containing 50 ml of medium consisting of 13 g of glucose, 7.0 g of corn starch, 7.0 g of NZ amine YTT (casein hydrolysate; Quest International Flavors, Norwich, N.Y.), 3.0 g of corn steep solid (spray-dried corn steep liquor; Roquette Corp., Gurnee, Ill.), 1.3 g of MgSO4 · 7H2O, 0.7 g of (NH4)2PO4, 0.7 g of KH2PO4, 2 mg of CoCl2 · 6H2O, 2 drops of soybean oil, and 1 drop of P2000 (polypropylene glycol; Fluka) in 1 liter of tap water, pH 7.0. This seed culture was incubated for 2 days at 30°C and 280 rpm and used to provide a 4% inoculum for 100 ml of production medium consisting of 50 g of glucose, 10 g of soy flour, 5.0 g of NZ amine YTT, 5.0 g of NaCl, 1.0 g of CaCO3, 2 drops of soybean oil, and 1 drop of P2000 in 1 liter of tap water (pH 7.0). This hygromycin A production culture was then incubated in a 500-ml baffled flask for 6 days at 30°C and 280 rpm.
Hygromycin A purification.
The culture filtrate was adsorbed by 1/20 volume of Amberlite XAD-4 (Sigma) at 5°C for 2 h. The column was washed with water and then eluted with methanol. The pooled fractions containing hygromycin A were further purified by preparative high-pressure liquid chromatography with a C18 reverse-phase column (250 by 10.0 mm; Phenomenex, Torrance, Calif.) using 20% acetonitrile-80% water as the eluent (2 ml/min). Hygromycin A was detected by monitoring at 272 nm with a UV detector. The hygromycin A eluate was freeze-dried and analyzed by 1H and 13C nuclear magnetic resonance (NMR) analyses (using previously published 13C assignments [33]). As previously observed (10, 33), the hygromycin A obtained from the fermentations typically contained a minor C-4" epihygromycin (Fig. 1) for which only the 13C resonances associated with the furanoside moiety were different.
Incorporation of isotope-labeled precursors into hygromycin A.
The labeled compounds d-[1,2-13C2]glucose, d-[1-13C]mannose, l-[methyl-13C]methionine, sodium [2,3-13C2]propionate, and [3-13C]tyrosine were all obtained from Cambridge Isotopes Laboratories, Inc. [3-13C]phenylalanine was obtained from CDN Isotopes Inc. [carboxy-13C]4-hydroxybenzoic acid and [carboxy-13C]benzoic acid were obtained from Isotec, Inc. All compounds were 99% labeled and separately pulse fed in four equal batches to a 100-ml culture of a hygromycin A producing culture of S. hygroscopicus at 3, 3.5, 4, and 4.5 days. The final concentration of the labeled materials added was 0.6 mg/ml of the production medium. After a total of 6 days of fermentation, hygromycin A was purified as described above. Approximately 25 to 50 mg of purified hygromycin A was obtained from each separate incorporation experiment. The 13C NMR spectrum of purified labeled hygromycin A in CD3OD was recorded on a 300 MHz NMR spectrometer (Varian Inc., Palo Alto, Calif.) and compared with standard hygromycin A provided by Pfizer and analyzed under the same conditions.
Inhibition of hygromycin A biosynthesis.
N-(Phosphonomethyl)-glycine was added to the production medium at 24 h of fermentation at a final concentration of 0.5 mg/ml. In another experiment, same concentration of N-(phosphonomethyl)-glycine and 0.25 mg of 4-hydroxybenzoic acid (Aldrich) per ml were added to the production medium at 24 h of fermentation. A control experiment was carried out at the same time. All fermentations were carried out in duplicate, and a time course of hygromycin A was determined by using a high-pressure liquid chromatography assay.
13C NMR analyses.
A region selective sensitivity enhanced heteronuclear single quantum coherence (HSQC) experiment (30) was performed with the hygromycin A sample from the d-[1,2-13C2]glucose incorporation experiment. This experiment allows observation of 13C-13C scalar coupling in the indirectly detected dimension and offers significant sensitivity advantages over 13C-13C INADEQUATE experiments and direct observation of 13C scalar couplings in conventional one-dimensional 13C experiments. The data set was obtained on a Varian Unity+ 500 MHz spectrometer equipped with a quadruple resonance probe and consisted of 1,024 complex t1 increments. Each t1 increment consisted of 1,024 complex points averaged for 16 transients with a 1.7-s repetition rate. Data were processed for phase-sensitive display with FELIX v97.2 (Accelrys, San Diego, Calif.) using cosine bell weighting functions in both dimensions.
RESULTS AND DISCUSSION
Hygromycin A production time course.
In order to determine the best time to add 13C-labeled biosynthetic precursors, we established a time course for hygromycin A production under our laboratory fermentation conditions (data not shown). This analysis showed that during the exponential growth phase (0 to 48 h), a low level of hygromycin A was produced. Hygromycin A production continued during the stationary phase and reached a maximal level of approximately 700 μg/ml by day 6. Based on these results, we chose to add biosynthetic precursors in a batchwise process between 72 and 108 h, during the period when the rate of hygromycin production was maximal.
Biosynthesis of the 6-deoxy-5-keto-d-arabino-hexofuranose moiety.
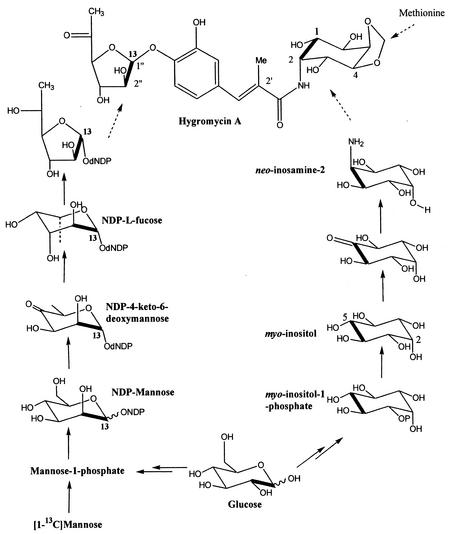
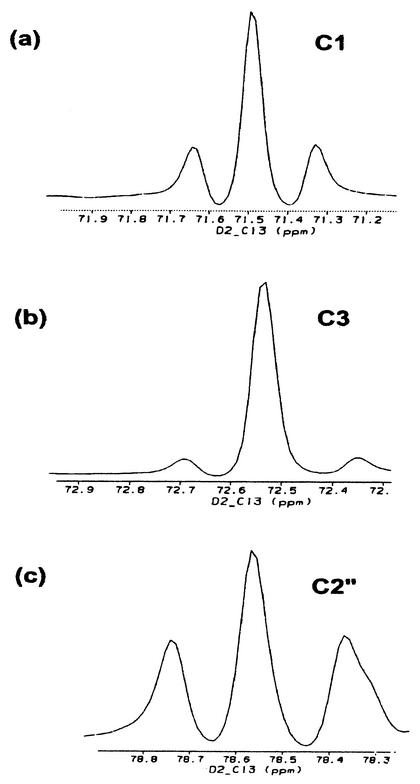
The stereochemical configuration at C-2"of hygromycin A indicates that mannose rather than glucose is the more immediate precursor of the 6-deoxy-5-keto-d-arabino-hexofuranose-derived moiety (Fig. 2). To test this hypothesis, [1-13C]mannose was fed to producing cultures. The resulting hygromycin A (34 mg) was shown by NMR analyses to be specifically enriched (threefold) at C-1" (103.7 ppm). In this experiment we were unable to detect any labeling of the aminocyclitol moiety of hygromycin A from mannose. As the aminocyclitol is efficiently labeled from d-[1,2-13C2]glucose (see below), this result indicates that under the conditions of this experiment the carbon flux is from glucose towards mannose. Consistent with this hypothesis was the efficient intact labeling of the 6-deoxy-5-keto-d-arabino-hexofuranose-derived moiety from glucose. In our incorporation study with d-[1,2-13C2]glucose, analyses of cross-peak volumes in the HSQC revealed a clear and unambiguous 1% intact labeling of hygromycin A (40 mg) (J = 50 Hz) at C-1" (103.7 ppm) and C-2" (78.6 ppm) (Fig. 3c). This labeling study also clearly demonstrated that the published assignments (33) for C-2" (78.2 ppm) and C-5 (78.6 ppm) should be reversed. Intact labeling was also observed for the C-1" and C-2" signals associated with epihygromycin A. No enrichments of other positions in the furanose moiety were observed.
FIG. 2.
Proposed pathway for production of the 6-deoxy-5-keto-d-arabino-hexofuranose and aminocyclitol moieties of hygromycin A from glucose. The bold bonds indicate intact labeling of hygromycin A from d-[1,2-13C2]glucose (low-level intact labeling of C-3 and C-4 of hygromycin was also observed). The “13” indicates labeling of the furanose ring of hygromycin A by [1-13C]mannose.
FIG. 3.
13C NMR analyses revealing efficient intact labeling of C-1-C-2 (A) and C-1"-C-2" (C) and lower intact labeling of C-3-C-4 (B) of hygromycin A from d-[1,2-13C2]glucose. Spectra are one-dimensional 13C slices from the HSQC data set for the hygromycin A sample from the d-[1,2-13C2]glucose labeling experiment.
These results are consistent with a pathway in which d-glucose-6-phosphate is converted via d-fructose-6 phosphate to d-mannose-1-phosphate (Fig. 2). In subsequent steps, this intermediate is likely converted to a nucleoside diphosphate (NDP)-activated l-fucose. l-Fucose is found in glycoconjugates of plants, animals, and bacteria (26, 31), including streptomycetes (34), and plays a role in maintaining structural integrity and molecular recognition. This pathway would involve elimination of water to generate an NDP-4-keto-6-deoxy-d-mannose, a step catalyzed by an enzyme such as GDP-d-mannose 4,6-dehydratase. This intermediate would then be converted to GDP-l-fucose by GDP-l-fucose synthetase, which catalyzes epimerization at the C-3 and C-5 positions of the hexose ring, and an NADPH-dependent reduction at the C-4 position (11, 26). It is unclear whether any of the genes required for this formation of the proposed NDP-activated l-fucose are located within the hygromycin A biosynthetic gene cluster. It is worth noting that genes encoding enzymes with homology to GDP-d-mannose 4,6-dehydratase have recently been identified in the nystatin, candicidin, and antibiotic A201A biosynthetic gene clusters (3, 5, 24). A mutation of the NDP-l-fucose (a pyranose) to the furanose and oxidation of the 5-keto group would provide the activated 6-deoxy-5-keto-d-arabino-hexofuranose for attachment to the C-4 hydroxyl of the 3,4-dihydroxy-α-methylcinnamic acid-derived moiety of hygromycin A. The mutase responsible for the ring contraction of GDP-l-fucose likely follows the mechanism recently established for the primary metabolic UDP-galactopyranomutase (23, 35). To the best of our knowledge, no rearrangements of this type have been reported previously for antibiotic-biosynthetic pathways. In this regard we note that antibiotic A201A, which has significant structural similarities to hygromycin A, contains a moiety that is likely derived from 5-keto-d-arabino-hexofuranose (Fig. 1). Biosynthetic experiments have not been reported for antibiotic A201A, and a partial sequence of the biosynthetic gene cluster (24) has not yet identified genes involved in formation of the furanose moiety.
Biosynthesis of the aminocyclitol moiety.
It has been proposed that the unusual neo-inosamine-2 of hygromycin A is presumably formed via a pathway which involves myo-inositol as an intermediate (18). This pathway has been studied for the scyllo-inosamine-derived moiety of streptomycin (1). In the latter case, oxidation of the C-2 hydroxyl of myo-inositol (derived from C-5 of glucose) and subsequent transamination (catalyzed by l-glutamine:scyllo-inosose amino transferase) (1) yields scyllo-inosamine. A similar process occurring at C-5 of myo-inositol (derived from C-2 of glucose) would be predicted to generate neo-inosamine-2 (Fig. 2). To test this possibility, we carried out an incorporation study with d-[1,2-13C2]glucose. Consistent with our predictions, the HSQC analysis of the hygromycin A obtained from this study demonstrated clear and unequivocal intact labeling at C-1 (71.6 ppm) (Fig. 3A) and C-2 (50.7 ppm) (J = 39.5 Hz) (0.7%). There was no detectable labeling at C-5 or C-6, consistent with divergent pathways leading from myo-inositol to neo-inosamine-2 and scyllo-inosamine. However, a low level of intact labeling (J = 44.8 Hz) (0.2%) at C-3 (72.6 ppm) (Fig. 3B) and C-4 (78.2 ppm) of hygromycin A was observed. This observation suggests that an alternative process or pathway must contribute, albeit to a lesser degree, to formation of neo-inosamine-2 from glucose.
The C-4 and C-5 hydroxyl groups in hygromycin A are bridged by a methylene group. We hypothesized that S-adenosylmethionine was the source of this methylene group and carried out an incorporation study with l-[methyl-13C]methionine. Consistent with our predictions, the 13C NMR analysis of the resulting hygromycin A (50 mg) exhibited a single 25-fold enrichment at 96.2 ppm, the resonance assigned to the methylene bridge. It remains to be determined at which stage during the biosynthetic process the methylene group is introduced onto the aminocyclitol ring. The high level of enrichment of hygromycin A by methionine in this experiment suggested that under our fermentation conditions methionine may be a limiting factor in fermentation titers. While a comprehensive fermentation study is needed to test this hypothesis, it is interesting that the highest level of hygromycin A in this series of studies was obtained in the methionine feeding study.
Biosynthesis of the 3,4-dihydroxy-α-methylcinnamic acid moiety.
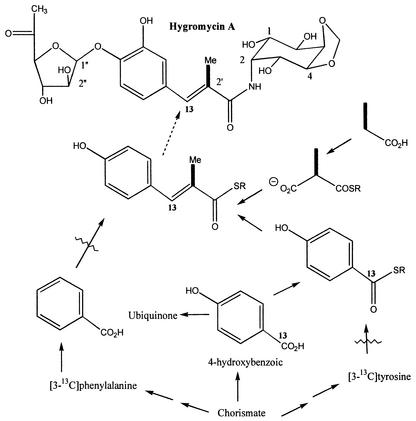
Hygromycin A, antibiotic A201A, and puromycin are all inhibitors of protein synthesis (albeit with specific differences in their mechanisms of action) and have been observed to have some similarities in structure (Fig. 1) (10). In particular, the 3,4-dihydroxy-α-methylcinnamic acid moiety of hygromycin A and the 4-hydroxy-α-methylcinnamic acid of antibiotic A201A are similar to the tyrosine-derived moiety of puromycin A (28). Despite these similarities, it is clear that a more complex process must provide the moieties of hygromycin A and antibiotic A201A. No incorporation at the C-2′ CH3 residue of hygromycin A was observed in the methionine incorporation experiment, ruling out an S-adenosylmethionine-dependent methylation process. Thus, we hypothesized that in the case of hygromycin A and antibiotic A201A, these moieties are derived from a 4-hydroxybenzoyl coenzyme A (CoA) or 3,4-dihydroxybenzoyl-CoA starter unit which is elongated by a methylmalonyl-CoA-derived extender unit (Fig. 4). Support for this hypothesis was obtained in an incorporation experiment with [2,3-13C2]propionate, where 13C NMR analysis of the resulting hygromycin A (31 mg) revealed a specific 1% intact incorporation (J = 44.1 Hz) of the two 13C labels at the predicted sites, C-2′ (132.2 ppm) and the C-2′ CH3 (14.6 ppm).
FIG. 4.
Proposed pathway for the production of the 3,4-dihydroxy-α-methylcinnamic acid moiety of hygromycin A. The bold bonds indicate intact labeling of hygromycin A from [2,3-13C2]propionate. The “13” indicates labeling of the hygromycin A by [carboxy-13C]4-hydroxybenzoic acid. Wavy lines indicate no detectable incorporation of 13C label from phenylalanine, tyrosine, and benzoic acid. SR is a CoA or acyl carrier protein thioester.
We hypothesized that the remainder of the 3,4-dihydroxy-α-methylcinnamic acid moiety may be derived from degradation of phenylalanine or tyrosine. It has recently been established that the benzoyl-CoA starter unit for the biosynthesis of the polyketide enterocin in Streptomyces maritimus is derived from β-oxidation of trans-cinnamic acid (derived from phenylalanine by a phenyl ammonia lyase) (17, 32). Certain bacteria can form benzoyl-CoA via successive rounds of α-oxidative decarboxylations of phenylpyruvate (formed by transamination of phenylalanine) (25). Recent evidence has also shown that the 3-dimethylallyl-4-hydroxybenzoic acid moiety of clorobiocin is formed from phenylalanine in a process that involves a prenylation and a retro-aldol reaction (20). We carried out two separate incorporation experiments with [3-13C]tyrosine and [3-13C]phenylalanine and obtained 31 and 20 mg of hygromycin A, respectively. In neither case was there any detectable level of 13C enrichment at the predicted C-3′ (135.0 ppm) of hygromycin A. However, we did observe a strong specific sixfold enrichment at this carbon when we analyzed hygromycin A (25 mg) obtained from an incorporation study using [carboxy-13C]-4-hydroxybenzoic acid. When this experiment was repeated with [carboxy-13C]benzoic acid, analysis of the hygromycin A (15 mg) by 13C NMR revealed no enrichment at C-3′. These analyses suggest that it is 4-hydroxybenzoyl-CoA which is coupled with a methylmalonyl-CoA to generate the 3,4-dihydroxy-α-methylcinnamic acid moiety of hygromycin A. The addition of the 3-hydroxyl group may occur prior to or after this elongation step. The data also suggest that neither hydroxylation of benzoyl-CoA nor degradation of tyrosine or phenylalanine plays a significant role in the formation of 4-hydroxybenzoyl-CoA. In bacteria, 4-hydroxybenzoic acid is derived from the shikimic acid pathway intermediate chorismic acid by the action of chorismate lyase (2, 21). This 4-hydroxybenzoic acid is then converted to ubiquinone by a series of steps, including decarboxylation and prenylation. Activation of this ubiquinone biosynthetic pathway intermediate, 4-hydroxybenzoic acid, to the appropriate thioester could provide the appropriate precursor for hygromycin A biosynthesis (Fig. 4).
The formation of the 3,4-dihydroxy-α-methylcinnamic acid moiety of hygromycin A most likely is catalyzed by a polyketide synthase. In this process, the 3-keto group of the product, generated by condensation of the methylmalonyl-CoA and either 4-hydroxybenzoyl-CoA or 3,4-dihydroxybenzoyl-CoA, would be processed by reduction and dehydration. Consistent with our prediction is a finding in a recent analysis of the portion of the antibiotic A201A gene cluster of the gene ataPKS1, which encodes a protein with some homology to the acyltransferase domains of modular type I polyketide synthases (24).
Inhibition of hygromycin A biosynthesis by N-(phosphonomethyl)-glycine.
Our incorporation studies indicate that the 4-hydroxybenzoic acid-derived moiety of hygromycin A is derived from the shikimic acid pathway intermediate chorismic acid. To test this hypothesis, we carried out a fermentation in the presence of N-(phosphonomethyl)-glycine, an inhibitor of 5-enol-pyruvylshikimic acid 3-phosphate synthase (27), which catalyzes a key step in chorismic acid biosynthesis. The inhibitor was added to the production medium after 24 h of fermentation, allowing the cells to grow and thus avoiding inhibitory effects on ubiquinone biosynthesis. Under these conditions we saw normal cell growth but 20 and 40% decreases in the titers of hygromycin A for inhibitor concentrations of 20 and 50 mg/100 ml, respectively. Higher inhibitor concentrations did not yield additional decreases in fermentation titers. A complete loss of hygromycin A was not expected in these experiments. Our time course for hygromycin A production indicated that approximately 15% of the overall hygromycin A is generated by 24 h of fermentation (prior to addition of the inhibitor), and there is likely to be some 4-hydroxybenzoic acid available to support additional hygromycin A biosynthesis.
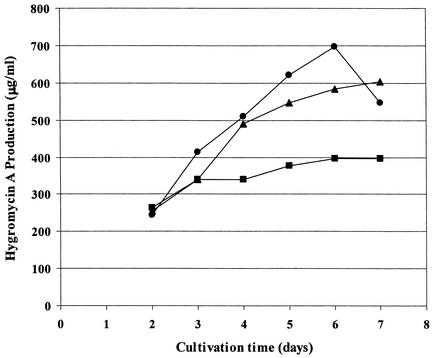
We also carried out a fermentation time course experiment to determine if addition of 4-hydroxybenzoic acid can restore hygromycin A to cultures treated with N-(phosphonomethyl)-glycine. As shown in Fig. 5, production of hygromycin A in the control continued through day 6, whereas in the cultures treated with the inhibitor, production stopped essentially by day 3. Continued production of hygromycin A through day 7, with levels comparable to the control, were observed when cultures were treated with a mixture of the inhibitor and 4-hydroxybenzoic acid. This series of experiments together with the labeling experiments described above are all consistent with the proposed pathway to the 4-hydroxybenzoyl-CoA precursor for hygromycin A biosynthesis (Fig. 4).
FIG. 5.
Inhibition of hygromycin A production by N-(phosphonomethyl)-glycine can be overcome by addition of exogenous 4-hydroxybenzoic acid. Circles, control fermentation; squares, addition of inhibitor at 24 h; triangles, addition of inhibitor and exogenous 4-hydroxybenzoic acid at 24 h. See the text for details.
Summary.
This study has clearly established the biosynthetic origins of hygromycin A from mannose, glucose, 4-hydroxybenzoic acid, propionate, and methionine. The incorporation results indicate that there are unique features for each of the pathways providing the three moieties of hygromycin A. These moieties are likely assembled separately and then joined by using an amide synthase and glycosyl transferase. Such a convergent biosynthetic route offers tremendous potential for structural modification via techniques of mutasynthesis and combinatorial biosynthesis. The first step in this process, the identification, cloning, and sequencing of the hygromycin A gene cluster, is under way in our laboratory, using appropriate probes designed based on knowledge gained through the biosynthetic studies described herein.
Acknowledgments
We thank Hamish McArthur and Maria Brown (Pfizer Incorporated, Central Research Division) for providing fiscal support.
We thank Hamish McArthur and Maria Brown for details on growth of S. hygroscopicus (NRRL 2388) and isolation and purification of hygromycin A. We also thank Ben Liu (University of Texas, Austin) for helpful discussions on the biosynthetic origins of the 6-deoxy-5-keto-d-arabino-hexofuranose moiety of hygromycin A.
REFERENCES
- 1.Ahlert, J., J. Distler, K. Mansouri, and W. Piepersberg. 1997. Identification of stsC, the gene encoding the l-glutamine:scyllo-inosose aminotransferase from streptomycin-producing Streptomycetes. Arch. Microbiol. 168:102-113. [DOI] [PubMed] [Google Scholar]
- 2.Barker, J. L., and J. W. Frost. 2001. Microbial synthesis of p-hydroxybenzoic acid from glucose. Biotechnol. Bioeng. 76:376-390. [DOI] [PubMed] [Google Scholar]
- 3.Brautaset, T., O. N. Sekurova, H. Sletta, T. E. Ellingsen, A. R. Strom, S. Valla, and S. B. Zotchev. 2000. Biosynthesis of the polyene antifungal antibiotic nystatin in Streptomyces noursei ATCC 11455: analysis of the gene cluster and deduction of the biosynthetic pathway. Chem. Biol. 7:395-403. [DOI] [PubMed] [Google Scholar]
- 4.Brodersen, D. E., W. M. Clemons, Jr., A. P. Carter, R. J. Morgan-Warren, B. T. Wimberly, and V. Ramakrishnan. 2000. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103:1143-1154. [DOI] [PubMed] [Google Scholar]
- 5.Campelo, A. B., and J. A. Gil. 2002. The candicidin gene cluster from Streptomyces griseus IMRU 3570. Microbiology 148:51-59. [DOI] [PubMed] [Google Scholar]
- 6.Ganoza, M. C., and M. C. Kiel. 2001. A ribosomal ATPase is a target for hygromycin B inhibition on Escherichia coli ribosomes. Antimicrob. Agents Chemother. 45:2813-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guerrero, M. D., and J. Modolell. 1980. Hygromycin A, a novel inhibitor of ribosomal peptidyltransferase. Eur. J. Biochem. 107:409-414. [DOI] [PubMed] [Google Scholar]
- 8.Hansen, J. L., J. A. Ippolito, N. Ban, P. Nissen, P. B. Moore, and T. A. Steitz. 2002. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol. Cell 10:117-128. [DOI] [PubMed] [Google Scholar]
- 9.Hayashi, S. F., L. J. Norcia, S. B. Seibel, and A. M. Silvia. 1997. Structure-activity relationships of hygromycin A and its analogs: protein synthesis inhibition activity in a cell free system. J. Antibiot. (Tokyo) 50:514-521. [DOI] [PubMed] [Google Scholar]
- 10.Hecker, S. J., M. L. Minich, and K. W. Werner. 1992. Semisynthetic modification of hygromycin A. 1. Synthesis and antibacterial activity of vinyl methyl and amide analogs. Bioorg. Med. Chem. Lett. 2:533-536. [Google Scholar]
- 11.Jarvinen, N., M. Maki, J. Rabina, C. Roos, P. Mattila, and R. Renkonen. 2001. Cloning and expression of Helicobacter pylori GDP-l-fucose synthesizing enzymes (GMD and GMER) in Saccharomyces cerevisiae. Eur. J. Biochem. 268:6458-6464. [DOI] [PubMed] [Google Scholar]
- 12.Kiel, M. C., and M. C. Ganoza. 2001. Functional interactions of an Escherichia coli ribosomal ATPase. Eur. J. Biochem. 268:278-286. [DOI] [PubMed] [Google Scholar]
- 13.Mann, R. L., and W. W. Bromer. 1958. The isolation of a second antibiotic from Streptomyces hygroscopicus. J. Am. Chem. Soc. 80:2714-2716. [Google Scholar]
- 14.Mann, R. L., R. M. Gale, and R. F. Van Abeele. 1953. Hygromycin. II. Isolation and properties. Antibiot. Chemother. 3:1279-1282. [PubMed] [Google Scholar]
- 15.Nakagawa, A., T. Fujimoto, S. Omura, J. C. Walsh, R. L. Stotish, and B. George. 1987. Hygromycin A, an antitreponemal substance. II. Therapeutic effect for swine dysentery. J. Antibiot. (Tokyo) 40:1627-1635. [DOI] [PubMed] [Google Scholar]
- 16.Omura, S., A. Nakagawa, T. Fujimoto, K. Saito, K. Otoguro, and J. C. Walsh. 1987. Hygromycin A, an antitreponemal substance. I. Screening method and therapeutic effect for Treponema hyodysenteriae-caused infection in CF-1 mice. J. Antibiot. (Tokyo) 40:1619-1626. [DOI] [PubMed] [Google Scholar]
- 17.Piel, J., C. Hertweck, P. R. Shipley, D. M. Hunt, M. S. Newman, and B. S. Moore. 2000. Cloning, sequencing and analysis of the enterocin biosynthesis gene cluster from the marine isolate Streptomyces maritimus: evidence for the derailment of an aromatic polyketide synthase. Chem. Biol. 7:943-955. [DOI] [PubMed] [Google Scholar]
- 18.Piepersberg, W. 1997. Molecular biology, biochemistry and fermentation of aminoglycoside antibiotics, p. 81-164. In W. E. Strohl (ed.), Biotechnology of antibiotics. Marcel Dekker, New York, N.Y.
- 19.Pittenger, R. C., R. N. Wolfe, P. N. Hoehn, W. A. Daily, and J. M. McGuire. 1953. Hygromycin. I. Preliminary studies in the production and biologic activity on a new antibiotic. Antibiot. Chemother. 3:1268-1278. [PubMed] [Google Scholar]
- 20.Pojer, F., S. M. Li, and L. Heide. 2002. Molecular cloning and sequence analysis of the clorobiocin biosynthetic gene cluster: new insights into the biosynthesis of aminocoumarin antibiotics. Microbiology 148:3901-3911. [DOI] [PubMed] [Google Scholar]
- 21.Poon, W. W., D. E. Davis, H. T. Ha, T. Jonassen, P. N. Rather, and C. F. Clarke. 2000. Identification of Escherichia coli ubiB, a gene required for the first monooxygenase step in ubiquinone biosynthesis. J. Bacteriol. 182:5139-5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poulsen, S. M., C. Kofoed, and B. Vester. 2000. Inhibition of the ribosomal peptidyl transferase reaction by the mycarose moiety of the antibiotics carbomycin, spiramycin and tylosin. J. Mol. Biol. 304:471-481. [DOI] [PubMed] [Google Scholar]
- 23.Sanders, D. A., A. G. Staines, S. A. McMahon, M. R. McNeil, C. Whitfield, and J. H. Naismith. 2001. UDP-galactopyranose mutase has a novel structure and mechanism. Nat. Struct. Biol. 8:858-863. [DOI] [PubMed] [Google Scholar]
- 24.Saugar, I., E. Sanz, M. Rubio, A., J. C. Espinose, and A. Jimenez. 2002. Identification of a set of genes involved in the biosynthesis of the aminonucleoside moiety of antibiotic A201A from Streptomyces capreolus. Eur. J. Biochem. 269:5527-5535. [DOI] [PubMed] [Google Scholar]
- 25.Schneider, S., M. E. Mohamed, and G. Fuchs. 1997. Anaerobic metabolism of l-phenylalanine via benzoyl CoA in the denitrifying bacterium Thauera aromatica. Arch. Microbiol. 168:310-320. [DOI] [PubMed] [Google Scholar]
- 26.Somers, W. S., M. L. Stahl, and F. X. Sullivan. 1998. GDP-fucose synthetase from Escherichia coli: structure of a unique member of the short-chain dehydrogenase/reductase family that catalyzes two distinct reactions at the same active site. Structure 6:1601-1612. [DOI] [PubMed] [Google Scholar]
- 27.Stalker, D. M., W. R. Hiatt, and L. Comai. 1985. A single amino acid substitution in the enzyme 5-enolpyruvylshikimate-3-phosphate synthase confers resistance to the herbicide glyphosate. J. Biol. Chem. 260:4724-4728. [PubMed] [Google Scholar]
- 28.Tercero, J. A., J. C. Espinosa, R. A. Lacalle, and A. Jimenez. 1996. The biosynthetic pathway of the aminonucleoside antibiotic puromycin, as deduced from the molecular analysis of the pur cluster of Streptomyces alboniger. J. Biol. Chem. 271:1579-1590. [DOI] [PubMed] [Google Scholar]
- 29.Uyeda, M., M. Mizukami, K. Yokomizo, and K. Suzuki. 2001. Pentalenolactone I and hygromycin A, immunosuppressants produced by Streptomyces filipinensis and Streptomyces hygroscopicus. Biosci. Biotechnol. Biochem. 65:1252-1254. [DOI] [PubMed] [Google Scholar]
- 30.Willker, W., U. Flögel, and D. Liebfritz. 1997. Ultra-high resolved HSQC spectra of multiple-13C-labeled biofluids. J. Magn. Reson. 125:216-219. [DOI] [PubMed] [Google Scholar]
- 31.Wu, B., Y. Zhang, and P. G. Wang. 2001. Identification and characterization of GDP-d-mannose 4,6-dehydratase and GDP-l-fucose synthetase in a GDP-l-fucose biosynthetic gene cluster from Helicobacter pylori. Biochem. Biophys. Res. Commun. 285:364-371. [DOI] [PubMed] [Google Scholar]
- 32.Xiang, L., and B. S. Moore. 2002. Inactivation, complementation, and heterologous expression of encP, a novel bacterial phenylalanine ammonia-lyase gene. J. Biol. Chem. 277:32505-32509. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida, M., E. Takahashi, T. Uozumi, and T. Beppu. 1986. Hygromycin A and methoxyhygromycin A, novel inhibitors of K88 antigen synthesis of enterotoxic Escherichia coli strain. Agric. Biol. Chem. 50:143-149. [Google Scholar]
- 34.Zaretskaia, M. S. H., M. V. Nefelova, L. A. Barativa, and A. N. Polin. 1984. Composition of cell walls of 2 mutant strains of Streptomyces chrysomallus. Antibiotiki 29:902-906. [PubMed] [Google Scholar]
- 35.Zhang, Q., and H. Liu. 2001. Mechanistic investigation of UDP-galactopyranose mutase from Escherichia coli using 2- and 3-fluorinated UDP-galactofuranose as probes. J. Am. Chem. Soc. 123:6756-6766. [DOI] [PubMed] [Google Scholar]