Abstract
Fluoroquinolone use in poultry production may select for resistant Escherichia coli that can be transmitted to humans. To define the prevalence and virulence potential of poultry-associated, quinolone-resistant E. coli in the United States, 169 retail chicken products from the Minneapolis-St. Paul area (1999 to 2000) were screened for nalidixic acid (Nal)-resistant E. coli. Sixty-two (37%) products yielded Nal-resistant E. coli. From 55 products that yielded both Nal-resistant and susceptible E. coli, two isolates (one resistant, one susceptible) per sample were further characterized. Twenty-three (21%) of the 110 E. coli isolates (13 resistant, 10 susceptible) satisfied criteria for extraintestinal pathogenic E. coli (ExPEC), i.e., exhibited ≥2 of pap (P fimbriae), sfa/foc (S/F1C fimbriae), afa/dra (Dr binding adhesins), iutA (aerobactin receptor), and kpsMT II (group 2 capsule synthesis). Compared with other isolates, ExPEC isolates more often derived from virulence-associated E. coli phylogenetic groups B2 or D (74% versus 32%; P < 0.001) and exhibited more ExPEC-associated virulence markers (median, 10.0 versus 4.0; P < 0.001). In contrast, the Nal-resistant and -susceptible populations were indistinguishable according to all characteristics analyzed, including pulsed-field gel electrophoresis profiles. These findings indicate that Nal-resistant E. coli is prevalent in retail poultry products and that a substantial minority of such strains represent potential human pathogens. The similarity of the Nal-resistant and -susceptible populations suggests that they derive from the same source population, presumably the avian fecal flora, with Nal resistance emerging by spontaneous mutation as a result of fluoroquinolone exposure.
The use of fluoroquinolone (FQ) agents in food animal production is suspected of selecting for FQ-resistant gram-negative bacteria, such as Salmonella enterica, Campylobacter jejuni, and Escherichia coli, that can be transmitted to humans via the food supply (7, 14, 15, 41). The epidemiological link between poultry consumption and human disease due to FQ-resistant C. jejuni, combined with the high prevalence of FQ-resistant C. jejuni in retail poultry products, has prompted the U.S. Food and Drug Administration to propose the withdrawal of FQs from use in poultry (8, 41). In contrast, evidence implicating the food supply, and specifically poultry products, as a source for FQ-resistant E. coli in humans is scant (14, 43). However, the burden of human disease due to E. coli is considerably greater than that due to Campylobacter (36). In addition, FQ resistance among clinical E. coli isolates, which has already reached alarming levels in many locales (5, 13, 14, 42), undermines current treatment algorithms for urinary tract infection. These presume that FQs can be relied on as a fallback option when resistance contraindicates the use of traditional agents, such as trimethoprim-sulfamethoxazole (44). Thus, clarification of the origins of FQ resistance in E. coli is urgently needed.
Retail poultry products are routinely heavily contaminated with avian fecal E. coli (40, 43). Such E. coli, which can be antibiotic resistant (including to FQs) (14), widely contaminates kitchen surfaces during meal preparation, is not readily removed from these surfaces by standard cleaning procedures, and can subsequently be isolated from the feces of persons preparing the meals (10, 30). Thus, the possibility of food-borne transmission of FQ-resistant E. coli from poultry to humans is highly plausible.
Unknowns include the prevalence of FQ-resistant E. coli in retail poultry products in the United States (3, 45) and the intrinsic virulence potential of such organisms for humans. The latter point is critical to the likelihood that such strains' resistance could complicate human infections. Plasmid- or transposon-associated drug resistance elements are a potential threat even if present in a low-virulence host strain, since they can be readily transmitted to a more virulent pathogen (12, 39), In contrast, FQ resistance is usually due to point mutations within the quinolone resistance determining regions of gyrA and/or parC, which are fixed on the bacterial chromosome (17, 38). Thus, to pose a significant infectious threat to noncompromised hosts, FQ-resistant E. coli bacteria presumably must themselves possess the diverse virulence factors (VFs), including adhesins, siderophores, capsules, toxins, etc., that characterize the distinctive extraintestinal pathogenic E. coli (ExPEC) strains that cause most episodes of urinary tract infection, sepsis, and neonatal meningitis due to E. coli (23, 37). Likewise, they presumably must derive, as do most human ExPEC isolates, from extraintestinal virulence-associated E. coli phylogenetic groups B2 and D, as defined by multilocus enzyme electrophoresis and sequence typing (16, 28), rather than from commensal and diarrheagenic E. coli-associated phylogenetic groups A and B1 (23, 33).
To address these unknowns, we surveyed retail chicken products purchased throughout the Minneapolis-St. Paul metropolitan area for quinolone-resistant E. coli. We used Nal resistance, which can result from a single point mutation within gyrA and is a precursor to full FQ resistance (38), as a marker for incipient FQ resistance. We then extensively characterized selected Nal-resistant and Nal-susceptible isolates with respect to virulence markers, phylogenetic origin, pulsed-field gel electrophoresis (PFGE) profiles, and O antigens.
MATERIALS AND METHODS
Isolation of Nal-resistant E. coli
From 18 April through 29 November 2000, approximately every 2 weeks, the Minnesota Department of Agriculture purchased 10 chicken products (cut-up parts) from diverse arbitrarily selected retail grocery stores in the Minneapolis-St. Paul, Minnesota, metropolitan area. Samples representing a total of 14 different brand names were purchased from 24 different stores located in 17 cities. Chicken samples were transferred aseptically to prelabeled zip-closure bags. To each bag was added 100 ml of buffered peptone water, and the bag was manually massaged for approximately 3 min. One milliliter of this chicken rinse was then used to inoculate lauryl sulfate-tryptose broth. After overnight incubation at 42°C, the broth was inoculated onto four MacConkey agar plates, one each containing no antibiotics, Nal at 8 μg/ml, and ciprofloxacin at 2 and 4 μg/ml. After overnight incubation at 42°C, these primary plates were inspected for colonies of presumptive E. coli, i.e., that were lactose-positive, flat, and nonmucoid. From each plate that exhibited such growth, three representatives of each E. coli-like colonial variant were subcultured to a blood agar plate, triple sugar iron agar slant, motility-indole-lysine agar tube, citrate agar slant, and Voges-Proskauer/methyl red tube (29). Colonies that exhibited reactions consistent with E. coli were defined as E. coli (29), whereas the rare isolates that exhibited questionable reactions were identified by using the API-20E system (bio-Merieux).
All selected colonies were screened for Nal resistance by placing a Nal-impregnated paper disk (30 μg of Nal; BBL) on a blood agar plate that had been streaked for confluence with a colony from the initial blood agar plate, followed by overnight incubation at 42°C. Confirmed E. coli isolates that exhibited a large zone of inhibition (≥20 mm) around the Nal disk were presumptively defined as Nal susceptible. All 23 of the presumed-susceptible isolates that underwent confirmatory testing were susceptible according to standardized susceptibility testing, which was done using Etest strips (AB Biodisk) according to NCCLS-defined methods and interpretive criteria (32), with E. coli strain ATCC 25922 used as a control. Isolates that exhibited a <20-mm zone or no zone of inhibition also underwent standardized susceptibility testing with Nal and ciprofloxacin Etest strips. From the 55 poultry samples that yielded both Nal-resistant and Nal-susceptible E. coli, one colony each of Nal-S and Nal-R E. coli per sample was arbitrarily selected for further analysis.
Phylogenetic analysis and virulence genotyping
Isolates were assigned to one of the four main phylogenetic groups of E. coli (A, B1, B2, and D), as originally defined according to multilocus enzyme electrophoresis (16), by using the multiplex PCR-based method of Clermont et al. (9). For virulence typing, all isolates initially were screened for five virulence markers, i.e., papA and papC (which were analyzed collectively), sfa/foc, afa/dra, iutA, and kpsMT II, to permit their classification as ExPEC or non-ExPEC. These five virulence markers were identified as independently predictive of ExPEC status by statistical analyses of virulence typing results from three strain collections, within which ExPEC status could be inferred based on epidemiological source, e.g., symptomatic urinary tract infection versus fecal, or on observed extraintestinal virulence in animal challenge experiments (not shown) (19, 33; J. R. Johnson, M. Kuskowski, E. Denamur, J. Elion, and B. Picard, Letter, Infect. Immun. 68:424-425, 2000; M. A. Khan, T. O'Bryan, N. Kaster, F. Lebahn, and J. R. Johnson, Abstr. 37th Annu. Meet. Infect. Dis. Soc. Am., p. 70, 1999). Since optimal discrimination between ExPEC and non-ExPEC isolates within these three collections was provided by a criterion of the presence of ≥2 of the five markers, this was used as the operational definition of ExPEC among the poultry-source isolates. All ExPEC isolates and a randomly selected control group of non-ExPEC isolates were then tested for 35 virulence markers of ExPEC and, if positive for any pap element, for the 13 papA alleles, using established PCR and dot blot-based assays, as described elsewhere (24, 27). All testing was done in duplicate using independently prepared boiled lysates of each isolate, with appropriate positive and negative controls included within each run.
An aggregate ExPEC marker score was calculated as the number of the five cardinal ExPEC markers for which an isolate tested positive. For isolates that underwent extended virulence genotyping, an aggregate virulence score was calculated as the sum of all virulence markers for which the isolates tested positive, with adjustment for multiple detection of the pap, sfa, and kps operons. Cluster analysis of VF data was done using the unweighted pair group method with averaging (UPGMA), as previously described (20).
Serotyping
O antigens were determined by the Gastroenteric Disease Research Center (University Park, Penn.) using 180 specific O antisera, according to standard methods.
PFGE
Selected isolates were subjected to PFGE using XbaI-restricted total DNA according to a protocol developed by the Centers for Diseases Control and Prevention (4). Reference E. coli O157:H7 strain G5244, provided by the Minnesota Department of Health laboratory, was included in each gel as a positive control and size standard to facilitate cross-gel comparisons. Profiles were captured and analyzed digitally, with operator input. Dendrograms were constructed according to UPGMA with the use of band-based Dice similarity coefficients, within the application Molecular Analyst-DNA Fingerprinting (Bio-Rad).
RAPD analysis
Isolates that sheared during PFGE analysis, plus selected other isolates and their derivatives from Nal-supplemented or nonsupplemented agar plates, were compared according to random amplified polymorphic DNA (RAPD) profiles. These were generated by using (separately) arbitrary decamer primers 1254, 1281, and/or 1283, with boiled lysates used as template DNA, followed by agarose gel electrophoresis and visual inspection, as previously described (2).
Statistical methods
Comparisons of proportions were analyzed using Fisher's exact test or McNemar's test for unpaired and paired comparisons, respectively (both two-tailed). Comparisons of ExPEC scores and aggregate virulence scores were analyzed using the Mann-Whitney U test. The threshold for statistical significance was a P value of <0.05.
RESULTS
Isolation of Nal-resistant and -susceptible E. coli.
Of the 169 chicken samples cultured, 150 (89%) yielded E. coli on nonselective media. Nal-resistant E. coli was recovered from 62 (41%), and Nal-susceptible E. coli was recovered from 143 (95%), of the 150 E. coli-positive samples, i.e., from 37% and 85%, respectively, of all 169 samples. Fifty-five samples yielded both susceptible and resistant E. coli, seven yielded only Nal-resistant isolates, and 88 yielded only susceptible isolates. One (1.6%) of the 62 products with Nal-resistant E. coli yielded an E. coli isolate that was also resistant to ciprofloxacin, with an MIC of 8 μg/ml (resistant, ≥4 μg/ml). From the 55 items that yielded both Nal-resistant and Nal-susceptible E. coli, one Nal-resistant isolate and one Nal-susceptible isolate per sample were analyzed further.
Nal MICs.
The distribution of Nal MICs observed among the 55 Nal-resistant isolates was as follows: ≥256 μg/ml (42 isolates), 128 μg/ml (1 isolate), 96 μg/ml (3 isolates), 64 μg/ml (6 isolates), 48 μg/ml (2 isolates), and 32 μg/ml (1 isolate). The 23 Nal-susceptible isolates that underwent MIC testing exhibited Nal MICs of 1.0 (6 isolates), 1.5 (11 isolates), or 2.0 (6 isolates).
Distribution of ExPEC markers in relation to Nal phenotype.
The 55 Nal-resistant and 55 Nal-susceptible chicken source E. coli isolates first were screened by PCR for five ExPEC-defining virulence markers, which were detected, in order of descending frequency, as follows: iutA (57%), papA and/or papC (14%), kpsMT II (13%), sfa/foc (1%), and afa/dra (none) (Table 1). Twenty-three (21%) of the isolates exhibited two (n = 18) or three (n = 5) of the markers, so they were defined as ExPEC. These 23 ExPEC isolates represented 21 (38%) of the poultry products from which E. coli isolates were screened for ExPEC status. (Two products yielded two ExPEC isolates each, whereas 19 yielded a single ExPEC isolate.) The remaining 87 (79%) screened isolates contained one (n = 42) or none (n = 45) of the ExPEC markers, so they were defined as non-ExPEC. Nal-resistant and Nal-susceptible isolates were similar with respect to the prevalence of each of the five ExPEC markers (Table 1), the proportion qualifying as ExPEC (Table 1), and aggregate ExPEC marker score (median score, 1.0 in both groups; P > 0.10).
TABLE 1.
Distribution of ExPEC-defining virulence markers and phylogenetic groups among poultry-source E. coli isolates according to Nal susceptibility status
| Bacterial trait | Prevalence of trait, no. (%)b
|
||
|---|---|---|---|
| Nal susceptible (n = 55) | Nal resistant (n = 55) | Total (n = 110) | |
| papA and/or papC | 6 (11) | 9 (16) | 15 (14) |
| afa/dra | 0 | 0 | 0 |
| sfa/foc | 1 (2) | 0 | 1 (1) |
| iutA | 28 (51) | 35 (64) | 63 (57) |
| kpsMT II | 6 (11) | 8 (15) | 14 (13) |
| ExPECa | 10 (18) | 13 (24) | 23 (21) |
| Phylogenetic group A | 21 (38) | 14 (25) | 35 (32) |
| Phylogenetic group B1 | 12 (22) | 18 (33) | 30 (26) |
| Phylogenetic group B2 | 6 (11) | 8 (15) | 14 (13) |
| Phylogenetic group D | 16 (29) | 15 (27) | 31 (25) |
≥2 markers present: papA and/or papC, afa/dra, sfa/foc, iutA, kpsMT II.
P > 0.10 (by Fisher's exact test) for all comparisons, Nal susceptible versus resistant.
Phylogenetic background.
According to PCR-based phylotyping, E. coli phylogenetic groups A, B1, and D each accounted for 25 to 32% of the isolates overall, whereas group B2 accounted for 14% of isolates. The Nal-susceptible and -resistant isolates were similar with respect to the relative prevalences of the four phylogenetic groups (Table 1).
Phylogenetic distribution of ExPEC markers.
In contrast to their indifferent associations with Nal phenotype, the ExPEC markers exhibited a striking phylogenetic distribution (Table 2). papC was associated negatively with phylogenetic group B1, iutA was associated negatively with groups A and B1 and positively with group D, and kpsMT II was associated negatively with groups A and B1 and positively with groups B2 and D (Table 2). Overall, ExPEC status was negatively associated with group B1 and positively associated with group D (Table 2). The prevalence of ExPEC status was highest within group B2 and declined progressively through groups D and A to group B1. However, because of its greater prevalence overall, group D contributed slightly more ExPEC isolates than did group B2 (Table 2). ExPEC isolates were significantly more likely to derive from phylogenetic groups B2 or D (17 of 23, 74%) than were non-ExPEC isolates (28 of 87, 32%; P < 0.001). The sole ciprofloxacin-resistant isolate was a non-ExPEC isolate from group A (not shown).
TABLE 2.
Phylogenetic distribution of selected virulence markers among 110 poultry-source E. coli isolates
| Bacterial trait | Total no. (% of 110) with trait | Prevalence of trait within phylogenetic group, no. (%)a
|
|||
|---|---|---|---|---|---|
| A (n = 35) | B1 (n = 30) | B2 (n = 14) | D (n = 31) | ||
| papA | 3 (3) | 1 (3) | 0 | 0 | 2 (6) |
| papC | 15 (14) | 7 (20) | 0 (**) | 1 (7) | 7 (23) |
| sfa/foc | 1 (1) | 0 | 0 | 1 (7) | 0 |
| iutA | 63 (57) | 14 (40)(**) | 10 (33)(**) | 9 (64) | 30 (97)*** |
| kpsMT II | 14 (13) | 0(*) | 0(**) | 6 (43)** | 8 (26) |
| ExPECb | 23 (21) | 6 (17) | 0(***) | 6 (43) | 11 (35)* |
P value symbols (for comparison of indicated group versus all others): ∗, P < 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (Parentheses around asterisk(s) indicate negative association.)
ExPEC, defined as presence of ≥2 of the following markers: papA and/or papC, afa/dra (none detected), sfa/foc, iutA, and kpsMT II.
Extended virulence genotypes.
The 23 ExPEC isolates and 29 randomly selected non-ExPEC isolates (stratified to give 15 and 14 each with an ExPEC score of 0 and 1, respectively) were next tested for the presence of 29 additional ExPEC-associated virulence markers and, among isolates positive for any pap element, the 13 papA alleles. Twenty-five of the 35 virulence markers sought (Table 3) and the F11 papA allele (three isolates) were detected in at least one isolate each. Markers not detected among the 52 isolates included papG alleles I and III, focG (F1C fimbriae), afa/dra (Dr binding adhesins), hlyD (hemolysin), cnf1 (cytotoxic necrotizing factor), cdtB (cytolethal distending toxin), the K2 kpsMT variant, and rfc (O4 lipopolysaccharide synthesis).
TABLE 3.
Prevalence of virulence markers according to Nal phenotype and ExPEC status among 52 poultry-derived E. coli isolates
| Bacterial traitb | Prevalence of trait, no. (%)
|
P valuec | |||
|---|---|---|---|---|---|
| Nal phenotypea
|
ExPEC status
|
||||
| Susceptible (n = 24) | Resistant (n = 38) | Not ExPEC (n = 29) | ExPEC (n = 23) | ||
| papA | 2 (8) | 1 (4) | 1 (3) | 2 (9) | |
| papCb | 6 (25) | 9 (32) | 2 (7) | 13 (57) | <.001 |
| sfa/foc | 1 (4) | 0 | 0 | 1 (4) | |
| sfaS | 1 (4) | 0 | 0 | 1 (4) | |
| gafD | 3 (13) | 3 (11) | 0 | 6 (26) | .005 |
| bmaE | 3 (13) | 3 (11) | 0 | 6 (26) | .005 |
| iha | 1 (4) | 0 | 1 (3) | 0 | |
| fimH | 24 (100) | 28 (100) | 29 (100) | 23 (100) | |
| fyuA | 7 (29) | 11 (39) | 4 (14) | 14 (61) | .001 |
| iutA | 16 (67) | 20 (71) | 13 (45) | 23 (100) | <.001 |
| iroN | 14 (58) | 20 (71) | 16 (55) | 18 (78) | |
| ireA | 8 (33) | 6 (21) | 3 (10) | 11 (49) | .004 |
| kpsMT II | 6 (25) | 8 (29) | 0 | 14 (61) | <.001 |
| K1 | 1 (4) | 5 (18) | 0 | 6 (26) | .005 |
| kpsMT III | 0 | 2 (7) | 2 (7) | 0 | |
| H7fliC | 0 | 3 (11) | 3 (10) | 0 | |
| ompT | 15 (63) | 16 (57) | 12 (41) | 19 (83) | .004 |
| ibeA | 2 (8) | 4 (14) | 0 | 6 (26) | .004 |
| cvaC | 9 (38) | 7 (25) | 6 (21) | 10 (43) | |
| traT | 15 (63) | 22 (79) | 29 (66) | 18 (78) | |
| iss | 14 (58) | 20 (71) | 16 (55) | 18 (78) | |
| malX | 2 (8) | 4 (14) | 0 | 6 (26) | .005 |
P > 0.10 for all comparisons, nalidixic acid susceptible versus resistant.
Results for papEG, papG, and papG allele II approximated those for papC. Not detected were afa/dra, focG, hlyD, cnf1, cdtB, and the K2 kpsMT II variant.
P values (by Fisher's exact test) for comparisons of non-ExPEC versus ExPEC shown only where values are <0.05.
Compared with the 29 non-ExPEC control isolates, the 23 ExPEC isolates exhibited a significantly higher prevalence of numerous virulence markers other than those included in the definition of ExPEC (Table 3) and had significantly higher aggregate virulence scores (median scores, 10.0 versus 4.0; P < 0.001). In contrast, virulence genotypes did not differ according to Nal phenotype. The Nal-resistant (n = 28) and -susceptible (n = 24) isolates were similar with respect to the prevalence of all of the virulence markers analyzed (Table 3) and to aggregate virulence score (median scores, 6.75 versus 6.0; P > 0.10). Similar findings were observed when these comparisons between Nal phenotypes were limited to the 23 ExPEC isolates, both for individual virulence markers (not shown) and for aggregate virulence scores (median scores, 10.0, Nal-resistant, versus 9.5, Nal-susceptible; P > 0.10).
As observed for the five defining ExPEC markers, many of the additional virulence markers included in the extended virulence genotyping assay exhibited a significant phylogenetic distribution, whether this was analyzed among the 23 ExPEC and 29 non-ExPEC isolates combined or among the 23 ExPEC isolates alone (Table 4). Aggregate virulence scores also varied significantly according to phylogenetic group, both among the combined 52 (ExPEC and non-ExPEC) isolates and among just the 23 ExPEC isolates. In the combined ExPEC and non-ExPEC population, aggregate virulence scores were highest within phylogenetic groups A and B2 (median score, 8.0 in both groups), slightly lower within group D (median, 7.0), and lowest within group B1 (median score, 4.0; versus all other isolates, P = 0.001). Among the 23 ExPEC isolates alone, similar virulence score trends were observed for groups A (median score, 10.75), B2 (median score, 10.0), and D (median, 7.0; for group A versus all others, P = 0.002; for group D versus all others, P < 0.001).
TABLE 4.
Phylogenetic distribution of virulence markers among poultry-source E. coli isolates
| Bacterial trait | Prevalence of trait by phylogenetic group, no. (%)
|
||||||
|---|---|---|---|---|---|---|---|
| ExPEC plus non-ExPEC (n = 52)
|
ExPEC only (n = 23)a
|
||||||
| A (n = 15) | B1 (n = 11) | B2 (n = 11) | D (n = 15) | A (n = 6) | B2 (n = 6) | D (n = 11) | |
| papCb | 7 (47) | 0(*)c | 1 (9) | 7 (47) | 6 (100)* | 1 (17) | 6 (55) |
| gafD | 6 (40)*** | 0 | 0 (0) | 0 (0) | 6 (100)*** | 0 (0) | 0(*) |
| bmaE | 6 (40)*** | 0 | 0 (0) | 0 (0) | 6 (100)*** | 0 (0) | 0(*) |
| fyuA | 9 (60)* | 0(**) | 6 (55) | 3 (20) | 6 (100)* | 6 (100)* | 2 (18)(***) |
| iutA | 11 (73) | 3 (27)(**) | 8 (73) | 14 (93)* | 6 (100) | 6 (100) | 11 (100) |
| ireA | 6 (40) | 0(*) | 1 (9) | 7 (47) | 6 (100)** | 0(**) | 5 (45) |
| kpsMT II | 0(**) | 0(*) | 6 (55)* | 8 (53)* | 0(***) | 7 (100)* | 8 (72) |
| K1 | 0 | 0 | 4 (36)* | 2 (13) | 0 | 4 (67)* | 2 (18) |
| ompT | 12 (80) | 2 (18)(**) | 7 (64) | 10 (67) | 6 (100) | 6 (100) | 7 (64)(*) |
| ibeA | 0 (0) | 0 | 5 (45)*** | 1 (7) | 0 | 5 (83)*** | 1 (9) |
| cvaC | 9 (60)** | 1 (9) | 3 (27) | 3 (20) | 6 (100)** | 2 (33) | 2 (18)(*) |
| traT | 7 (47)(*) | 8 (73) | 9 (82) | 13 (87) | 2 (33)(**) | 6 (100) | 10 (91) |
| malX | 0 (0) | 0 (0) | 5 (45)*** | 1 (7) | 0 | 5 (83)*** | 1 (9) |
Only groups A, B2, and D are shown (n = 24), since no B1 isolate was ExPEC.
Results for papEG, papG, and papG allele II approximated those for papC. Only those traits that yielded at least one association at the P < 0.05 level are included in the table.
P value symbols (for comparison of indicated group versus all others): ∗, P < 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. (Parentheses indicated negative associations.)
Cluster analysis of VF profiles.
The extended virulence genotype data for these 52 isolates were subjected to cluster analysis according to UPGMA, stratified by phylogenetic group. Within the resulting four dendrograms, which collectively comprised 36 unique phylo-pathotypes, the Nal-susceptible and -resistant isolates were considerably intermixed. Overall, isolates were significantly more likely to have as their nearest neighbor in these trees an isolate of the alternate Nal phenotype (n = 11) or a mixed cluster containing both Nal phenotypes (n = 24) than an isolate of the same Nal phenotype (n = 17; P = 0.018, McNemar's test). In contrast, all but two isolates had as their nearest neighbor an isolate, or a homogenous cluster of isolates, of the same ExPEC status (for having a similar ExPEC-status isolate or cluster as a nearest neighbor, versus a dissimilar isolate or cluster; P < 0.001, McNemar's test).
O antigens.
O antigens were assessed for 22 of the 23 ExPEC isolates (12 Nal-resistant, 10 Nal-susceptible) and for 10 non-ExPEC isolates. (The latter were selected from among the non-ExPEC isolates that underwent extended virulence genotyping so as to include five with no ExPEC markers and five with a single ExPEC marker, and five each of Nal-resistant and Nal-susceptible isolates.) Overall, 16 unique O antigens were detected among the 35 isolates. These included, in descending order of prevalence (number of isolates), O78 (six), O120 and O7 (three each), O25 (two), and O1, O2, O6, O8, O18, O23, O29, O33, O46, O53, O73, and O77 (one each). Six isolates were O nontypeable. No differences in O antigen distribution were evident between the Nal-susceptible and -resistant isolates, and two of the four multiply encountered serogroups (i.e., O7 and O78) each were represented by both susceptible and resistant isolates. In contrast, compared with the non-ExPEC isolates, the ExPEC isolates were more likely to be O typeable (20 of 22, versus 6 of 10; P = 0.06) and to exhibit the O7, O78, or O120 antigen (27, 14, and 14%, respectively, ExPEC, versus 0%, non-ExPEC; P = 0.004).
PFGE and RAPD analysis.
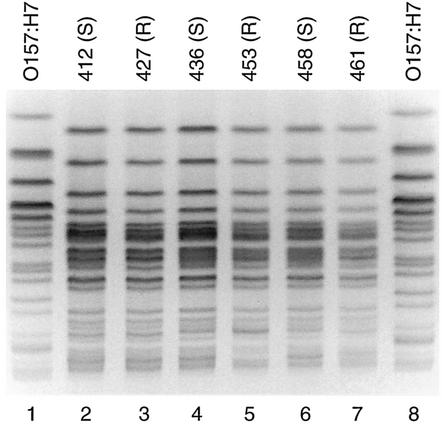
The 52 isolates that underwent extended virulence genotyping also were subjected to PFGE analysis. Forty-four isolates yielded interpretable PFGE profiles with XbaI, which were compared by creating an UPGMA-based similarity dendrogram for each phylogenetic group. In these dendrograms the Nal-susceptible and -resistant isolates were considerably intermixed (not shown). Overall, isolates were more likely to have as their nearest neighbor in the PFGE-based dendrogram an isolate of the alternate Nal phenotype (n = 12) or a mixed cluster containing representatives of both Nal phenotypes (n = 13) than an isolate of the same Nal phenotype (n = 19). Six of the isolates (three resistant, three susceptible), all O78 and ExPEC-positive, yielded indistinguishable PFGE profiles (e.g., Fig. 1). These six isolates derived from diverse poultry items representing the same brand name but processed on six different dates, from 12 August through 3 December, in two different plants, and purchased at five different stores during five separate weeks.
FIG. 1.
PFGE profiles of E. coli isolates from retail chicken products. Lane numbers are shown below image. XbaI PFGE profiles reveal identity between Nal-susceptible (S) isolates 412, 436, and 458 (lanes 2, 4, and 6) and Nal-resistant (R) isolate 427, 453, and 461 (lanes 3, 5, and 7). Lanes 1 and 8, reference E. coli O157:H7 strain G5244.
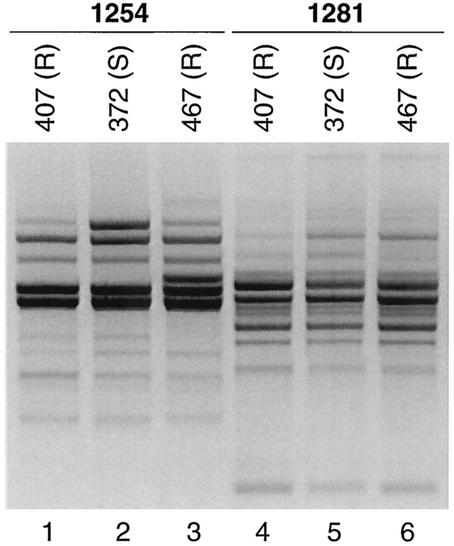
The eight isolates (five resistant, three susceptible) that consistently sheared during PFGE with XbaI were subjected to RAPD analysis using the primer 1254. This revealed five unique RAPD profiles. Four profiles were specific to a single isolate each (not shown), whereas one was shared among three isolates (two resistant, one susceptible) from phylogenetic group D (Fig. 2).
FIG. 2.
RAPD profiles of poultry-source E. coli isolates refractory to macrorestriction analysis. Lane numbers are shown below image. According to analysis with RAPD primers 1254 and 1281 (as listed above the gel lanes), Nal-resistant (R) isolates (isolates 407, lanes 1 and 4; and 467, lanes 3 and 6) are indistinguishable from Nal-susceptible (S) isolate 372 (lanes 2 and 5), within the reproducibility limits of RAPD analysis.
Spontaneous in vitro mutation to Nal resistance.
To assess the degree to which in vitro mutation to Nal resistance during sample processing might have contributed to the Nal-resistant population studied, 59 Nal-susceptible isolates (i.e., the 55 from samples yielding both Nal-resistant and -susceptible isolates, plus four more from samples yielding only susceptible isolates) were analyzed. Susceptible isolates, picked as single colonies from primary nonselective plates, were amplified overnight in nonselective broth and then plated (10 μl) to MacConkey's agar with and without Nal supplementation (8 μg/ml). Whereas on the nonsupplemented plates all samples yielded confluent growth, on the Nal-MacConkey plates only nine samples (15%) yielded colonies that corresponded with the parent according to RAPD analysis (data not shown). Standardized susceptibility testing by Etest showed that eight of the nine isolates from Nal-MacConkey plates were resistant, whereas one was intermediately susceptible, to Nal. Thus, the estimated overall frequency of putative in vitro mutation to Nal resistance was 8 of 59, or 14%.
DISCUSSION
Our findings support three main conclusions. First, nearly 40% of retail chicken products contain Nal-resistant E. coli. Second, although most poultry-derived Nal-resistant E. coli strains are nonpathogens by molecular epidemiological criteria, a substantial minority (24%) represent ExPEC and derive from virulence-associated E. coli phylogenetic groups B2 and D (42%) and hence are of potential health significance to humans. Third, among these poultry-derived strains, Nal resistance is independent of virulence genotype and phylogenetic background, consistent with nonspecific selection for Nal-resistant mutants within the avian fecal flora by exposure to FQs in the production environment.
According to current understandings of the genetics of quinolone and FQ resistance in E. coli, the Nal-resistant isolates detected here would be expected to become FQ resistant if they were to acquire one or two additional mutations in gyrA and/or parC (17, 38). These additional mutational steps conceivably could occur either on the farm or after acquisition of the strains by humans. In either scenario, these isolates clearly represent a high-risk population for progression to full FQ resistance if exposed to further selection pressure. That 37% of retail chicken products contained Nal-resistant E. coli indicates that these products are a plausible vehicle for transmission of such high-risk strains to humans, particularly in view of the ease with which E. coli strains from chicken carcasses can contaminate kitchen surfaces (10) and appear in the feces of meal preparers (30).
We found that 21% of the E. coli isolates that were screened for virulence markers qualified as ExPEC and that 38% of the corresponding chicken products contained at least one ExPEC isolate. If the same rate of positivity were assumed also for the products that yielded only Nal-susceptible or Nal-resistant E. coli, the overall estimated prevalence of ExPEC among all 165 products sampled would be 34%. Of note, this represents only a minimum estimate, since only two colonies were examined for ExPEC status per chicken product, whereas multiple strains are typically present in individual retail chicken products (J. R. Johnson, P. Delavari, and S. R. Tatini, Abstr. 100th Gen. Meet. Am. Soc. Microbiol., abstr. P-42, p. 521, 2000.).
Many characteristics of the ExPEC strains recovered from the chicken samples, including virulence profiles, phylogenetic background, and O antigens, resemble those of clinical isolates from humans with diverse extraintestinal infections (19, 22, 24, 26, 31; J. R. Johnson, T. T. O'Bryan, A. R. Manges, and L. W. Riley, abstract from the 39th Annual Meeting of the Infectious Diseases Society of America 2001, Clin. Infect. Dis. 33:1234, 2001.). This suggests that these chicken-derived strains may be pathogenic for humans. There is ample precedent for cross-species pathogenicity of E. coli. In experimental models of extraintestinal infection. such as urinary tract infection, pneumonia, neonatal meningitis, and systemic sepsis, human-source E. coli isolates routinely cause significant and sometimes fatal disease in diverse animal hosts (18, 33-35). Likewise, identity at the level of phylogenetic group and extended virulence profiles has been extensively documented among human and canine-source clinical ExPEC isolates (21, 25), and one pair of isolates (dog-human) has been shown to exhibit nearly indistinguishable genomic macrorestriction profiles by PFGE (21). Commonality among clinical isolates from diseased farm animals (including poultry) and humans also has been documented, specifically for E. coli O2:K1 and O78 (1, 6). Of note, O78 was the single most commonly encountered O antigen in the present study, accounting for 19% of the isolates that underwent O typing. Thus, poultry and poultry products may represent a reservoir and vehicle for dissemination of diverse drug-resistant ExPEC clonal groups. We are currently analyzing additional retail poultry products to further evaluate this hypothesis (Johnson et al., Abstr. 100th Gen. Meet. Am. Soc. Microbiol. 2000).
The observation that the Nal-resistant and -susceptible poultry isolates were essentially indistinguishable with respect to all parameters analyzed strongly suggests that these two groups derive from the same source population, with the resistant isolates presumably having arisen from susceptible ancestors by random point mutations in gyrA and/or parC without regard for other bacterial characteristics. This supports a model whereby FQ use in poultry production provides generalized selection pressure within the avian fecal flora for spontaneous resistant mutants (11). It refutes the counter-hypotheses that the resistant population either is derived from an alternative source, including possibly (antibiotic-consuming) humans, or represents a distinctive subset of the avian fecal E. coli population with an enhanced propensity for mutation to resistance. It also suggests that the extent of exposure of poultry flocks to FQs can be expected to determine the prevalence of quinolone and FQ resistance within, but not the phylogenetic or pathotypic composition of, the birds' fecal flora. Thus, although FQ use in poultry production does not specifically select for ExPEC, it presumably tends to render any preexisting ExPEC strains more highly antibiotic resistant.
We documented a low frequency of presumed spontaneous mutation to Nal resistance in vitro during specimen processing. Thus, a substantial fraction (conceivably, 32%) of the resistant population studied may have arisen artifactually within the laboratory rather than deriving from the primary samples per se, as assumed. This phenomenon would tend to obscure any preexisting differences between the resistant and susceptible populations, thereby biasing the study toward the observed “no difference” result. However, the magnitude of this effect would not be expected to completely obliterate biologically significant between-population differences, whereas we found no suggestion of such a difference. Thus, it seems unlikely that important differences between the Nal-resistant and Nal-susceptible populations were missed because of in vitro mutation to resistance.
Another potential limitation of the study is the use of multiple comparisons, which increases the likelihood of a type I error, i.e., of falsely identifying a chance difference as significant. However, most of the differences identified yielded P values of ≤0.01, reducing the chances of a type I error, and were consistent with previous findings from other populations. Conversely, the study's power for correctly detecting between-group differences is statistically limited by the sample size, with the attendant possibility of type II errors, i.e., of falsely concluding against a difference when one actually exists. However, for the key comparisons between the Nal-resistant and Nal-susceptible isolates, not even suggestive trends toward differences were evident, whereas numerous significant differences were detected between ExPEC and non-ExPEC and among the four phylogenetic groups. Finally, sampling was limited to one geographical area, albeit with a diversity of stores and of brand names, many of which are nationally distributed.
In summary, we found that nearly 40% of retail chicken products contained Nal-resistant E. coli, that approximately 20% of Nal-resistant and Nal-susceptible isolates alike represented ExPEC, and that except for the Nal phenotype these two populations were indistinguishable according to all of the molecular and phenotypic methods used. This suggests that retail chicken products are a potential source of antibiotic-resistant pathogenic E. coli for acquisition by humans and that the resistant and susceptible strains derive from the same source population, presumably the avian fecal flora.
Acknowledgments
This material is based upon work supported by Office of Research and Development, Medical Research Service, Department of Veterans Affairs (J.R.J.), National Institutes of Health grant DK-47504 (J.R.J.), and National Research Initiative (NRI) Competitive Grants Program/United States Department of Agriculture grant 00-35212-9408 (J.R.J.).
Ann Emery helped with manuscript preparation.
REFERENCES
- 1.Achtman, M., M. Heuzenroeder, B. Kusecek, H. Ochman, D. Caugant, R. K. Selander, V. Vaisanen-Rhen, T. K. Korhonen, S. Stuart, F. Orskov, and I. Orskov. 1986. Clonal analysis of Escherichia coli O2:K1 isolated from diseased humans and animals. Infect. Immun. 51:268-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg, D. E., N. S. Akopyants, and D. Kersulyte. 1994. Fingerprinting microbial genomes using the RAPD or AP-PCR method. Methods Mol. Cell. Biol. 5:13-24. [Google Scholar]
- 3.Boothe, D. H., and J. W. Arnold. 2003. Resistance of bacterial isolates from poultry products to therapeutic veterinary antibiotics. J. Food Prot. 66:94-102. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1998. Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis, training course. Centers for Disease Control and Prevention, Atlanta, Ga.
- 5.Cheong, H.-J., C.-W. Yoo, J.-W. Sohn, W.-J. Kim, M.-J. Kim, and S.-C. Park. 2001. Bacteremia due to quinolone-resistant Escherichia coli in a teaching hospital in South Korea. Clin. Infect. Dis. 33:48-53. [DOI] [PubMed] [Google Scholar]
- 6.Cherifi, A., M. Contrepois, B. Picard, P. Goullet, I. Orskov, and F. Orskov. 1994. Clonal relationships among Escherichia coli serogroup O78 isolates from human and animal infections. J. Clin. Microbiol. 32:1197-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiu, C.-H., T.-L. Wu, L.-H. Su, C. Chu, J.-H. Chia, A.-J. Kuo, M.-S. Chien, and T.-Y. Lin. 2002. The emergence in Taiwan of fluoroquinolone resistance in Salmonella enterica serotype choleraesuis. N. Engl. J. Med. 346:413-419. [DOI] [PubMed] [Google Scholar]
- 8.Cimons, M. 2001. FDA planning to halt ag use of two antibiotics. ASM News 67:9-10. [Google Scholar]
- 9.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cogan, T. A., S. F. Bloomfield, and T. J. Humphrey. 1999. The effectiveness of hygiene procedures for prevention of cross-contamination from chicken carcasses in the domestic kitchen. Lett. Appl. Microbiol. 29:354-358. [DOI] [PubMed] [Google Scholar]
- 11.Feinman, S. I. 1998. Antibiotics in animal feed—drug resistance revisited. ASM News 64:24-30. [Google Scholar]
- 12.Fierer, J., and D. Guiney. 1999. Extended-spectrum B-lactamases. A plague of plasmids. JAMA 281:563-564. [DOI] [PubMed] [Google Scholar]
- 13.Gales, A. C., R. N. Jones, K. A. Gordon, H. S. Sader, W. W. Wilki, M. L. Beach, M. A. Pfaller, G. V. Doern, and The SENTRY Study Group (Latin America). 2000. Activity and spectrum of 22 antimicrobial agents tested against urinary tract infection pathogens in hospitalized patients in Latin America: report from the second year of the SENTRY Antimicrobial Surveillance Program (1998). J. Antimicrob. Chemother. 45:295-303. [DOI] [PubMed] [Google Scholar]
- 14.Garau, J., M. Xercavins, M. Rodriguez-Carballeira, J. Gomez-Vera, I. Coll, D. Vidal, T. Llovet, and A. Ruiz-Bremon. 1999. Emergence and dissemination of quinolone-resistant Escherichia coli in the community. Antimicrob. Agents Chemother. 43:2736-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorbach, S. 2001. Antimicrobial use in animal feed—time to stop. N. Engl. J. Med. 345:1201-1203. [DOI] [PubMed] [Google Scholar]
- 16.Herzer, P. J., S. Inouye, M. Inouye, and T. S. Whittam. 1990. Phylogenetic distribution of branched RNS-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172:6175-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hooper, D. C. 2000. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin. Infect. Dis. 31(Suppl. 2):S24-S28. [DOI] [PubMed] [Google Scholar]
- 18.Huang, S.-H., Z.-S. Wan, Y.-H. Chen, A. Y. Jong, and K. S. Kim. 2001. Further characterization of Escherichia coli brain microvascular endothelial cell invasion gene ibeA by deletion, complementation, and protein expression. J. Infect. Dis. 183:1071-1078. [DOI] [PubMed] [Google Scholar]
- 19.Johnson, J. R., P. Delavari, M. Kuskowski, and A. L. Stell. 2001. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J. Infect. Dis. 183:78-88. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, J. R., T. T. O'Bryan, M. A. Kuskowski, and J. N. Maslow. 2001. Ongoing horizontal and vertical transmission of virulence genes and papA alleles among Escherichia coli blood isolates from patients with diverse-source bacteremia. Infect. Immun. 69:5363-5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, J. R., T. T. O'Bryan, D. A. Low, G. Ling, P. Delavari, C. Fasching, T. A. Russo, U. Carlino, and A. L. Stell. 2000. Evidence of commonality between canine and human extraintestinal pathogenic Escherichia coli that express papG allele III. Infect. Immun. 68:3327-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, J. R., E. Oswald, T. T. O'Bryan, M. A. Kuskowski, and L. Spanjaard. 2002. Phylogenetic distribution of virulence-associated genes among neonatal meningitis isolates of Escherichia coli from The Netherlands. J. Infect. Dis. 185:774-784. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, J. R., and T. A. Russo. 2002. Extraintestinal pathogenic Escherichia coli (ExPEC): the “other bad E. coli.” J. Lab. Clin. Med. 139:155-162. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, J. R., A. L. Stell, P. Delavari, M. A. C., M. Kuskowski, and W. Gaastra. 2001. Phylogenetic and pathotypic similarities between Escherichia coli isolates from urinary tract infections in dogs and extraintestinal infections in humans. J. Infect. Dis. 183:897-906. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, J. R., A. L. Stell, T. T. O'Bryan, M. Kuskowski, B. Nowicki, C. Johnson, J. N. Maslow, A. Kaul, J. Kavle, and G. Prats. 2002. Global molecular epidemiology of extraintestinal pathogenic Escherichia coli clonal group O15:K52:H1: evidence of distribution beyond Europe. J. Clin. Microbiol. 40:1913-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, J. R., C. Van der Schee, M. A. Kuskowski, W. Goessens, and A. Van Belkum. 2002. Phylogenetic background and virulence profiles of fluoroquinolone-resistant clinical Escherichia coli isolates from The Netherlands. J. Infect. Dis. 186:1852-1856. [DOI] [PubMed]
- 28.Lecointre, G., L. Rachdi, P. Darlu, and E. Denamur. 1998. Escherichia coli molecular phylogeny using the incongruence length difference test. Mol. Biol. Evol. 15:1685-1695. [DOI] [PubMed] [Google Scholar]
- 29.Lennette, E. H., A. Balows, W. J. Hausler, and H. J. Shadomy. 1985. Manual of clinical microbiology, 4th ed. American Society for Microbiology, Washington, D.C.
- 30.Linton, A. H., K. Howe, P. M. Bennett, and M. H. Richmond. 1977. The colonization of the human gut by antibiotic resistant Escherichia coli from chickens. J. Appl. Bacteriol. 43:465-469. [DOI] [PubMed] [Google Scholar]
- 31.Manges, A. R., J. R. Johnson, B. Foxman, T. T. O'Bryan, K. E. Fullerton, and L. W. Riley. 2001. Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N. Engl. J. Med. 345:1007-1013. [DOI] [PubMed] [Google Scholar]
- 32.NCCLS. 2001. Performance standards for antimicrobial susceptibility testing. M100-S11. Vol. 21, no. 1. NCCLS, Wayne, Pa.
- 33.Picard, B., J. Sevali Garcia, S. Gouriou, P. Duriez, N. Brahimi, E. Bingen, J. Elion, and E. Denamur. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roberts, J. A., M. B. Kaack, G. Baskin, and L. N. Martin. 1993. Events leading to septic death from experimental acute pyelonephritis in the monkey. J. Urol. 150:1030-1033. [DOI] [PubMed] [Google Scholar]
- 35.Russo, T. A., U. B. Carlino, and J. R. Johnson. 2001. Identification of ireA, a novel iron regulated virulence gene in an extraintestinal pathogenic isolate of Escherichia coli. Infect. Immun. 69:6209-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russo, T. A., and J. R. Johnson. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: an overlooked epidemic. Microbes Infect. 5:449-456. [DOI] [PubMed]
- 37.Russo, T. A., and J. R. Johnson. 2000. A proposal for an inclusive designation for extraintestinal pathogenic Escherichia coli: ExPEC. J. Infect. Dis. 181:1753-1754. [DOI] [PubMed] [Google Scholar]
- 38.Sanders, C. 2001. Mechanisms responsible for cross-resistance and dichotomous resistance among the quinolones. Clin. Infect. Dis. 32(Suppl. 1):S1-S8. [DOI] [PubMed] [Google Scholar]
- 39.Shoemaker, N. B., H. Vlamakis, K. Hayes, and A. A. Salyers. 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shooter, R. A., E. M. Cooke, S. O'Farrell, K. A. Bettelheim, M. E. Chandler, and F. M. Bushrod. 1974. The isolation of Escherichia coli from a poultry packing station and an abattoir. J. Hyg. 73:245-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, K. E., J. M. Besser, C. W. Hedberg, F. T. Leano, J. B. Bender, J. H. Wicklund, B. P. Johnson, K. A. Moore, M. T. Osterholm, and T. I. Team. 1999. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992-1998. N. Engl. J. Med. 340:1525-1532. [DOI] [PubMed] [Google Scholar]
- 42.van Belkum, A., W. Goessens, C. van der Schee, N. Lemmens-den Toom, M. C. Vos, J. Cornelissen, E. Lugtenburg, S. de Marie, H. Verbrugh, B. Lowenbuerg, and H. Endtz. 2001. Rapid emergence of ciprofloxacin-resistant enterobacteriaceae containing multiple gentamicin resistance-associated integrons in a Dutch hospital. Emerg. Infect. Dis. 7:862-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van den Bogaard, A. E., N. London, C. Driessen, and E. E. Stobberingh. 2001. Antibiotic resistance of faecal Escherichia coli in poultry, poultry farmers and poultry slaughterers. J. Antimicrob. Chemother. 47:763-771. [DOI] [PubMed] [Google Scholar]
- 44.Warren, J. W., E. Abrutyn, J. R. Hebel, J. R. Johnson, A. J. Schaffer, and W. E. Stamm. 1999. Guidelines for antimicrobial therapy of uncomplicated acute bacterial cystitis and acute pyelonephritis in women. Clin. Infect. Dis. 29:745-758. [DOI] [PubMed] [Google Scholar]
- 45.White, D., L. J. V. Piddock, J. J. Maurer, S. Zhao, V. Ricci, and S. Thayer. 2000. Characterization of fluoroquinolone resistance among veterinary isolates of avian Escherichia coli. Antimicrob. Agents Chemother. 44:2897-2899. [DOI] [PMC free article] [PubMed] [Google Scholar]