Abstract
We introduce a mathematical framework describing static response of networks occurring in molecular biology. This formalism has many similarities with the Laplace–Kirchhoff equations for electrical networks. We introduce the concept of graph boundary and we show how the response of the biological networks to external perturbations can be related to the Dirichlet or Neumann problems for the corresponding equations on the interaction graph. Solutions to these two problems are given in terms of path moduli (measuring path rigidity with respect to the propagation of interaction along the graph). Path moduli are related to loop products in the interaction graph via generalized Mason–Coates formulae. We apply our results to two specific biological examples: the lactose operon and the genetic regulation of lipogenesis. Our applications show consistency with experimental results and in the case of lipogenesis check some hypothesis on the behaviour of hepatic fatty acids on fasting.
Keywords: systems biology, gene and metabolic networks, steady-state shifts, Dirichlet-to-Neumann map
1. Introduction
Network-based representations are widely used to describe gene regulation or metabolic pathways at cellular level; graph theoretical properties of these biological networks, dynamical properties, as well as their inference from experimental data, were the subject of intensive studies (Wagner 2001; de Jong 2002; Kaminski & Friedman 2002; Kholodenko et al. 2002; de Jong et al. 2004; Thieffry & Sanchez 2004; Yamanishi et al. 2004). However, network inference techniques need huge amounts of data which are not always available or often have poor accuracy. Similarly, dynamical studies need accurate data.
In this paper, our goal is different. Instead of using data and gene perturbations for building networks from scratch, we develop mathematical techniques in order to refine the analysis of incomplete models or to compare models and data. Our mathematical results connect network topology and the response to steady-state shift experiments. Steady-state shift experiments are useful tools in chemistry allowing in principle to recover the reaction mechanisms (Chevalier et al. 1993). We argue that similar approaches are well adapted to differential microarray experiments which compare gene expressions between two different states (Kaminski & Friedman 2002).
A theory of steady-state shifts is, in fact, a theory of static response, linear when the shifts are small and nonlinear when the shifts are big. Theories of linear response were developed in various contexts such as condensed matter and in statistical physics (Kubo et al. 1998), electrical, physical and mechanical systems (MacFarlane 1970), chemical and biochemical systems (Oster et al. 1973; Perelson & Oster 1974), complex fluids (Larson 1988), metabolic networks (Kacser & Burns 1973; Heinrich & Rapoport 1974) and gene networks (Kholodenko et al. 2002; Vlad et al. 2004). The basic quantities in such theories are the susceptivities, representing derivatives (generically functional derivatives) of outputs with respect to inputs. Susceptivities (and also response in general) are obtained from constitutive equations that must be compatible with thermodynamics (Oster et al. 1973; Grmela 2001). Nonlinear response theories that apply to large perturbations are generally more difficult to handle (Larson 1988; Grmela 2001). Vlad et al. (2004) showed that under special ‘neutrality’ conditions the response of reaction–diffusion systems to special perturbations consisting in changing the abundance of some marked individuals is linear even at large perturbations.
In the static case, structural stability (Smale 1980; Ruelle 1989) suggests a simple approach to nonlinear response. Like in classical thermodynamics (Callen 1985), finite static response can be obtained by integrating the (state dependent) differential forms whose coefficients are the susceptivities, provided that the integration path does not contain singularities (phase transitions).
Susceptivities depend on the network topology and, therefore, can be used in reverse engineering. This method is complementary to already well-developed correlation analysis of gene and metabolic networks (Kaminski & Friedman 2002; Yamanishi et al. 2004) or of time-series in chemical reaction systems (Arkin & Ross 1995). Vlad et al. (2004) showed that under the neutrality condition the dynamical susceptivity of a system of chemical reactions with respect to abundance perturbations is the exponential of a connectivity matrix, which contains information about the chemical reaction mechanism. It is known (Chevalier et al. 1993) that small concentration shifts measurements allow to calculate the Jacobian of a chemical kinetics evolution equations. Very related to this is the work of Sontag (Kholodenko et al. 2002) and the older Kacser–Burns connectivity property from metabolic networks (Cornish-Bowden 1995), meaning that the matrix of elasticities (the Jacobian) is the inverse of the matrix of control coefficients (static susceptivities). The dynamical Jacobian is also important in gene networks dynamics. It defines the interaction graph (Soulé 2003), which is the natural mathematical candidate to represent the biologist's gene regulation graph (Thomas 1981). Systems of chemical reactions allow for other graphical representations (the interested reader could refer to the comprehensive reviews of Oster et al. 1973; Perelson & Oster 1974).
In this paper we generalize Mason–Coates type formulae from electrical circuits (Mason 1953; Coates 1959), providing graphical representations of static susceptivities of gene networks.
We also exploit useful connections between the static response of gene networks and the Dirichlet-to-Neumann map (Curtis & Morrow 2000), which is a relatively new field in graph theory. This field originated from a problem by Calderon (Calderon 1980) who asked whether it is possible to reconstruct the conductivity of a conducting plate from measurements of injected currents and voltages on its boundary. This problem gave rise to non-trivial developments in graph theory. Our most important result is related to the Dirichlet-to-Neumann map and consists in a systematic use of the notion of boundary. Boundaries serve to delimitate subgraphs or to couple subgraphs one with another like in Kholodenko et al. (2002). We define response of subgraphs as solutions of the Dirichlet or the Neumann problems. The novelty of our method is that boundaries delimitating analysed subgraphs are chosen according to the missing piece of information.
This paper is organized as follows. In §2, we present the interaction graph, the subgraphs and their boundaries. In §3, we define linear response and discuss an electrical network analogy. In §4, we construct Mason–Coates solutions to the linear Dirichlet and Neumann problems and we extend the applicability of the results to nonlinear response. Finally, we give two biological applications in §5.
2. Steady-state shifts, interaction graph
The state of a biological network is given by a vector , whose components are concentrations of various interacting actors, such as DNA regions coding for genes, RNA transcripts, various produced and regulating proteins, metabolites.
Here we consider a differential dynamics for X, dX/dt=F(X, P), , where stems for a set of parameters. This assumption is rather general, because Boolean and piecewise deterministic dynamics can be approximated by differential inclusions (Gouzé & Sari 2003). Although not proven in full generality but used in practice, dynamical systems with delays can be approximated by differential systems either by introducing extra variables (Belych 1998) or by centre manifold techniques (Wang & Hu 2001). The main restriction in our theory is that we consider non-degenerate stable steady states, which are hyperbolic fixed points of the dynamics. Steady states are solutions of the nonlinear system of equations:
| (2.1) |
Changes in the control parameters P produce steady-state shifts. If , where is the Jacobian calculated at a non-degenerate stable steady-state X0, then from the implicit function theorem equation (2.1) defines X as a function of P locally, in a neighbourhood of (X0, P0). This local dependence represents the static linear response of the system. In §4.4, we shall give a global existence theorem for such a function.
2.1 Interaction graph
The Jacobian J calculated in a state X introduces in a natural way a signed oriented graph, called the interaction graph which is a triplet (, , σ). The set of nodes ={1, …, n} consists of the interacting actors. The set of oriented edges ⊂× is defined by (j, i)∈, iff Jij≠0. The edge sign is a function , . An edge (j, i) is called positive whenever σ(j, i)>0 and negative whenever σ(j, i)<0.
From a biological point of view there is an arc from j to i if the actor j has a direct influence on the dynamics of i. This influence can be positive (activation) or negative (repression). Many examples can be considered. For instance, j may be a transcription factor regulating the expression of i, or a protein involved in the phosphorylation or methylation of i, or an enzyme involved in the production of i, etc.
When F is nonlinear, the signs of its partial derivatives may change, therefore the interaction graph generally depends on the state X where it is calculated. This is a weakness of the interaction graph compared with other more complex graphical representations such as ‘bond graphs’ (Oster et al. 1973; Perelson & Oster 1974). Nonetheless, this weakness is largely compensated by the fact that many qualitative properties of the dynamics and of steady states depend on topological conditions on the interaction graph (Thomas 1981; Gouzé 1998; Snoussi 1998; Soulé 2003).
2.2 Boundary
Boundary is a key concept of any modular approach trying to reduce the overwhelming complexity of large networks to understandable behaviour of simpler subsystems. Furthermore, data in molecular biology can be extensive, but it is rarely complete. Our modular approach to data analysis can be used to fill in missing information, to check existing data, or to correct eventual errors.
Because there is no precise criterion to fix the boundary between the actors that we consider and those that we forget, we shall allow a free choice of the boundary. We shall see that instead of becoming a handicap, this freedom of choice becomes a handy tool.
The construction by which we freely choose a boundary in an interaction graph is as follows. Let G be the set of nodes that we isolate in the larger set . The entrance boundary of G, denoted by ℸinG is the set of nodes of G that have incoming arcs from the exterior, i.e. from nodes of which are not nodes of G. The other nodes of G that are not directly influenced by the exterior are in the interior of G which is denoted .
2.3 Paths, loops, loop partitions
A path c=i⇝j is a sequence of nodes (i1=i, i2, …, ip=j) such that (ik, ik+1) is an edge of the graph and such that all the nodes in the sequence are visited just once. Note that such a path exists in the interaction graph if . A path i⇝j in the interaction graph is called positive if the path product of the Jacobian elements is positive (negative).
A loop l=i⇝i is a sequence of nodes (i1=i, i2, …, ip=i) such that (ik, ik+1) is an edge of the graph and such that all the nodes in the sequence are visited just once with the exception of the two terminals that coincide. A loop is called positive (negative) if the product of the Jacobian elements along it is positive (negative). Self-interaction loops (i, i) contain only one node and correspond to diagonal elements of the Jacobian Jii.
A loop partition of a graph is a partition of the nodes into disjoint loops (including self-interaction loops). The set of loop partitions of a subgraph G is denoted (G). For L∈(G), |L| denotes the number of loops in L. For instance, if G={1, 2, 3} and the oriented edges are {(1, 1), (2, 2), (3, 3), (1, 2), (2, 3)}, (G) consists of two loop partitions L1={(1, 1), (2, 2), (3, 3)}, |L1|=3 and L2={(1, 2, 3)}, |L2|=1.
3. Linear response of a network to influences from the exterior
3.1 Electrical networks, flowgraphs, Markov chains
Classical examples of linear networks are the linear electrical networks. Let us consider an electrical network with n nodes. Nodes i and j are connected by wires having admittances Yij=Yji. Injecting currents Ii in some or all of the nodes we produce the steady-state potentials Vi. Applying Ohm's law and Kirchhoff's first law to the node i it follows that . In matrix form, the linear relation between node voltages and node current sources reads
| (3.1) |
is the node admittance matrix and is obtained from the edge admittances
| (3.2) |
In order to express the voltages for a given current configuration we need to solve the system of Kirchhoff–Laplace equation (3.2).
Note that the matrix is singular: . This comes from a special symmetry of electrical networks meaning that voltages are determined up to a constant and that the only measurable quantities are voltage differences.
There are many other examples coming from different fields of science where linear equations are interpreted as graphs.
Markov or semi-Markov processes on multistate stochastic networks occur frequently in biology and physics and can be used for data analysis. For instance, the evolution of a disease can be treated as a series of stochastic transitions between various grades of disease (Yau & Huzurbazar 2002). The possible transitions and transition probabilities can be gathered on a flowgraph. A flowgraph is a weighted oriented graph. Each edge is labelled by a transition rate (or transmittance, or gain) which is the probability pji to perform the jump from the state j to the state i divided by the mean waiting time τji from a state j to i: tji=pji/τji. The probabilities πi of being in a state i satisfy the equilibrium equation which is same as equation (3.1) with zero currents I=0.
3.2 Analogy between steady-state shifts and electrical networks
In order to obtain an analogy between linear response of electrical and biological networks, let us consider that there is a non-degenerated stable state X0 satisfying equation (2.1) for parameters P0. Supposing that dynamics is structurally stable, the theorem on persistence of hyperbolic sets (Smale 1980; Ruelle 1989) implies that small variations of the parameters in equation (2.1) produce shifts of this state without loss of stability. Differentiating the equilibrium equations for the nodes in a subgraph G (including the boundary nodes) we obtain
| (3.3) |
Let us also consider that all the exterior influences on G are transmitted via its boundary. This means that
| (3.4) |
The condition in equation (3.4) is a modelling assumption, that can be lifted with minor changes. From a statical point of view the parameters P play the same role as the other variables. If the modeller judges that equation (3.4) is not reasonable, it is better to consider P as a variable and to connect it to the other variables.
Noting that for all k∈I\G and , it follows that
| (3.5) |
where denote the ‘forcing variations’ which are non-vanishing only on the boundary, and are defined by
| (3.6) |
In matrix form, equation (3.5) reads
| (3.7) |
Equations (3.1) and (3.7) are analogous. They represent the linear response of a network to influence from the exterior:
the opposite Jacobian −JG restricted to G and calculated at a non-degenerated stable steady state is analogous to the node admittance matrix ,
the concentration variations δX are analogous to the voltages V,
the forcing variations δXf are analogous to the injected currents I.
Equation (3.4) implies that the forcing variations are vanishing in the interior and are non-zero on the boundary of the subgraph G. For electrical networks this means that the influence of the exterior on a subnetwork can be represented by current sources on its boundary.
Electrical networks are different from biological networks because they have much more symmetries, such as the following.
The matrix −JG is not necessarily symmetric, while the matrix is always symmetric. The interaction graph is intrinsically oriented, while an electrical network is not oriented (a wire conducts in both directions).
The diagonal elements of the matrix are obtained from the non-diagonal ones: . The diagonal elements of the matrix JG are independent of the non-diagonal ones.
4. Topological expressions for the linear response of networks
Throughout this section we consider that the Jacobian JG is calculated at a non-degenerate steady state (stability can be imposed, but it is not compulsory). For subgraphs, steady states are constrained by imposed boundary variations or forcings. To simplify notation we drop the index G in the Jacobian; it will be supposed that the J is restricted to a subset G, unless otherwise stated. Supposing that J is invertible, solutions of equation (3.7) can be obtained from the inverse matrix J−1. In this section, the elements of J−1 are connected to the topology of the interaction graph.
4.1 Moduli and loops
We start with the following well-known relation (Bloom 1979)
| (4.1) |
where Δji is the minor obtained by deleting row j and column i in J, and Δ=det(J).
Next, we develop the minor Δji into a sum of principal minors (Bloom 1979)
| (4.2) |
where j⇝i is any path in the subgraph G leading from j to i, is the number of edges in the path, is the product of elements of J along this path, is the principal minor obtained by deleting all the rows and columns whose indices are included in the path, Δj is the principal minor obtained by deleting the line and the column j. Finally, principal minors of the Jacobian can be related to loops in the subgraph G
| (4.3) |
| (4.4) |
| (4.5) |
where lp(L) is the loop product defined as the product of elements of J along the loops of L. is the subgraph obtained by deleting from G all nodes in the path j⇝i, while Gi is obtained by deleting node i. dim denotes the dimension of the minor.
We now introduce a quantity later referred to as modulus which measures rigidity of the network and can be seen as the inverse of sensibility, as shown later.
Let G be a subgraph of , let Δ, Δi, be minors of the Jacobian restricted to G and defined as above. The modulus of a path j⇝i is
(4.6) The modulus of a node i is
(4.7)
We shall usually consider non-degenerate steady states and Δ≠0, therefore moduli never vanish; they can instead diverge when minors of J vanish.
4.2 Neumann and Dirichlet problems
In electrical networks, fulfilment of Kirchhoff–Laplace equation (3.1) implies a unique set of interior voltages (up to a constant) due to imposed boundary voltages (Dirichlet problem) or a unique set of voltages (up to a constant) everywhere due to imposed boundary currents (Neumann problem). For biological networks, the Neumann and the Dirichlet problems have the following equivalents.
- Linear Dirichlet problem: determine the variations δXi, of the interior nodes, when the values of the variations are imposed (or known) on the boundary δXi, i∈ℸinG. For biological networks the Dirichlet problem shows its utility when one possesses only a partial knowledge of the system (Kuipers 1994; Siegel et al. in press). We can define the corresponding Dirichlet static susceptivities
(4.8) - Linear Neumann problem: determine the variations δXi, i∈G everywhere, when the forced variations are imposed on the boundary , i∈ℸinG. Unlike injected currents forced variations (defined by equations (3.6)) cannot be measured directly. In electrical networks, currents can be measured as voltages across calibrated resistors of apparatuses coupled to the network. We cannot see what can be a calibrated molecular apparatus for biological networks. In spite of this drawback, the Neumann problem is important in theory of control of biological networks: it describes shifts of equilibria under constraints imposed by the change of external conditions. The corresponding static susceptivities are defined as
(4.9)
The following theorem states the existence and gives a solution to the Neumann problem.
Let G be a subgraph of . Let J be the restriction of the Jacobian of F to G, calculated at a non-degenerated, Neumann constrained (given forcings) steady state: , i, j∈G. Let G denote the set of paths included in G. If det(J)≠0 and if there is no direct influence of the parameters on the nodes of G, i.e. , ∀i∈G, then the response of i∈G to small changes of the exterior of G satisfies
(4.10)
(4.11)
We have supposed that Δ=det(J)≠0. According to equations (4.6) and (4.7), we can supplement equations (4.10) and (4.11) with the following formal rule coping with diverging moduli: 1/∞=0.
Using a language coming half from mechanics, half from flowgraphs we can say that a ‘force’ propagates from the boundary node j along the path j⇝i. This force is bigger when the product of interaction coefficients along the path is bigger. is the ratio force/response and, therefore, can be called ‘path modulus’. A large path modulus implies a small response at the end of the path, even if the force is big. Therefore, the modulus is the inverse of sensitivity.
Equation (4.10) expresses interior δXi as functions of the boundary forcings . Equation (4.11) gives the relation between forcings and variations on the boundary. The corresponding linear mapping (m=#ℸinG, ) is the equivalent of the Neumann-to-Dirichlet map for electrical networks (Curtis & Morrow 1991). The inverse (if it exists) of this mapping is the Dirichlet-to-Neumann map (Curtis & Morrow 1991).
Theorem 4.2 implies the following.
If there are no paths in G connecting two different nodes on the boundary (boundary is not necessarily disconnected, because connections passing through the exterior are allowed), and if for all nodes i∈ℸinG, moduli Ci are finite and non-vanishing, then the Neumann-to-Dirichlet mapping is diagonal and its inverse exists. In this case, the forcings are proportional to the variations on the boundary
(4.12)
By using the Dirichlet-to-Neumann map we can associate to any solution of the Neumann problem, a solution of the Dirichlet problem. We can thus obtain formulae for the solutions of the Dirichlet problem. In Siegel et al. (in press), we have used a direct method to obtain solutions to the Dirichlet problem. Briefly, we have used equilibrium equations for the interior nodes in the same way as in theorem 4.2 (which uses equilibrium equations for all the nodes including the boundary). Let us state the result.
Let G be a subgraph of . Let be the interior of G. Let be the restriction of the Jacobian of F to calculated in a non-degenerated, Dirichlet constrained (given boundary values, free interior) steady state: , . Let us suppose that and that there is no direct influence of the parameters on the interior nodes of G, i.e. .
Then, the response of to small changes on the boundary of G is given by
(4.13)
G denotes the set of paths included in G, starting on the boundary and that do not return to the boundary.
denotes the path modulus of the internal path k(j)⇝i where is the second node after j of the path j⇝i. If k(j)=i, then .
Theorems 4.2 and 4.6 make no assumption on the connectivity of the subgraph G. For disconnected internal nodes we should have δXi=0, meaning that at steady states Xi does not depend on the parameters. For such a node the stationarity equations read Fi(Xi)=0.
Equations (4.6) and (4.7) give the loop products expression of moduli. They generalize Mason and Coates gain formulae (Mason 1953; Coates 1959) from electrical networks. Susceptivities are related to moduli according to the following equations
| (4.14) |
| (4.15) |
Let us note that the restriction of the Neumann susceptivity matrix to the boundary is the Neumann-to-Dirichlet map.
4.3 Signs of moduli
The signs of moduli are important for qualitative discussions of the transmission of influences (Siegel et al. in press). Equations (4.14) and (4.15) lead to an interesting possibility. Even if all the paths from j to i correspond to globally positive regulation (path products are all positive), it is possible to have negative susceptivity χij, which means negative correlation between Xi and Xj. This possibility occurs when the moduli are negative (see §5.2) and is a feedback effect. Indeed, from equations (4.6) and (4.7) it follows:
If the subgraph G contains no positive loops for a given non-degenerate constrained steady state, then all its path moduli and node moduli Ci are positive for that state.
4.4 Significance and limits of the approach
Equations (4.10), (4.11) and (4.13) should be interpreted as differential constraints connecting small variations of the concentrations at steady state, when this steady state is shifted as a result of the change of parameters. The significance of these constraints is the propagation of influence along the interaction graph: for any analysed subset of the interacting species concentration variations of internal species are obtained from concentration variations of boundary species. Mathematically, these constraints express a local functional relation between the internal variables and the boundary variables, which follows from the implicit function theorem. Let us note that we have not addressed yet the question of the global existence of a unique implicit function expressing the internal variables as functions of the boundary variables (the nonlinear Dirichlet problem). The existence of such a function depends on the nonlinearities of the system and cannot be proven without further additional hypothesis. Without being exhaustive, let us give here some results applying to particular cases.
Let us consider that for certain subsets G⊂ we are able to specify global interiors, i.e. sets of species that are never directly influenced by external species, whatever the concentrations. The steady-state equations for internal species constrained by boundary species read
| (4.16) |
where X, X′ have coordinates in , ℸinG, respectively.
The implicit function theorem implies that X is a function of X′ in a neighbourhood of any non-degenerate steady state satisfying . The global existence of the implicit function follows from an existence result for the solution of the system (4.16) and from the following global univalence theorem ensuring the uniqueness of this solution.
Let us consider that , where λi>0 and Φi(X, X′) are differentiable, bounded and satisfy
(4.17) Then for any X′ the system (4.16) has at least a solution X such that all concentrations Xi, are positive.
Let us consider that the interaction subgraph corresponding to the interior of G has no positive loop, whatever the concentrations may be. Then, the solution of the system (4.16) is unique for any X′.
We shall only give a brief outline of the proof. The first part of theorem 4.9 is based on the following standard mathematical lemma which is a consequence of Poincaré–Hopf formula.
Let D be a smooth ball in a metric space, let S be the boundary of D. Let F be a vector field defined on a neighbourhood of D. If F points inward D at any point of S, then F admits a zero in the interior of D.
The second part of the theorem is known as the Thomas's conjecture (Thomas 1981; Gouzé 1998; Snoussi 1998; Soulé 2003) and can be proven as follows. When the subgraph G has no positive loops whatever the concentrations, equations (4.3)–(4.5) imply that all the minors of the opposite Jacobian −J are positive. From this and from the Gale–Nikaido–Inada theorem (Parthasarathy 1983) it follows the uniqueness of the equilibrium. The complete proof of Thomas's conjecture can be found in Soulé (2003).
The conditions of the first part of theorem 4.9 are fulfilled by gene networks. Indeed, the terms λiXi correspond to degradation or to dilution produced by cell growth. The production terms Φi are bounded because they saturate at large concentrations. Equation (4.17) is natural, meaning that concentrations can never become negative.
We come now to the experimental significance of our approach. The type of experiment that we consider is the following: the set of parameters is changed such that we start and we end in a non-degenerate stable steady state. If the conditions of theorem 4.9 are fulfilled then the nonlinear Dirichlet problem has a unique solution and the internal variables are constrained to be univoque functions of the boundary variables. The finite variations of these functions between the start and the end of the experiment can be obtained by integrating the differential form in equation (4.13) along any differentiable path connecting the two steady states and made of non-degenerate constrained steady states (non-degenerate solutions of equation (4.16))
| (4.18) |
The uniqueness of the implicit function ensures the path independence of the result. We must emphasize that equation (4.18) is not destined to provide numerical values. It serves to identify the signs of different contributions to the concentration variations and leads to qualitative equations in sign algebra (Kuipers 1994; details can be found in Siegel et al. in press). Similar results hold for shifts of Neumann constrained steady states (nonlinear Neumann problem).
This result fails when the steady-state shift meets a singular point, defined by vanishing moduli (infinite sensitivity). Then, the steady state loses stability. The new attractor is either a distance apart (for instance in the cusp catastrophe), or it has a different type (for instance in the Hopf bifurcation a point attractor becomes a stable limit cycle) from the old one.
The cusp catastrophe occurs rather often in molecular biology. It occurs in the well-studied lactose operon, that we shall present in §5. This situation is represented in figure 1. The boundary variables , influence the internal variable X3. Suppose that changes. Then, the state of the system moves along the trajectories starting from P. For small changes ending in P1 there is a differentiable path of stable steady states connecting P and P1 and we can apply equation (4.18). For large changes ending in P2 there is no such path. The steady-state shift is discontinuous: the state jumps from the attractor branch to the branch . Equation (4.18) has to be changed by adding a finite jump .
Figure 1.
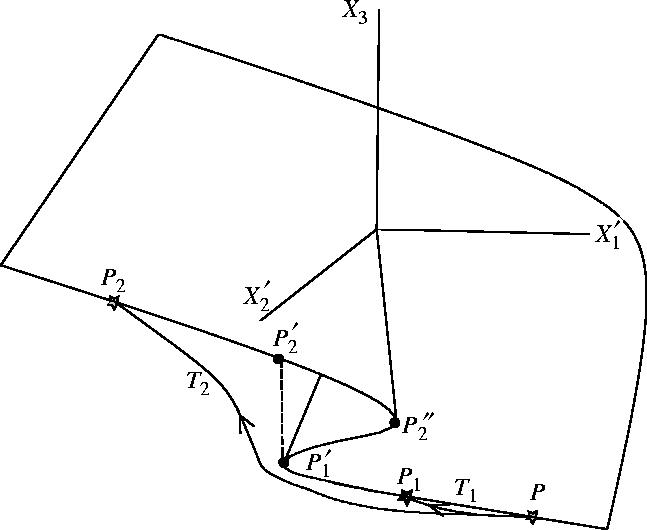
Cusp catastrophe. Two trajectories T1 and T2 start in the steady state P and end in the steady states P1 and P2, respectively. In order to find the variation of the internal variable X3 one could integrate the differential response along the smooth branch PP1 (for T1) and along the branches and (for T2). For T2 the finite jump should be added.
5. Examples
5.1 Lactose operon
The main enzymes for the lactose (L) metabolism in Escherichia coli are LacY (lactose permease) allowing the uptake of lactose, LacZ (β-galactosidase) catalysing the degradation of lactose to glucose (Yildirim & Mackey 2003; Mackey et al. 2004).
The transcriptional regulators for the genes lacY and lacZ are an activator (cAMP receptor protein, CRP) and a repressor (LacI). An inducer (cAMP) binds to the activator and triggers it. Lactose, under an isomeric form named allolactose, binds to the repressor and inhibits it. The glucose inhibits the activator. The interaction graph for this system is represented in figure 2. The exterior and interior lactose are denoted as Le, Li, respectively.
Figure 2.
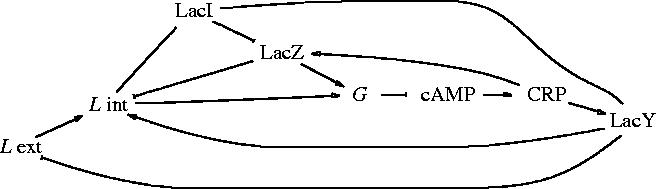
Operon lactose interaction network. L int and L ext stand for Li and Le, respectively.
Negative self-interaction loops should be added to each node, in order to take into account degradation processes or dilution produced by cell growth. These correspond to negative diagonal elements for the Jacobian (self-interactions).
There are four loops in the interaction graph: two positive (even number of negative regulations) ones , and three negative (odd number of negative regulations) ones , , . The existence of positive loops is a necessary condition (Soulé 2003) for the observed bistability of the operon lactose.
Experiments show that when Le is increased, the operon switches from a transcriptionally blocked, lactose poor state to a transcriptionally active, lactose rich state. We intend to check whether the linear response of the model given in figure 2 is coherent to observed variations, namely whether the Dirichlet susceptivity is positive.
From theorem 4.6, , where from the construction of the model (see figure 2). This means that the influence of Le on Li is positive provided that the modulus of the path Le⇝Li is positive. The modulus is given by
| (5.1) |
where is the absolute value of the self-interaction on Li, is the loop product divided by the product of absolute values of self-interactions for nodes in the loop. is positive for positive loops and negative for negative loops.
The condition for a monotonic response is given by the following.
The dependence of Li on Le is monotonically increasing provided that the modulus is positive, i.e.
(5.2)
The monotonicity condition (5.2) is valid for small positive loops products. The typical scenario for strong positive loops is the cusp catastrophe: the operon becomes bistable and its response is discontinuous and hysteretic (Mackey et al. 2004). Bistability requires the existence of a branch of states with negative moduli on which the condition (5.2) is broken, but this branch is usually unstable (see also §5.2).
The susceptivity approach can predict the effect of gene knock-outs. For instance, a knock-out of the inducer or of the activator cancels the loops , , . According to equation (5.1), the modulus increases, therefore the uptake of lactose decreases, provided that . Similarly, a knock-out of the repressor cancels the loops , . The uptake of lactose decreases provided that .
5.2 Negative moduli and non-monotonic response
For the lactose operon the input/output relation relating outside and inside lactose is monotonic and the moduli for stable branches are positive. We build here an artificial example of switch with non-monotonic response.
Let us suppose that X1 regulates positively X2 and that the modulus . Then increasing X1 produces a decrease in X2. In order to create an example of this kind one should consider at least another component X3 of the system. Indeed, stability asks for eigenvalues of the Jacobian to be in the left half of the complex plane; in dimension 2 this is equivalent to the absence of positive loops, hence moduli are positive at steady states (property 4.8). A simple example illustrating the possibility of having negative moduli is
| (5.3) |
with . The steady state is shifted by changing the parameter λ, which at equilibrium is equivalent to changing the boundary value x1=1+λ.
The solution of the Dirichlet problem is , , where
| (5.4) |
| (5.5) |
The model has unique equilibrium for a<3/2 and is bistable for a>3/2. In the monostable regime (a<3/2) everywhere; when x3 is inside the interval and is positive outside the interval I1. For and, therefore, the susceptivity vanishes: the response curve for x2 represented in figure 3 have a maximum and a minimum at these points.
Figure 3.
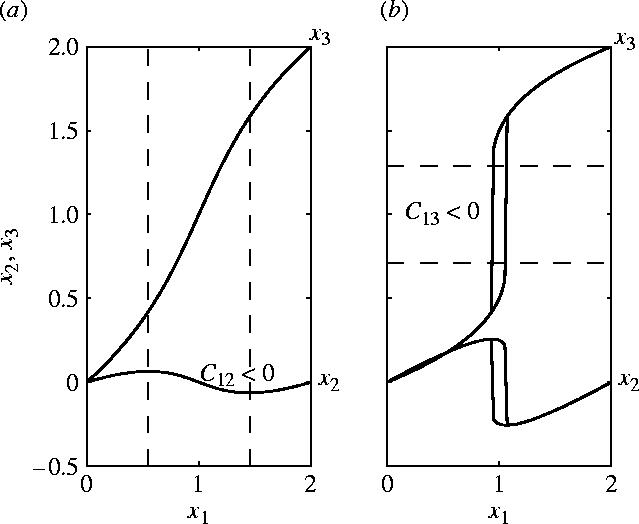
Solution of the Dirichlet problem for the model in equation (5.3): stable equilibrium values of x2 and x3 as a function of the boundary value x1. (a) Monostable (a=0.5) with a region (between dashed vertical lines) of negative modulus . (b) Bistable (a=2) with discontinuous, hysteretic response; the response of x2 is non-monotonic, the response of x3 is monotonic. can be negative when x3 is between the two dashed horizontal lines, but the corresponding states are unstable.
For a>3/2, the system is bistable. We have unless when and unless when . The region I3 where is in fact unstable and is never reached. When x1 increases x3 jumps discontinuously from one attractor to another without taking values inside the interval I3. The jump occurs at different values of x1 on increase and decrease of the control parameter. Thus, the response of x3 is discontinuous and hysteretic, but monotonic. The response of x2 is also discontinuous and hysteretic, but non-monotonic. There is a small region on the response curve of x2 where ; this region corresponds to the intersection between the stability domain for x3 and I2.
5.3 Regulation of lipogenesis in hepatocytes
5.3.1 Interaction model
Two ways of production of fatty acids coexist in liver. Saturated and mono-unsaturated fatty acids are produced from citrates thanks to a metabolic pathway composed of four enzymes, namely ATP citrate lyase (ACL), acetyl-Coenzyme A carboxylase (ACC), fatty acid synthase (FAS) and Stearoyl-CoA desaturase 1 (SCD1). Polyunsaturated fatty acids (PUFA) are synthesized from essential fatty acids provided by nutrition; Delta-5 Desaturase (D5D) and Delta-6 Desaturase (D6D) catalyse the key steps of the synthesis of PUFA.
PUFA plays pivotal roles in many biological functions; among them, they regulate the expression of genes that impact on lipid, carbohydrate and protein metabolism. The effects of PUFA are mediated either directly through their specific binding to various nuclear receptors (PPARα—peroxisome proliferator activated receptors, LXRα—Liver-X-Receptor α, HNF-4α) leading to changes in the trans-activating activity of these transcription factors; or indirectly as the result of changes in the abundance of regulatory transcription factors (SREBP-1c—sterol regulatory element binding protein, ChREBP—carbohydrate response element binding protein, etc.; Jump 2004).
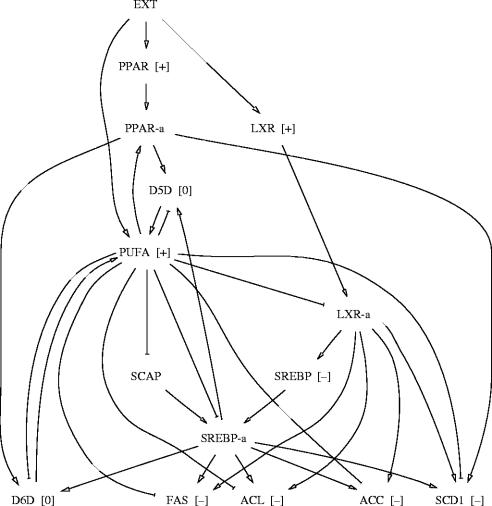
We consider in our model nuclear receptors PPARα, LXRα, SREBP-1c (denoted by PPAR, LXR, SREBP, respectively, in the model), as they are synthesized from the corresponding genes and the trans-activating active forms of these transcription factors, i.e. LXR-a (denoting a complex LXRα:RXRα), PPAR-a (denoting a complex PPARα:RXRα) and SREBP-a (denoting the cleaved form of SREBP-1c). SCAP—(SREBP cleavage activating protein) is a key enzyme involved in the cleavage of SREBP-1c. We also include in the model ‘final’ products, i.e. enzymes ACL, ACC, FAS, SCD1 (implied in the fatty acid synthesis from citrate), D5D, D6D (implied in PUFA synthesis) as well as PUFA themselves.
Relations between the variables are the following. SREBP-a is an activator of the transcription of ACL, ACC, FAS, SCD1, D5D and D6D (Jump 2004). LXR-a is a direct activator of the transcription of SREBP and FAS, it also indirectly activates ACL, ACC and SCD1 (Steffensen & Gustafsson 2004). Note that these indirect actions are kept in the model because we do not know whether they are only SREBP-mediated. PUFA activates the formation of PPAR-a from PPAR, and inhibits the formation of LXR-a from LXR as well as the formation of SREBP-a (by inducing the degradation of mRNA and inhibiting the cleavage; Jump 2004). SCAP represents the activators of the formation of SREBP-a from SREBP, and is inhibited by PUFA. PPAR directly activates the production of SCD1, D5D, D6D (Miller & Ntambi 1996; Matsuzaka et al. 2002; Tang et al. 2003).
The interaction graph for this model is shown in figure 4. Like in the lactose operon example, for each node we have supposed the existence of negative self-interaction loops.
Figure 4.
Interaction graph for a model of regulation of the synthesis of fatty acids. The node EXT represents the exterior world. Negative self-interaction loops on nodes are omitted for the sake of clarity. Labelled loops: , , , , l+={PUFA, PPAR-a, D6D, PUFA}. Labelled paths p={PUFA, LXR-a, SREBP}, p′={LXR, LXR-a, SREBP}, p1={PUFA, D6D}, p2={PUFA, SCAP, SREBP-a, D6D}, p3={D6D, PUFA}, p4={PUFA, LXR-a, SREBP, SREBP-a, D6D}, p5={LXR, LXR-a, SREBP, SREBP-a, D6D}, p6={PPAR, PPAR-a, D6D}, p7={PUFA, PPAR-a, D6D}, p8={PUFA, SREBP-a, D6D}, p9={PPAR, PPAR-a, D6D, PUFA}, p10={LXR, LXR-a, SREBP, SREBP-a, D6D, PUFA}.
5.3.2 Fasting-refeeding protocols
The fasting-refeeding protocols are suited for studying lipogenesis regulation; during an experimentation, animals (rodents or chicken) were kept fasting during several hours and then refed. Hepatic mRNA of LXR, SREBP, PPAR, ACL, ACC and SCD1 were quantified by DNA microarray analysis. PUFA variations were determined by biochemical measurements.
A compilation of recent literature on lipogenesis regulation provides results of such protocols: SREBP, ACL, ACC, FAS and SCD1 decline in liver during fasting (Liang et al. 2002); this state is characterized by an inhibition of fatty acid synthesis and an activation of the fatty acid oxidation. However, Tobin et al. (2000) showed that fasting rats for 24 h increased the hepatic LXR mRNA and Matsuzaka et al. (2002) observe no difference in either the hepatic D5D or D6D mRNA level between fasting and refeeding in normal mouse livers.
5.3.3 Static response and topology
One of the advantages of the approach that we present here is that we have no difficulty in focusing on subgraphs of a large network. In order to illustrate this possibility we considered the following biological questions:
Question 1. Lee et al. studied the fatty acids profiles in triglycerides TG, which are the predominant (more than 50%) hepatic lipids and also in phospholipids PL which go into cellular membranes. Moreover PL contribute much less than TG to the total lipid mass. These authors (Lee et al. 2004) show that after 72 h of fasting fatty acids profiles do not change significantly in PL, but there is a strong increase of TG and of their fatty acids constituents, in particular PUFA. Let us recall that TG are transient storage forms of fatty acids. Based on these experimental findings, we make the hypothesis of a mass increase of regulating PUFA in the hepatic cell. Can we prove this hypothesis indirectly by using the incomplete model, i.e. considering that all variations excluding PUFA are known?
Question 2. The dual regulation of desaturases SCD1, D5D and D6D by SREBP and PPAR is paradoxical, because SREBP transactivates genes for fatty acid synthesis in liver, while PPAR induces enzymes for fatty acid oxidation. Furthermore, all three desaturases have similar regulation. Nonetheless, on fasting SCD1 decreases, while D5D, D6D have non-significant variations. How does the model cope with this?
In order to answer Question 1, let us consider the Dirichlet problem for the subgraph G1={PUFA, LXR, LXR-a, SREBP}. From theorem 4.6 and the paths labels from figure 4 it follows that
| (5.6) |
Because , , δSREBP<0, δLXR>0, the only possible variation compatible with equation (5.6) is δPUFA>0. Applying a similar reasoning to the subgraph G2={PUFA, LXR, LXR-a, SREBP, SREBP-a, SCAP, FAS} we have the following.
The only possible variation compatible with δSREBP<0 and δLXR<0 is δPUFA>0. The only possible variation compatible with δFAS<0 and δLXR<0, is δPUFA>0. This proves our hypothesis that regulating PUFA increase during fasting.
In order to answer to the Question 2, let us see how the model connects the variations of PUFA and D6D. PUFA being on the boundary we need to consider the Neumann problem. Let us consider the subgraph G3={X=PUFA, W=PPAR, PPAR-a, Z=D6D, SREBP-a, SREBP, LXR-a, U=LXR, SCAP}. Information from literature suggests that G3 has the boundary ℸinG3={U, W, X}, meaning that all external interaction on G3 acts via LXR, PPAR, PUFA. Nevertheless, in order to understand the differential behaviour of D6D and SCD1, let us suppose that D6D has unknown extra regulation that is not represented. This can be taken into account easily by including D6D in the boundary: ℸinG3={U, W, X, Z}.
From theorem 4.2 and the paths labels from figure 4 it follows that
| (5.7) |
where are path products divided by the products of the absolute values of the self-interactions χ of nodes in the paths. The modulus is
| (5.8) |
Equations (5.7) and (5.8) reveal the importance of the dual regulation of D6D by PPAR and SREBP. The existence of the positive loop l+ involving PUFA, D6D and PPAR-a decreases the modulus in equation (5.8) therefore, according to equation (5.7), it boosts PUFA variations. The biological significance of this has been pointed out by Nakamura & Nara (2003) who refer to this phenomenon as a compensatory reaction to the increased demand of PUFA caused by oxidation at fasting.
In equation (5.7), the D6D variation δZ is the sum of different contributions of different signs: the influences transmitted along the paths p1, p2, p4, p8 are negative and those transmitted along the paths p7, p5, p6 are positive. The model is, therefore, compatible with compensated variations δZ=0, even in absence of unknown interaction whose effect is the forcing δZf. Nevertheless, it is difficult to explain the difference between D6D and SCD1 without the extra forcing, because this forcing is the only difference between the two products in the model. Nakamura & Nara (2003) arrive at a similar conclusion in their analysis of the regulation of D6D. To our knowledge, the nature of this extra regulation is still unknown.
6. Discussion
Static response provides quantitative and qualitative information on how the steady state is changed by forcings.
We have shown here how to relate static response of a biological network to the topology of its interaction graph. The key ingredient is the use of moduli that express the rigidity (opposed to sensitivity) of the network. Mason–Coates formulae from electrical networks computes moduli from loop products and were applied to biological networks. A general consequence of these formulae is that positive (negative) loops decrease (increase) moduli and rigidity. Static susceptivity was related to moduli by using path expansions. An interesting possibility arises when moduli and susceptivities have opposite signs: in this case the sign of the correlation between two nodes can be opposite to the sign of the regulation paths connecting them.
Susceptivities characterize linear response which generally applies to small variations of the concentrations. We showed that in the absence of bifurcations, local linear response can be integrated in order to obtain nonlinear response. This argument justifies qualitative analysis techniques applying to situations when the interactions have constant signs and when the constrained steady states of analysed subgraphs do not bifurcate under forcings. Our formalism is well adapted for qualitative analysis of differential transcriptional, metabolic or proteomic data. It allows several types of analysis: checking the consistency between model and data, or between different types of data, filling in missing information, and predicting the effects of gene knock-outs or of genetic deficiencies. Qualitative analysis of biological networks has been described in detail in Siegel et al. (in press).
An important ingredient in our approach is the systematic use of boundaries. This allows to separate subgraphs for independent study. The type of biological problem dictates the choice of the subgraph and the type of boundary conditions (Neumann or Dirichlet).
Boundaries could be useful in further developments such as hierarchical connection of subgraphs. Subgraphs can be connected in series (the entrance boundary of one is included in the exit boundary of another) or in parallel (boundary subsets are common) and their equivalent response can be obtained from equations (4.10) and (4.11). Another promising direction is the reduction of complexity of biological networks. One would like to classify all the graph contractions that preserve the response of the system. This has been done for planar electrical networks (Curtis et al. 1998) in relation to the Dirichlet-to-Neumann map. There is no similar work for biological networks.
Acknowledgments
This research was supported by ACI IMPBio, a French Ministry for Research program on interdisciplinarity. We thank M. Crouzeix, A. Gorban, C. Soulé and J. Scott for inspiring discussions. We thank E. Pécou for suggesting us to use Lemma 4.10, and the reviewers for constructive comments.
Footnotes
One contribution of 8 to a themed supplement ‘Statistical mechanics of molecular and cellular biological systems’.
References
- Arkin A, Ross J. Statistical construction of chemical reaction mechanisms from measured time-series. J. Phys. Chem. 1995;99:970–979. doi:10.1021/j100003a020 [Google Scholar]
- Belych L. On the problem of replacing an ordinary differential equation with constant time delay by the system of ordinary differential equations. Russ. J. Numer. Anal. Math. Model. 1998;13:169–190. [Google Scholar]
- Bloom D. Cambridge University Press; Cambridge: 1979. Linear algebra and geometry. [Google Scholar]
- Calderon A. Seminar on Numerical Analysis and its Applications to Continuum Physics. Soc. Brasileira de Mathematica; Rio de Janeiro: 1980. On an inverse boundary value problem; pp. 65–73. [Google Scholar]
- Callen H. Wiley; New York: 1985. Thermodynamics and a introduction to thermostatics. [Google Scholar]
- Chevalier T, Schreiber T, Ross J. Toward a systematic determination of complex reaction mechanisms. J. Phys. Chem. 1993;97:6776–6787. doi:10.1021/j100128a006 [Google Scholar]
- Coates C. Flow-graph solutions of linear algebraic equations. IRE Trans. Circuit Theory. 1959;CT-6:170–187. [Google Scholar]
- Cornish-Bowden A. Portland Press; London: 1995. Fundamentals of enzyme kinetics. [Google Scholar]
- Curtis E, Morrow J. The Dirichlet to Neumann map for a resistor network. SIAM J. Appl. Math. 1991;51:1011–1029. doi:10.1137/0151051 [Google Scholar]
- Curtis E, Morrow J. World Scientific; Singapore: 2000. Inverse problems for electrical networks. [Google Scholar]
- Curtis E, Ingerman D, Morrow J. Circular planar graphs and resistor networks. Linear Algebra Appl. 1998;283:115–150. doi:10.1016/S0024-3795(98)10087-3 [Google Scholar]
- de Jong H. Modeling and simulation of genetic regulatory systems: a literature review. J. Comput. Biol. 2002;9:69–105. doi: 10.1089/10665270252833208. doi:10.1089/10665270252833208 [DOI] [PubMed] [Google Scholar]
- de Jong H, Gouzé J.-L, Hernandez C, Page M, Sari T, Geiselmann J. Qualitative simulation of genetic regulatory networks using piecewise-linear models. Bull. Math. Biol. 2004;66:301–340. doi: 10.1016/j.bulm.2003.08.010. doi:10.1016/j.bulm.2003.08.010 [DOI] [PubMed] [Google Scholar]
- Gouzé J.-L. Positive and negative circuits in dynamical systems. J. Biol. Syst. 1998;6:11–15. [Google Scholar]
- Gouzé J.-L, Sari T. A class of piecewise linear differential equations arising in biological models. Dyn. Syst. 2003;17:299–316. [Google Scholar]
- Grmela M. Complex fluids subjected to external influences. J. Non-Newtonian Fluid Mech. 2001;96:221–254. doi:10.1016/S0377-0257(00)00186-5 [Google Scholar]
- Heinrich R, Rapoport T. A linear steady-state treatment of enzymatic chains. General properties, and effector strength. Eur. J. Biochem. 1974;42:89–95. doi: 10.1111/j.1432-1033.1974.tb03318.x. doi:10.1111/j.1432-1033.1974.tb03318.x [DOI] [PubMed] [Google Scholar]
- Jump D. Fatty acid regulation of gene transcription. Crit. Rev. Clin. Lab. Sci. 2004;41:41–78. doi: 10.1080/10408360490278341. doi:10.1080/10408360490278341 [DOI] [PubMed] [Google Scholar]
- Kacser H, Burns J. The control of flux. Symp. Soc. Exp. Biol. 1973;27:65–104. [PubMed] [Google Scholar]
- Kaminski N, Friedman N. Practical approaches to analyzing results of microarray experiments. Am. J. Respir. Cell Mol. Biol. 2002;27:125–113. doi: 10.1165/ajrcmb.27.2.f247. [DOI] [PubMed] [Google Scholar]
- Kholodenko B, Kiyatkin A, Bruggeman F, Sontag E, Westerhoff H, Hoek J. Untangling the wires: a strategy to trace functional interactions in signaling and gene networks. Proc. Natl Acad. Sci. USA. 2002;99:12841–12846. doi: 10.1073/pnas.192442699. doi:10.1073/pnas.192442699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo R, Toda M, Hashitsume N. Springer; Berlin: 1998. Statistical physics II. Nonequilibrium statistical mechanics. [Google Scholar]
- Kuipers B. MIT Press; Cambridge, MA: 1994. Qualitative reasoning. Modeling and simulation with incomplete knowledge. [Google Scholar]
- Larson R. Butterworths; Boston: 1988. Constitutive equations for polymer melts and solutions. [Google Scholar]
- Lee S, Chan W, Lo C, Wan D, Tsang D, Cheung W. Requirement of PPARα in maintaining phospholipid and triacylglycerol homeostasis during energy deprivation. J. Lipid Res. 2004;45:2025–2037. doi: 10.1194/jlr.M400078-JLR200. doi:10.1194/jlr.M400078-JLR200 [DOI] [PubMed] [Google Scholar]
- Liang G, Yang J, Horton J, Hammer R, Goldstein J, Brown M. Diminished hepatic response to fasting/refeeding and liver×receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J. Biol. Chem. 2002;277:9520–9528. doi: 10.1074/jbc.M111421200. doi:10.1074/jbc.M111421200 [DOI] [PubMed] [Google Scholar]
- MacFarlane A. Harap; London: 1970. Dynamical systems models. [Google Scholar]
- Mackey M, Santillán M, Yildirim N. Modeling operon dynamics: the tryptophan and lactose operons as paradigms. C. R. Biol. 2004;327:211–224. doi: 10.1016/j.crvi.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Mason S. Feedback theory—properties of signal flow graphs. Proc. IRE. 1953;41:1144–1156. [Google Scholar]
- Matsuzaka T, et al. Dual regulation of mouse delta(5)- and delta(6)-desaturase gene expression by srebp-1 and pparalpha. J. Lipid Res. 2002;43:107–114. [PubMed] [Google Scholar]
- Miller C, Ntambi J. Peroxisome proliferators induce mouse liver stearoyl-coa desaturase 1 gene expression. Proc. Natl Acad. Sci. USA. 1996;93:9443–9448. doi: 10.1073/pnas.93.18.9443. doi:10.1073/pnas.93.18.9443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Nara T. Essential fatty acid synthesis and its regulation. Prostglandins Leukot. Essent. Fatty Acids. 2003;68:145–150. doi: 10.1016/s0952-3278(02)00264-8. doi:10.1016/S0952-3278(02)00264-8 [DOI] [PubMed] [Google Scholar]
- Oster G, Perelson A, Katchalsky A. Network thermodynamics: dynamic modelling of biophysical systems. Q. Rev. Biophys. 1973;6:1–134. doi: 10.1017/s0033583500000081. [DOI] [PubMed] [Google Scholar]
- Parthasarathy T. Lecture Notes in Mathematics. vol. 977. Springer; Berlin: 1983. On global univalence theorems. [Google Scholar]
- Perelson A, Oster G. Chemical reaction dynamics. Part II. Reaction networks. Arch. Ration. Mech. Anal. 1974;57:31–98. doi:10.1007/BF00287096 [Google Scholar]
- Ruelle D. Academic Press; Boston: 1989. Elements of differentiable dynamics and bifurcation theory. [Google Scholar]
- Siegel, A., Radulescu, O., Le Borgne, M., Veber, P., Ouy, J. & Lagarrigue, S. In press. Qualitative analysis of the relation between DNA microarray data and behavioral models of regulation networks. Biosystems [DOI] [PubMed]
- Smale S. Springer; New York: 1980. Mathematics of time. [Google Scholar]
- Snoussi E. Necessary conditions for multistationarity and stable periodicity. J. Biol. Syst. 1998;6:3–9. doi:10.1142/S0218339098000042 [Google Scholar]
- Soulé C. Graphic requirements for multistationarity. Complexus. 2003;1:123–133. doi:10.1159/000076100 [Google Scholar]
- Steffensen K, Gustafsson J. Putative metabolic effects of the liver×receptor (l×r) Diabetes. 2004;53(Suppl. 1):36–52. doi: 10.2337/diabetes.53.2007.s36. [DOI] [PubMed] [Google Scholar]
- Tang C, Cho H, Nakamura M, Clarke S. Regulation of human delta-6 desaturase gene transcription: identification of a functional direct repeat-1 element. J. Lipid Res. 2003;44:686–695. doi: 10.1194/jlr.M200195-JLR200. doi:10.1194/jlr.M200195-JLR200 [DOI] [PubMed] [Google Scholar]
- Thieffry D, Sanchez L. Qualitative analysis of gene networks: towards the delineation of trans-regulatory modules. In: Schlosser G, Wagner G, editors. Modularity in development and evolution. University of Chicago Press; Chicago: 2004. pp. 222–243. [Google Scholar]
- Thomas R. On the relation between the logical structure of systems and their ability to generate multiple steady states or sustained oscillations. Springer Ser. Synerget. 1981;9:180–193. [Google Scholar]
- Tobin K, Steineger H, Alberti S, Spydevold O, Auwerx J, Gustafsson J, Nebb H. Cross-talk between fatty acid and cholesterol metabolism mediated by liver×receptor-alpha. Mol. Endocrinol. 2000;14:741–752. doi: 10.1210/mend.14.5.0459. doi:10.1210/me.14.5.741 [DOI] [PubMed] [Google Scholar]
- Vlad M, Arkin A, Ross J. Response experiments for nonlinear systems with application to reaction kinetics and genetics. Proc. Natl Acad. Sci. USA. 2004;101:7223–7228. doi: 10.1073/pnas.0402049101. doi:10.1073/pnas.0402049101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. How to reconstruct a large genetic network from n gene perturbations in fewer than n2 easy steps. Bioinformatics. 2001;17:1183–1197. doi: 10.1093/bioinformatics/17.12.1183. doi:10.1093/bioinformatics/17.12.1183 [DOI] [PubMed] [Google Scholar]
- Wang Z, Hu H. Dimensional reduction for nonlinear time-delayed systems composed of stiff and soft substructures. Nonlinear Dyn. 2001;25:317–331. doi:10.1023/A:1012981822882 [Google Scholar]
- Yamanishi Y, Vert J.-P, Kanehisa M. Protein network inference from multiple genomic data: a supervised approach. Bioinformatics. 2004;20:i363–i370. doi: 10.1093/bioinformatics/bth910. doi:10.1093/bioinformatics/bth910 [DOI] [PubMed] [Google Scholar]
- Yau C, Huzurbazar A. Analysis of censored and incomplete survival data using flowgraph models. Stat. Med. 2002;21:3727–3743. doi: 10.1002/sim.1237. doi:10.1002/sim.1237 [DOI] [PubMed] [Google Scholar]
- Yildirim N, Mackey M. Feedback regulation in the lactose operon: a mathematical modeling study and comparison with experimental data. Biophys. J. 2003;84:2841–2851. doi: 10.1016/S0006-3495(03)70013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]