Abstract
A monoclonal anti-rhodopsin antibody (B6–30N), characterized by Hargrave and coworkers [Adamus, G., Zam, Z. S., Arendt, A., Palczewski, K., McDowell, J. M. & Hargrave, P. (1991) Vision Res. 31, 17–31] as recognizing a short peptide sequence at the N terminus, failed to bind to rhodopsin when the latter was solubilized in dodecylmaltoside (DM). Of the detergents tested thus far, DM affords maximum stability to rhodopsin. Solubilization of rhodopsin in cholate allowed binding of the antibody, but the binding caused destabilization as evidenced by the accelerated loss of absorbance at 500 nm. The result provides support for the earlier conclusion that the N-terminal segment is an integral part of a tertiary structure in the intradiscal domain of native rhodopsin coupled to a tertiary structure in the transmembrane domain. Additional comparative studies on the stability of rhodopsin in different detergents were carried out after direct solubilization from rod outer segments and after extensive treatments to remove the endogenous phospholipids. Purification of rhodopsin in DM resulted in essentially quantitative removal of endogenous phospholipids. When rhodopsin thus purified was treated with the above antibody in DM and in cholate, enhanced destabilization (5-fold) was observed in the latter detergent.
Keywords: 11-cis retinal, transmembrane domain, retinitis pigmentosa, cholate, dodecylmaltoside
Structure function studies on rhodopsin require purification of mutants as expressed in mammalian cell lines. The anti-rhodopsin mAb rho1D4 with the C-terminal octapeptide sequence as the epitope (Fig. 1; ref. 1) has been useful in single-step purification of all the mutants in which the epitope has been left intact. Many of our current studies of rhodopsin require amino acid replacements at the extreme C terminus or even truncation of rhodopsin with removal of parts of the C-terminal segment (Fig. 1). For purification of such mutants, a complementary approach with an antibody recognizing a peptide sequence at the N terminus is desirable. A mAb (B6–30N) recognizing the N-terminal sequence G3–S14 (Fig. 1) has indeed been characterized by Hargrave and coworkers (2), and the antibody has proved useful in identification of rhodopsin fragments containing the N terminus (3, 4). When, in the present study, rhodopsin solubilized in dodecylmaltoside (DM) was tested, the antibody failed to bind. Solubilization of rhodopsin in deoxycholate allowed binding of the antibody, but the binding resulted in destabilization of rhodopsin. This result provided support for the previous important conclusion that a tertiary structure is present in the intradiscal domain of which the N-terminal segment forms an integral part. Additional comparative studies have been carried out on the stability of rhodopsin in different detergents; (i) after direct solubilization of rhodopsin from rod outer segments (ROS) and (ii) after extensive treatments to remove the endogenous phospholipids (PLs). Immunoaffinity purification of rhodopsin in DM resulted in practically quantitative removal of the endogenous PLs. The stability of purified rhodopsin in DM and in cholate was compared in the presence and absence of the B6–30N antibody. Again, enhanced destabilization was observed specifically in cholate.†
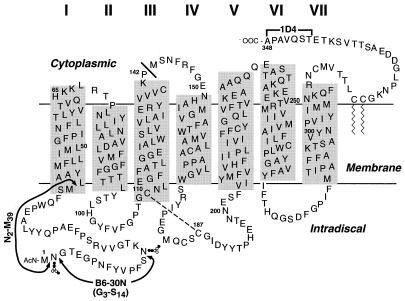
Figure 1.
The location of the epitopes of monoclonal anti-rhodopsin antibodies 1D4 and B6–30N on a secondary structure model of bovine rhodopsin. The lengths of the transmembrane (TM) helices are as deduced by Schertler and coworkers (21) from electron diffraction studies. The solid bar next to P142 shows the end of the N-terminal fragment, noncovalently linked to the remainder of the opsin sequence (10). The N-terminal fragment N2–M39 is also shown. The dashed line shows the conserved disulfide bond between C110 and C187.
Materials and Methods
Materials.
Frozen bovine retinae were from J. A. Lawson (Lincoln, NE). Cell culture media and supplements were from Irvine Scientific and Sigma. Radioactive [32P]Pi, [35S]-l-methionine, and [3H]acetic anhydride were from DuPont/NEN. DM was from Anatrace (Maumee, OH). Sodium cholate and octyl-β-d-glucoside (OG) were from Roche Molecular Biochemicals, and 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) and cetyltrimethylammonium bromide (CTAB) were from Sigma. Dodecyldimethylaminoxide (DDAO) was from Fluka, and dodecyltrimethylammonium bromide (DTAB) was from Aldrich Chem (Metuchen, NJ). PLs were from Avanti Polar Lipids. CNBr-activated Sepharose was from Amersham Pharmacia. Peptides corresponding to the C terminus (T340–A348) and N terminus (G3–S14) of rhodopsin were synthesized by the Biopolymers Laboratory (Massachusetts Institute of Technology) and purified by HPLC. The N2–M39 polypeptide (Fig. 1) was produced by CNBr digestion of rhodopsin and purified by using a Vydac C18 HPLC column (Hesperia, CA; ref. 5). The nonapeptide T340–A348 was used to elute rhodopsin from rho-1D4-Sepharose beads at a concentration of 100 μM throughout. Buffers used were as follows: buffer A, 20 mM 1,3-bis[tris(hydroxymethyl)methylamino]propane (BTP)/140 mM NaCl, pH 7.0; buffer B, buffer A containing 300 mM NaCl; buffer C, 1.5 mM KH2PO4/8 mM Na2HPO4/137 mM NaCl/2.7 mM KCl, pH 7.4; buffer D, 5 mM Hepes/140 mM NaCl, pH 7.5; buffer E, 20 mM BTP/140 mM NaCl, pH 6.0; buffer F, 10 mM BTP/150 mM NaCl/0.2% SDS/2 mM DTT pH 6.0; buffer G, 0.1 M triethylamine bicarbonate, pH 7.5; buffer H, 2 mM sodium phosphate, pH 7.0.
Antibodies and Their Coupling to Sepharose Beads.
Anti-rhodopsin mAb rho1D4 (2) was prepared by the National Cell Culture Center (Coonrapids, MN). The hybridoma producing the anti-rhodopsin mAb B6–30N was provided by P. Hargrave (University of Florida, Gainsville, FL). It was grown in DMEM or serum free HB-Gro medium (Irvine Scientific). Cells were removed from stationary phase cultures by centrifugation, and the supernatants were used for mAb purification by using protein A-Sepharose affinity chromatography. The purified mAb B6–30N and the mAb rho1D4 were coupled to CNBr-activated Sepharose beads as described (6), except that 6 and 10 mg of the purified mAb proteins, respectively, were bound per 1 ml of rehydrated beads. The resulting mAb rho1D4-Sepharose beads had capacity for binding of about 1 mg rhodopsin per ml of settled beads.
Solubilization of Rhodopsin in Different Detergents for Stability Studies.
ROS membranes were prepared (7) and urea-stripped (8). Samples of the membranes containing 0.5 nmol rhodopsin were solubilized in 0.5 ml of buffer C containing one of the following detergents at the concentrations indicated: DM (1% wt/vol), cholate (2% wt/vol), taurocholate (2% wt/vol), OG (2% wt/vol), CHAPS (2% wt/vol), DDAO (2% wt/vol), DTAB (2% wt/vol), or CTAB (2% wt/vol). The suspensions were centrifuged (100,000 × g for 30 min), and the supernatants were kept in the dark at 18°C. Decrease in A500 was recorded at time intervals indicated in the figures. UV-visible (UV-vis) absorption spectroscopy was performed with a Lambda 6 spectrophotometer (Perkin–Elmer) equipped with a temperature-regulated cuvette holder with a slit width of 2 nm and a response time of 1 s.
Purification and Delipidation of ROS Rhodopsin in Different Detergents.
Rhodopsin was purified from ROS, transfected COS-1 cells (6), or an HEK293S stable cell line (9) by immunoaffinity chromatography as follows. Rhodopsin (180 μg, as determined by light-dark difference spectroscopy; ref. 9) was solubilized in 8 ml of buffer A containing phenylmethylsulfonyl fluoride (0.5 mM) and one of the following detergents at the concentrations indicated: DM (1% wt/vol), cholate (7% wt/vol), OG (4% wt/vol), or CHAPS (4% wt/vol). After solubilization for 40 min, the clarified (centrifugation) supernatants were mixed with 0.2 ml (settled beads) of rho1D4-Sepharose for 1.5 h. The Sepharose beads were then packed into a minicolumn (7-mm inner diameter) and washed with buffer A containing the same detergents at reduced concentrations (0.1% for DM and 2% wt/vol for other detergents) by using 40 bed volumes (total 8.0 ml). Rhodopsin was eluted with the same buffered detergent solutions containing the C-terminal nonapeptide. In one experiment, rhodopsin solubilized with DM was bound to rho1D4-Sepharose and washed with 0.1% DM (40 bed volumes). The column was then washed further with buffer A containing 2% (wt/vol) cholate (40 bed volumes) before elution in the same buffer. The A500 remaining at different time points is expressed as the percentage of absorbance present at the start of the experiment. In other experiments (see Fig. 6), purified anti-rhodopsin mAb B6–30N was added to the detergent-washed rhodopsin samples at a concentration of 0.7 μM.
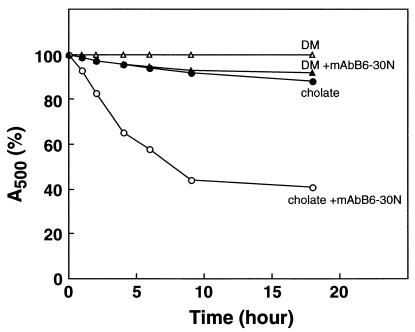
Figure 6.
Stability of purified rhodopsin in detergents in the absence and presence of mAb B6–30N. For details, see text.
Preparation of Rhodopsin Containing 32P-Labeled PLs.
The HEK293S cell line expressing wild-type opsin was used in this study (9). Cell cultures (500 ml) were inoculated and grown as described except that DMEM containing 10% (vol/vol) FBS supplemented with penicillin (100 units/ml), streptomycin (100 units/ml), and glutamine (0.29 mg/ml) was used. After 6 days growth, the cells were collected by centrifugation (4,000 × g for 10 min at 4°C) and resuspended gently in 300 ml of phosphate-free DMEM supplemented as before but with 5% (vol/vol) FBS that had been dialyzed against buffer D by using a 1-kDa cutoff membrane. The cell suspension was returned to a spinner flask, treated with 2.5 mCi of [32P]Pi (1 Ci = 37 GBq), and incubated for a further 24 h. Cells were harvested, treated with 11-cis retinal to constitute rhodopsin, and stored at −70°C as described (9). Cell pellets from 37.5 ml of culture each were solubilized (at 4°C for 3 h) in 4 ml of buffer A containing one of the following detergents at the concentrations indicated: DM (1% wt/vol), cholate (7% wt/vol), OG (4% wt/vol), or CHAPS (4% wt/vol). After centrifugation (6,000 × g for 15 min), the postnuclear fraction was clarified further by ultracentrifugation (120,000 × g for 15 min). The samples were applied by gravity flow to 200 μl of settled rho1D4-Sepharose beads packed in columns (8-mm internal diameter) equipped with upper and lower frits (Pierce). The flow through was reapplied to the column, and then the columns were washed as follows: wash 1, 30 ml of buffer B containing the corresponding detergent but at the concentrations indicated [DM (0.1%), cholate (2% wt/vol), OG (2% wt/vol), or CHAPS (2% wt/vol)] and wash 2, 10 ml of buffer A containing the same detergents at the same concentrations. Elution was performed with buffer A containing each detergent and the C-terminal nonapeptide. All samples were analyzed by UV-vis spectroscopy and direct Cerenkov counting for radioactivity. The specific activity of 32P-labeled phosphatidylcholine (PC, 9,030 cpm/nmol PC) was determined by phosphorous analysis after purification of the PL.
Silica Gel TLC Analysis of Residual PLs Associated with Purified Rhodopsin.
Purified rhodopsin samples and whole-cell PLs extracted from [32P]Pi-labeled HEK293S cells were applied directly to 20 × 20-cm glass support TLC plates (Silica gel 60, 2-mm or 0.2-mm thickness, EM Reagents, Gibbstown, NJ). PLs were separated with the solvent system, chloroform (260 ml), methanol (105 ml), and water (20 ml). Radioactive spots were identified by PhosphorImager analysis (Molecular Dynamics) and autoradiography. Nonradioactive control PLs were identified by charring with 9 M sulfuric acid.
Preparation of [35S]Met-Labeled Opsin and Rhodopsin.
COS-1 cells (1 × 107 to 2 × 107 cells per 15-cm dish) were transiently transfected with the opsin gene. At 24 h after transfection, the cell culture medium in each cell culture dish was replaced with 15 ml of DMEM deficient for both Met and Cys and supplemented with 10% (vol/vol) FBS that had been dialyzed against buffer C with a 1-kDa cutoff membrane. The COS-1 cells were then treated with 300 μCi of [35S]Met (20 μCi/ml) and incubated for a further 30 h before harvesting. For in vivo constitution of rhodopsin, COS-1 cells (1 × 107 to 2 × 107 cells per 15-cm dish) were resuspended in 2 ml of buffer C and treated with 11-cis retinal (5 μM) for 3 h at 4°C. COS-1 cells expressing the full-length wild-type gene were used for the preparation of 35S-labeled rhodopsin and 35S-labeled opsin essentially as described above in Silica Gel TLC Analysis of Residual PLs Associated with Purified Rhodopsin by using DM throughout (6).
Preparation of [35S]Met-Labeled N-Terminal Fragment M1–P142.
For the preparation of 35S-labeled fragment M1–P142, transfected and [35S]Met-labeled COS-1 cells coexpressing both gene fragments (corresponding to M1–P142 and M143–A348; refs. 10 and 11) were treated with 11-cis retinal (20 μM) and solubilized with buffer C containing DM (1% wt/vol). This pigment, comprising the two noncovalently linked opsin fragments, was purified with rho1D4-Sepharose essentially as described above. The C-terminal nonapeptide was removed by using Sephadex G-50, and the eluate was mixed with 10 μl of settled rho1D4-Sepharose beads for 20 min in the presence of buffer A containing 0.05% DM. The binding efficiency was >98% in this step. The beads were then washed two times with 20 μl of the same buffer, and the M1–P142 polypeptide was dissociated from the beads by the addition of 100 μl of buffer F for 20 min. The recovery was 91%. The sample was then diluted 2-fold with buffer A, and 20 μl of settled B6–30N beads (6 mg/ml) were added. After 3 h, SDS was exchanged by washing the beads three times with 0.1 ml of buffer A containing either 2% (wt/vol) cholate or 0.05% DM. The 35S-labeled M1–P142 polypeptide was eluted from the beads by using G3–S14 synthetic peptide (450 μM) in the same detergent solution. The elution peptide was removed subsequently by gel filtration with Sephadex G-50.
Preparation of [3H]Acetylated N-Terminal Peptides N2–M39.
The reaction mixture in a glass test tube (10 × 75-mm) on ice contained 0.1 μmol N2–M39 polypeptide and 50 μmol triethylamine in 0.1 ml of acetonitrile. [3H]Acetic anhydride (1 μl; 50 μmol; 10 mCi/mmol) was added in five 0.2-μl aliquots over 20 min. Completeness of reaction was confirmed by fluorescamine assay (12). The mixture was then lyophilized; the residues were dissolved in 80 μl of buffer G; and residual free acetic acid was removed with a Sephadex G-15 column (bed volume; 11 ml) in buffer G. Radioactive acetylated peptides were eluted at 5.5–6.5 ml. The samples were lyophilized and dissolved in 60 μl of buffer H. Samples were relyophilized and dissolved in 60 μl of distilled water.
Kinetic Analysis of Binding of Rhodopsin and Fragments to Anti-Rhodopsin mAb B6–30N.
The radioactively labeled polypeptides [3H]acetyl N2–M39 (10 μM) and [35S]-labeled M1–P142 (1 μM) were each dissolved separately in 40 μl of buffer C containing either DM (0.05%) or cholate (2% wt/vol). To each of the polypeptide solutions was added 20 μl of the antibody B6–30N-Sepharose beads with a total capacity of 800 pmol rhodopsin. At different time intervals, small portions of the matrix were pelleted by centrifugation (8,000 × g for 2 min), and the radioactivity in the supernatants was determined by scintillation counting. The beads were transferred to a Whatman GF/B filter under a vacuum and then washed rapidly five times with 500 μl of buffer C containing each detergent before scintillation counting. For the rhodopsin samples, A500 was also monitored by UV-vis absorbance spectroscopy.
Binding of Rhodopsin to Anti-Rhodopsin B6–30N-Sepharose and Elution.
Purified rhodopsin (80 μg each) in 0.05% DM or 2% (wt/vol) cholate was treated with 200 μl of the anti-rhodopsin B6–30N-Sepharose beads (see above) for 4 h in the dark at 4°C. The suspensions were packed into a column, and the latter was washed with 8 ml of buffer A containing DM or cholate detergent. Elution was carried out with buffer A containing G3–S14 synthetic peptide (450 μM) in the same detergent solution. Elution of rhodopsin was measured by UV-vis absorption spectroscopy.
Pi Determination.
Pi determination was carried out by the method of Ames and Dubin (13) by using a 0.3-ml total volume.
Quantitation of Peptides by Fluorescamine Assay.
The procedure of Lai et al. (12) was used at 0.2-ml scale. Fluorescence emission was monitored at 475 nm on 390-nm excitation by using a PTI spectrofluorometer (Photon Technology International, Princeton).
Results
Binding of Rhodopsin N-Terminal Fragments, Opsin, and Rhodopsin to mAb B6–30N in the Presence of DM and Cholate.
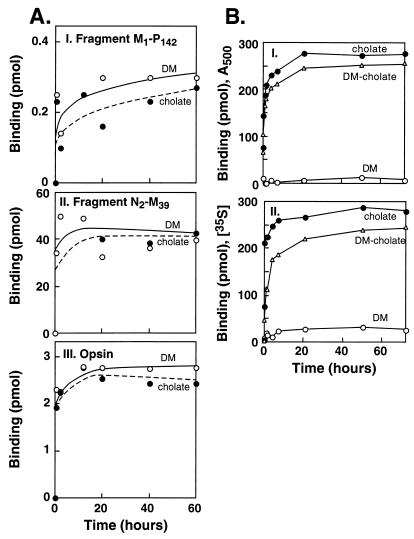
Initial experiments suggested the selection of two detergents, DM and cholate, that vary greatly in critical micellar concentration, for the antibody binding studies. First, in kinetic studies, we were able to confirm the previous conclusion that rhodopsin fragments starting from the N terminus are able to bind to mAb B6–30N. As shown in Fig. 2A, binding of the antibody occurred to both fragments, M1–P142 and N2–M39, (Fig. 2A I and II, respectively). Further, in both cases, binding was rapid, and there was no significant difference in the kinetics of binding in DM and cholate. As seen in Fig. 2A III, rapid binding of the antibody also occurred to opsin in both detergents.
Figure 2.
Kinetics of binding of opsin, N-terminal fragments of rhodopsin, and rhodopsin to the antibody B6–30N in different detergents. (A) Binding of opsin and N-terminal fragments: 35S-labeled M1-P142 (I), 3H-labeled N2-M39 (II), and opsin (III), respectively. (B) Binding of rhodopsin: binding of A500 (I) and binding of 35S-labeled rhodopsin (II). The detergents used were DM (open circles), cholate (closed circles), and a mixture of DM (0.1%) and cholate (2% vol/vol) (triangles).
The binding of intact rhodopsin to the antibody was monitored by recording the loss of A500 and of γ-35S from the medium on binding to the antibody beads (Fig. 2B I and II, respectively). A striking difference was observed between DM and cholate. Rhodopsin did not bind to B6–30N beads in DM; however, the binding of a rhodopsin sample solubilized in cholate to B6–30N beds was essentially complete in about 20 min. Similarly, another rhodopsin sample that was purified first by binding to rho1D4-Sepharose in DM and then eluted in the presence of cholate also showed complete binding to B6–30N beads (Fig. 2B).
Stability of Rhodopsin Solubilized from ROS in Different Detergents and After Purification from Endogenous PLs.
Incubation of rhodopsin in DM with B6–30N-Sepharose resulted in minimal binding to the beads. As a consequence, there was no rhodopsin recovered after elution with the G3–S14 peptide. In contrast, binding of rhodopsin to B6–30N-Sepharose in cholate was quantitative under the same conditions. However, the low recovery (32%) and the poor spectral ratio (3.3–6.0) of rhodopsin eluted from the beads indicated that the bound rhodopsin was destabilized during the procedure.
Solubilization of rhodopsin in different detergents has received a great deal of attention, particularly in the context of stability (14–16). The findings reported above on the binding of the antibody B6–30N in cholate and in DM led to a more comprehensive study of the stability of rhodopsin (i) as directly solubilized from ROS and (ii) after purification to remove extensively the endogenous ROS PLs, because solubilization and stability of membrane proteins in detergents is related, at least in part, to efficacy of removal of PLs associated with the proteins.
Stability after direct solubilization in different detergents.
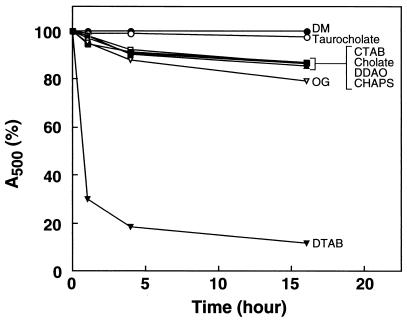
Rhodopsin solubilized in different detergents (see Materials and Methods) was kept at 18°C in the dark, and the decrease in A500 was followed as a function of time. The results are shown in Fig. 3. Rhodopsin was completely stable in DM and taurocholate for 16 h. It was less stable in CTAB, cholate, DDAO, CHAPS, or OG, with decrease of 15–20% over the 16-h period. Rhodopsin solubilized in DTAB was particularly unstable, and only 15% of the chromophore remained after 16 h.
Figure 3.
Kinetics of decrease of A500 in rhodopsin samples solubilized in different detergents. For details, see text.
Stability of rhodopsin in different detergents after purification and removal of PLs.
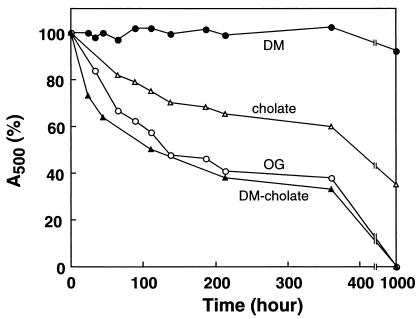
The results shown in Fig. 3, like those obtained previously (15, 16), do not take into account the stabilization effects of PLs that remain bound to rhodopsin or are present in mixed micelles resulting from solubilization from ROS. Therefore, in the next set of experiments, PLs were removed by extensive washing with the detergents of interest after binding solubilized rhodopsin to rho-1D4-Sepharose (see Materials and Methods). The stability of rhodopsin subsequently eluted from the immunoaffinity column again was measured spectrophotometrically, and the results are shown in Fig. 4 and Table 1. Rhodopsin was stable in DM (t1/2 > 6 months) but was less stable in the presence of cholate (t1/2 = 600 h) and OG (t1/2 = 187 h; Table 1). In another experiment, rhodopsin solubilized in DM was bound to rho-1D4-Sepharose, washed further with DM and cholate, and then eluted with the nonapeptide in the presence of cholate. As seen in Fig. 4, the stability of rhodopsin in cholate further decreased with this procedure (t1/2 = 110 h).
Figure 4.
Kinetics of decrease of A500 in rhodopsin samples purified and kept in different detergents. For details, see text.
Table 1.
Stability of ROS rhodopsin after purification in different detergents and quantitation of phospholipids remaining with rhodopsin
| Detergent (concentration, %) | Stability, t1/2 | mol Pi per mol rhodopsin |
|---|---|---|
| DM (0.1) | >6 months | 1.4 |
| Cholate (2) | 600 h | 7.6 |
| OG (2) | 187 h | 4.4 |
| CHAPS (2) | 140 h | 3.6 |
| DM (0.1) | 110 h | 1.4 |
| Cholate (2) |
For details, see text. The amount of PL remaining with rhodopsin was determined by Pi analysis (see Materials and Methods). The amount of rhodopsin was determined spectrophotometrically.
Retention of PLs by Rhodopsin Purified in Different Detergents.
To test whether the stabilities observed in Fig. 4 related to the amount of PL remaining in rhodopsin preparations delipidated by using the detergents, the amounts of residual PL were quantitated. As seen in Table 1, after washes with OG, 4.4 mol Pi per mol rhodopsin remained, but with cholate, 7.6 mol Pi per mol rhodopsin remained. The most effective detergent for PL removal, however, was DM in which 1.4 mol Pi per mol rhodopsin remained (Table 1). Thus, the reduced stability of rhodopsin in cholate after purification in DM is attributed to much lower PL content.
Analysis of PLs Remaining with Rhodopsin After Purification and Extensive Washing with Different Detergents.
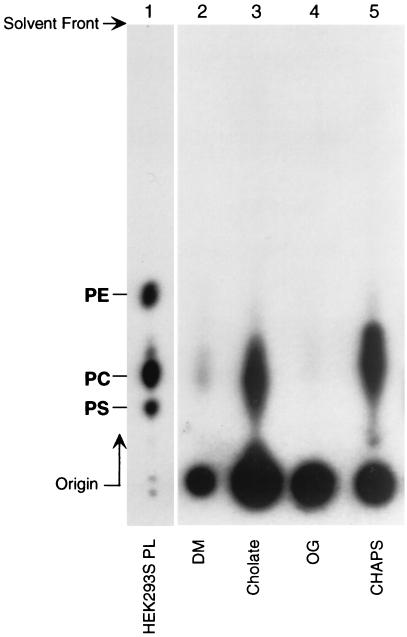
In the next experiments, the efficacy of different detergents to remove PLs from rhodopsin after extensive washing was investigated. An HEK293S cell line expressing the wild-type opsin gene was grown in the presence of [32P]Pi; rhodopsin was constituted with 11-cis retinal and was purified as described in Materials and Methods. The PL remaining with rhodopsin after more extensive washing (200 bed volume; 40 ml) was analyzed by TLC (Fig. 5). The residual PL was thus identified as PC. Determination of radioactivity in the spots corresponding to PC (Fig. 5 and Table 2) showed that rhodopsin was delipidated (>99%) with DM and OG. In contrast, delipidation with cholate or CHAPS was 10 times less efficient. The radioactivity in the spots at origin is concluded to be due to endogenous [32P]phosphorylation of rhodopsin.
Figure 5.
TLC analysis of residual PLs in rhodopsin after purification and extensive washing with different detergents (details in text). Lane 1, total PLs from HEK293S cells; lanes 2–5, residual PL after purification and extensive washings in the detergents shown. Mobilities of control samples of phosphatidylserine (PS), phosphatidylcholine (PC), and phosphatidylethanolamine (PE) are shown. Quantitation of radioactive PC remaining is shown in Table 2.
Table 2.
Characterization of residual 32P-labeled PL remaining with rhodopsin after purification in various detergents
| Detergent (concentration, %) | Rhodopsin, nmol | PL
|
mol PL per mol rhodopsin | |
|---|---|---|---|---|
| 32P recovered, cpm | nmol | |||
| DM (0.1) | 2.34 | 147 | 0.016 | 0.007 |
| Cholate (2) | 2.46 | 1,365 | 0.151 | 0.06 |
| OG (2) | 2.52 | 58 | 0.006 | 0.003 |
| CHAPS (2) | 1.92 | 1,196 | 0.132 | 0.07 |
The molar ratio of PL to rhodopsin was determined after calculating specific radioactivity of PC (9,030 cpm/nmol PC) as described in Materials and Methods.
Enhanced Destabilization of Rhodopsin in Cholate in the Presence of mAb B6–30N.
The results shown in Fig. 4 prompted further examination of the stability of rhodopsin in the detergents DM and cholate in the presence and absence of mAb B6–30N. As seen in Fig. 6, marked destabilization of rhodopsin in cholate was observed in the presence of mAb B6–30N.
Discussion
This study was performed with the aim of developing a general method for the purification of rhodopsin mutants expressed in mammalian cells that lacked the C-terminal epitope recognized by the mAb rho1D4. Attempts to use the previously characterized (2) mAb B6–30N that binds to a short N-terminal sequence in rhodopsin failed, and this failure stimulated further examination of detergents that have been studied and used previously in work with rhodopsin (14–16). Our comparative studies of selected detergents included, first, efficiency of solubilization and analysis of rhodopsin stability in them after direct solubilization. Second, because detergents vary widely in their efficacy in removing PLs and because the residual PLs bound to the membrane protein enhance stability, further experiments were carried out to determine stability in detergents after extensive washing of rho1D4-bound rhodopsin to remove the endogenous PLs. The stabilities thus obtained are shown in Fig. 4. DM provided complete stability over prolonged periods of time, whereas the stability decreased in cholate and was even lower in OG (Table 1). Quantitation of the PL remaining showed that DM was also the most efficient in delipidation (Table 1). Cholate retained about 8 mol PL per mol of rhodopsin, whereas OG retained about half that amount. Thus, the stabilities in the two detergents seemed to correlate with the amounts of the lipids retained. As might have been expected, the stability in cholate was decreased further when rhodopsin was first delipidated in DM (Table 1 and Fig. 4). Finally, efficacy of PL removal by detergents was quantitated more carefully after in vivo [32P]Pi labeling of an HEK293S cell line expressing the opsin gene. Extensive washing with any of the detergents tested was capable of producing virtually PL-free rhodopsin (Fig. 5 and Table 2).
The most significant accomplishment of the present work has been the direct support that it has provided for the involvement of the N-terminal segment in a tertiary structure in the intradiscal domain, which must be tightly coupled to a tertiary structure in the TM domain in which the retinal binding pocket lies. Fig. 2 shows clearly that, in DM where no binding of B6–30N antibody occurs, the N-terminal segment is firmly buried in an intradiscal tertiary structure. It is also noted that the binding of the antibody to opsin occurred in DM (Fig. 2), indicating that the binding of 11-cis retinal is required for stabilization of the coupled structures in the TM and the intradiscal domains. In contrast, rhodopsin in cholate allows the binding of the antibody, showing that, in this detergent, the intradiscal tertiary structure must be “breathing” for the N-terminal segment to bind. The important difference between DM and cholate in stabilization versus destabilization of rhodopsin is highlighted further in Fig. 6.
The presence of a tertiary structure in the intradiscal domain of rhodopsin that included all the three loops and the N-terminal segment was proposed in early mutagenic studies (17, 18). Extensive studies since then with a variety of retinitis pigmentosa mutants in both intradiscal and TM domains have consistently supported the presence of a tertiary structure in the intradiscal domain as well as its coupling to packing of the seven helices to form the TM domain (19). The present work has provided direct support for participation of the N-terminal segment in the intradiscal tertiary structure and the coupling of this structure to that in the TM domain.
Acknowledgments
We are grateful to Dr. K. D. Ridge (Center for Advanced Research in Biotechnology, Rockville, MD) for carefully reading the manuscript and making clarifying suggestions. We have benefited greatly from discussion with Prof. U. L. RajBhandary (Biology Department, Massachusetts Institute of Technology) and with Drs. Christophe Bruel, Jong Kim, and Kewen Cai. We thank Drs. Daniel Oprian and Masahiro Kono for providing M1–P142 and M143–A348 plasmids and Ms. Judy Carlin for assistance in the preparation of the manuscript. Research reported herein was supported by National Institutes of Health Grants EY11716 and GM28289 (to H.G.K.).
Abbreviations
- DM
dodecylmaltoside
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- OG
octyl-β-d-glucoside
- DTAB
dodecyltrimethylammonium bromide
- CTAB
cetyltrimethylammonium bromide
- DDAO
dodecyldimethylaminoxide
- BTP
1,3-bis[tris(hydroxymethyl)methylamino]propane
- ROS
rod outer segment
- UV-vis
UV-visible
- PL
phospholipid
- PC
phosphatidylcholine
- TM
transmembrane
Footnotes
This is paper 41 in the series “Structure and Function in Rhodopsin.” Paper 40 is ref. 20.
References
- 1.Molday R S, McKenzie D. Biochemistry. 1983;22:653–660. doi: 10.1021/bi00272a020. [DOI] [PubMed] [Google Scholar]
- 2.Adamus G, Zam Z S, Arendt A, Palczewski K, McDowell J M, Hargrave P. Vision Res. 1991;31:17–31. doi: 10.1016/0042-6989(91)90069-h. [DOI] [PubMed] [Google Scholar]
- 3.Ridge K D, Lee S S J, Abdulaev N G. J Biol Chem. 1996;271:7860–7867. doi: 10.1074/jbc.271.13.7860. [DOI] [PubMed] [Google Scholar]
- 4.Heymann J A W, Subramaniam S. Proc Natl Acad Sci USA. 1997;94:4966–4971. doi: 10.1073/pnas.94.10.4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laird D W, Wong S Y C, Molday R S. In: Membrane Proteins: Proceedings of the Membrane Protein Symposium. Goheen S C, editor. Richmond, CA: Bio-Rad Lab; 1987. [Google Scholar]
- 6.Oprian D D, Molday R S, Kaufman R J, Khorana H G. Proc Natl Acad Sci USA. 1987;84:8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papermaster D S. Methods Enzymol. 1982;81:240–246. doi: 10.1016/s0076-6879(82)81037-9. [DOI] [PubMed] [Google Scholar]
- 8.Shichi H, Somers R L. J Biol Chem. 1978;253:7040–7046. [PubMed] [Google Scholar]
- 9.Reeves P J, Thurmond R L, Khorana H G. Proc Natl Acad Sci USA. 1996;93:11487–11492. doi: 10.1073/pnas.93.21.11487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kono M, Yu H, Oprian D D. Biochemistry. 1998;37:1302–1305. doi: 10.1021/bi9721445. [DOI] [PubMed] [Google Scholar]
- 11.Yu H, Kono M, McKee T D, Oprian D D. Biochemistry. 1995;34:14963–14969. doi: 10.1021/bi00046a002. [DOI] [PubMed] [Google Scholar]
- 12.Lai C Y. Methods Enzymol. 1977;57:236–243. doi: 10.1016/0076-6879(77)47028-9. [DOI] [PubMed] [Google Scholar]
- 13.Ames B N, Dubin D T. J Biol Chem. 1960;235:769–775. [PubMed] [Google Scholar]
- 14.Knudsen P, Hubbell W L. Membr Biochem. 1978;1:297–322. doi: 10.3109/09687687809063853. [DOI] [PubMed] [Google Scholar]
- 15.Fong S, Tsin A T C, Bridges C P B, Liou G I. Methods Enzymol. 1982;81:133–140. doi: 10.1016/s0076-6879(82)81022-7. [DOI] [PubMed] [Google Scholar]
- 16.De Grip W J. Methods Enzymol. 1982;81:256–265. doi: 10.1016/s0076-6879(82)81040-9. [DOI] [PubMed] [Google Scholar]
- 17.Anukanth A, Khorana H G. J Biol Chem. 1994;269:19738–19744. [PubMed] [Google Scholar]
- 18.Doi T, Molday R S, Khorana H G. Proc Natl Acad Sci USA. 1990;87:4991–4995. doi: 10.1073/pnas.87.13.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwa J, Garriga P, Liu X, Khorana H G. Proc Natl Acad Sci USA. 1997;94:10571–10576. doi: 10.1073/pnas.94.20.10571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruel C, Cha K, Niu L, Reeves P J, Khorana H G. Proc Natl Acad Sci USA. 2000;97:3010–3015. doi: 10.1073/pnas.97.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Unger U M, Hargrave P A, Baldwin J M, Schertler G F X. Nature (London) 1997;389:203–206. doi: 10.1038/38316. [DOI] [PubMed] [Google Scholar]