Abstract
The uptake of iodide by epithelial thyroid cells requires the expression of a specific transporter, the Na+/I− symporter, NIS. Benign and malignant thyroid tumors of epithelial origin show a decrease up to a loss of iodide uptake activity. Previous studies of the human NIS (hNIS) gene expression in these tumors, based on the amplification of transcripts and/or immunohistochemical detection of the protein, have yielded divergent data; hNIS expression was found either increased or decreased. To get a new and integrated view of the alterations of hNIS expression in hypofunctioning thyroid tumors, we performed investigations of hNIS transcript and hNIS protein levels on the same tumors and paired normal tissue samples. HNIS, identified as a 75- to 80-kd species, was present in all normal tissue samples from euthyroid patients, but was undetectable, even at high membrane protein input, in all benign and malignant hypofunctioning thyroid tumors. By contrast, ∼50% of tumors contained hNIS transcripts. This dissociation between transcript and protein levels was not found for the transcript and protein encoded by the PDS gene assayed in the same tumors. The hNIS transcript-positive tumors contained small amounts of low-molecular mass hNIS-immunoreactive species identified as nonglycosylated hNIS. Tumors containing the nonmature form of hNIS exhibited a predominant intracellular immunolabeling. In conclusion, our data show that benign and malignant hypofunctioning thyroid tumors either no longer express hNIS protein or express only a very low amount of nonglycosylated hNIS and indicate that the impairment of hNIS gene expression might result from alterations at both transcriptional and posttranscriptional levels.
The transport and concentration of iodide into the thyroid gland represents the first step in the production of thyroid hormones. The iodide uptake across the basolateral membrane of polarized thyroid follicular cells brings into play a specific transporter, the Na+/I− symporter or NIS. NIS co-transports two Na+ ions along with one iodide ion; the transmembrane Na+ gradient serves as the driving force for iodide transport against its electrochemical gradient. The Na+ gradient is maintained by the Na+/K+-ATPase. Molecular tools generated from the NIS gene cloning1,2 have served to analyze different aspects of NIS expression in thyroid as well as in nonthyroid tissues from different species. There is now abundant literature on human NIS (hNIS) expression in normal and pathological thyroid tissues. A series of articles analyzing hNIS transcript levels by reverse transcriptase-polymerase chain reaction (RT-PCR)3–8 or tissue distribution of hNIS immunoreactivity on paraffin-embedded tissue sections9–12 report a general decrease in, sometimes a loss of hNIS expression in benign and malignant thyroid tumors. These data are in keeping with the fact that both benign [follicular adenomas (FAs)] and malignant (papillary and follicular carcinomas) thyroid tumors, with very few exceptions, exhibit a decrease up to a loss of iodide uptake activity. By contrast, other studies13–15 based on combined Northern blot analyses and immunodetection of the protein13 or only immunohistochemical staining14,15 suggest that hNIS is expressed or even overexpressed in most thyroid carcinomas (TCs)13,14 and thyroid adenomas.15 In the two latter reports, hNIS immunoreactivity was mostly, if not exclusively, found inside thyroid cells; this led the authors to postulate that the iodide uptake defect could result from a defective targeting of hNIS to the plasma membrane.
To try to get new information on this debated issue, we decided to investigate the alterations of hNIS gene expression occurring in thyroid tumors by analyzing hNIS transcript and hNIS protein levels in the same tumors. For this purpose, we set up a semiquantitative Western blot procedure, an approach not used in previous studies, to assess the hNIS protein content of tumors as compared to that of paired normal tissue, and we assayed hNIS transcript by a commonly used RT-PCR procedure coupled to Southern blot to increase the sensitivity of hNIS amplicon detection. Results from these combined approaches prompted us to examine the relationship between the expressed forms of hNIS and the cellular distribution of hNIS immunoreactivity in the same tumors and paired normal thyroid tissue. We confirm that the hNIS gene expression is impaired in all hypofunctioning thyroid tumors and we show that the impairment results from transcriptional alterations but also from alterations at posttranscriptional levels, including translation, maturation, and plasma membrane targeting of the protein.
Materials and Methods
Human Thyroid Tissues
Thyroid tissue samples were taken from the Lyon Thyroid Tumor Bank set up since 1998 at the Lyon University Hospital Center as previously described.16 Tissue samples weighing 50 to 200 mg, quickly frozen in liquid nitrogen and stored at −80°C, consisted of paired samples, benign or malignant thyroid tumors, and surrounding normal tissue (NT). Pathological and clinical annotations were anonymously associated with thyroid tissue samples classified according to World Health Organization recommendations. This study was performed on 22 unselected hypofunctioning tumors (3 FAs and 19 TCs) and paired NT samples. Among the carcinomas, 17 were papillary thyroid carcinomas (PTCs) and 2 were follicular thyroid carcinomas (FTCs). The 22 paired samples were available in duplicate or triplicate. Three toxic adenomas or TA and paired NT were used as positive (TA) or negative (paired NT) controls in the hNIS protein assay. Hypofunctioning tumor samples were obtained from euthyroid patients at the time of surgery except four patients with PTC known to be under T4 therapy before surgery. The three patients with TA presented a low serum thyroid-stimulating hormone (TSH) concentration (below 0.05 mU/L) and a hot nodule (extinctive for the adjacent NT) at thyroid scintigraphy. Thyroid scintigraphy was performed in 14 of the 22 patients with TC or FA; in all cases, the nodules appeared hypofunctioning. Patients with FA were two males and one female (mean age, 37 years) and patients with TC were 7 males and 12 females (mean age ± SD, 43 ± 15 years). According to the TNM classification, nine TCs were classified as grade I, six as grade II, and four as grade III. This study was approved by the supervision interdisciplinary committee of the Tumor Bank and performed in accordance with protocols previously approved by the local human studies committee.
Preparation and Testing of Antibodies
A peptide corresponding to the C-terminal sequence (amino acids 630 to 643) of hNIS (Swiss Prot Bank access no. Q92911) was generated by solid phase synthesis. The purified peptide, conjugated to keyhole limpet hemocyanin using glutaraldehyde, was used to immunize rabbits according to standard procedures. One immune serum, polyclonal antibody (pAb) 795, with a high anti-peptide antibody titer in enzyme-linked immunosorbent assay (higher than 1 × 106), was selected for subsequent studies. Immunological detection of hNIS by Western blot or immunohistochemistry was performed using the purified IgG fraction.
Western Blot Analyses
Frozen tissue samples were rapidly weighed and homogenized in ice-cold phosphate-buffered saline (PBS) (1 ml/100 mg tissue) supplemented with protease inhibitors (apoprotinin, peptasin, leupeptin, each at a concentration of 1 μg/ml) using a Teflon-glass Potter homogenizer. Homogenates were centrifuged at 100,000 × g for 30 minutes at 4°C to obtain the crude thyroid membrane fractions. The pelleted material was suspended in 1 ml of PBS containing protease inhibitors, and aliquots were frozen at −80°C. Protein was assayed by the Lowry method after solubilization in 0.1% sodium deoxycholate. The total amount of protein per mass of tissue and the proportion of membrane protein (100,000 × g pellet) obtained from tumors (FA, PTC, or TA) and paired NT samples were very similar;16 protein of the crude membrane fractions represented 25 to 35% of total protein. Membrane proteins (1 to 180 μg of protein) were subjected to electrophoresis on 7.5% acrylamide gel in the presence of sodium dodecyl sulfate (SDS) and electrotransferred onto Immobilon P membranes (Millipore Corp., Bedford, MA). Membranes were preincubated in PBS supplemented with 5% nonfat dry milk and 0.2% Tween 20 for 1 hour at room temperature and then incubated overnight at 4°C with the IgG fraction from pAb 795 immune serum (5 to 30 μg/ml) in the same buffer. After washes in 0.2% Tween PBS, membranes were incubated with a goat anti-rabbit IgG conjugated to horseradish peroxidase (Biorad Laboratoires Inc., Marnes la Coquette, France) for 1 hour at room temperature. Immune complexes were revealed by the enhanced chemiluminescence method using the enhanced chemiluminescent substrate from Amersham Biosciences (Orsay, France) and exposed to Kodak X-Omat AR films (Eastman Kodak Co., Rochester, NY). In a second step, the same membranes were incubated with a monoclonal antibody (mAb) directed against the α-subunit of Na+,K+-adenosine triphosphatase (Na+,K+-ATPase) at a final dilution of 1:25,00016 and then with a biotinylated goat anti-mouse Ig from Amersham Biosciences. Immune complexes were visualized using streptavidin conjugated to alkaline phosphatase and nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate as substrate. Western blot signals from the enhanced chemiluminescence (hNIS) and alkaline phosphatase (Na+,K+-ATPase α-subunit) reactions were quantified as previously reported16 and expressed in luminance values. The α-subunit of the Na+,K+-ATPase was used to control the amount of protein subjected to the hNIS Western blot assay. To compare signals obtained from the large number of Western blot membranes required for the serial analyses in duplicate or triplicate of more than 50 tissue samples, a NT membrane preparation (H2) was included on each Western blot and used as an internal reference.
Detection of hNIS Transcripts by RT-PCR and Southern Blot
Total RNA was isolated from thyroid tissues using the acid guanidinium thiocyanate-phenol-chloroform method according to Chomczynski and Sacchi.17 The RNA concentration was determined by absorbance measurements at 260 nm, and RNA purity and integrity were assessed by determination of the A260/A280 ratio and electrophoresis on 1% agarose, followed by ethidium bromide staining of 28S and 18S RNA. Reverse-transcription was performed on total RNA. Human NIS cDNAs were synthesized from 3 μg of total RNA using 200 U Moloney murine leukemia virus reverse transcriptase in 20 μl of the following buffer (7 mmol/L MgCl2, 40 mmol/L KCl, 10 mmol/L dithiothreitol, 100 μg/ml bovine serum albumin, 50 mmol/L Tris-HCl, pH 8.3) containing 2 mmol/L of each dNTP and 25 U RNAsin (Promega Corp., Madison, WI). The reverse transcription was initiated by the addition of 10 pmol of the downstream primer: 1064GAGCCGCTATACATTCTGGA1083 complementary to a 3′-region of the hNIS cDNA (anti-sense primer). After 60 minutes at 42°C, the reaction mixture was heated at 95°C for 5 minutes to inactivate the reverse transcriptase and to denature the cDNA-RNA hybrids. Reverse transcription products were then amplified by PCR with the sense primer: 630CTTCTGAACTCGGTCCTCAC649. The reaction mixture (100 μl) contained 10 μl of the reverse transcription solution and 90 μl of the PCR buffer (50 mmol/L KCl, 2.5 mmol/L MgCl2, 2 mmol/L dNTP, 2 mmol/L dithiothreitol, 10 mmol/L Tris-HCl, pH 8.3) supplemented with the sense primer and 0.5 U Thermus aquaticus DNA polymerase (Promega Corp.). Samples were then submitted to 35 cycles of amplification (1 minute at 94°C for denaturation, 1 minute at 58°C for annealing, and 2 minutes at 72°C for extension). Amplified products were analyzed by electrophoresis on 1% agarose gels, stained with 0.5 μl/ml ethidium bromide and transferred to positively charged nylon membranes (Amersham Biosciences). Amplicons were identified by hybridization with a hNIS[32P]-labeled cDNA probe. After hybridization, the nylon membrane was washed twice at 42°C with 2× standard saline citrate (SSC), twice at 60°C with 2× SSC and 0.5% SDS, and twice at the same temperature with 0.2× SSC and 1% SDS, and then exposed to X-ray film at −80°C with an intensifier screen. The 0.8-kb hNIS cDNA probe, generated by RT-PCR using a pair of hNIS-specific primers (sense primer: nucleotides 799 to 820; anti-sense primer: nucleotides 1551 to 1571), was cloned into the PGEMT vector (Promega Corp.). The 0.8-kb hNIS cDNA insert was radiolabeled with [α-32P] dCTP (3000 Ci/mmol) by the random oligonucleotide primed synthesis kit from Roche Diagnostics (Meylan, France). The specific radioactivity of the cDNA probe ranged from 7 to 10 × 108 counts/minute/μg.
Thyroglobulin (Tg) transcripts were reverse-transcribed from the same RNA preparations using the following primers: 570GCCTGTCCAGTGCAAATTTGTCAAC594 (sense primer) and 1086CTTGTGCCTCCGAAAGGCAGCAGG1111 (anti-sense primer). Reaction conditions were the same as those mentioned above with the following modifications: the reverse transcription was performed from 1.5 μg of total RNA and the PCR amplification was performed from 1 μl of a 1:75 dilution of the cDNA template solution using 3.5 mmol/L of MgCl2, and an annealing temperature of 60°C. Samples were submitted to 30 cycles of amplification. Amplicons were analyzed by electrophoresis on 2% agarose gel and stained with 0.5 μg/ml of ethidium bromide. Quantification of signals was performed as previously described.16
Immunohistochemical Analyses
Formalin-fixed tissue samples were embedded in paraffin. Morphological analyses of hematoxylin and eosin-stained tissue were performed on 4-μm-thick sections. Tissue sections in 0.1 mol/L citrate, pH 7.0, were treated for 40 minutes at 90°C for antigen retrieval. After washes in PBS, endogenous peroxidase activity was blocked by a 15-minute treatment in 5% H2O2 in water. Sections were then incubated with pAb 795 IgG (20 μg/ml) for 1 hour at room temperature. Immune complexes were detected using biotinylated goat anti-rabbit Ig antibodies and streptavidin-peroxidase from DAKO (Carpinteria, CA); the color reaction was developed using 3,3-diaminobenzidine as substrate. After nuclear staining with hemalun, glass slides were mounted and observed at magnification ranging from ×40 to ×1000. Negative controls, including nonimmune IgG or omission of the first antibody, were performed in parallel.
Results
Validation of the Procedures Used for Semiquantitative Measurements of hNIS Protein and hNIS Transcript
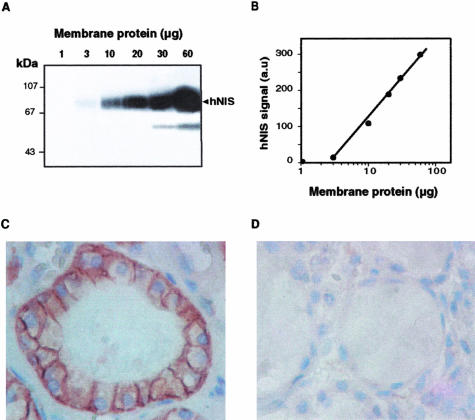
The Western blot analysis of membrane fractions from normal human thyroid tissue using pAb 795 IgG revealed a band migrating as a 75- to 80-kd species (Figure 1A), corresponding to glycosylated hNIS (as demonstrated farther in the article). An additional hNIS-immunoreactive species with an apparent molecular mass ranging from 50 to 55 kd was detected at high thyroid membrane protein input (Figure 1A). Neither the 75- to 80-kd band nor the low-molecular mass immunoreactive species were observed when pAb 795 IgG were replaced by control IgG. pAb 795 IgG detected the 75- to 80-kd hNIS from 3 to 10 μg of membrane protein prepared from normal thyroid tissue (NT). The hNIS signal intensity was proportional to the membrane protein input over a 10-fold protein concentration range (Figure 1B). The cellular location of the pAb 795 IgG-labeled material was examined on paraffin-embedded NT sections. The immunoreactivity was primarily located at the basolateral plasma membrane of polarized thyroid epithelial cells (Figure 1, C and D). A definite intracellular labeling, likely membrane-associated, was also observed. Nonepithelial cells (probably endothelial cells or fibroblasts) in the vicinity of the thyroid follicles were not stained.
Figure 1.
Semiquantitative measurements and immunodetection of hNIS using pAb 795 anti-peptide antibodies. A and B: Identification of hNIS by Western blot. Increasing amounts of membrane protein (1 to 60 μg) from a normal thyroid tissue sample were subjected to polyacrylamide gel electrophoresis in the presence of SDS and electrotransferred onto Immobilon P membrane. Transfer membranes were incubated with pAb 795 IgG (20 μg/ml). Western blot signals were quantified as reported in the Materials and Methods section. A: Image of the Western blot membrane. The position of proteins of known molecular mass (expressed in kd) is indicated on the left. B: Intensity of hNIS signal as a function of the membrane protein input. C and D: hNIS immunodetection on paraffin-embedded NT sections. Tissue sections were incubated with 200 μg/ml of pAb 795 IgG (C) or preimmune IgG (D). Immune complexes were detected as indicated in the Materials and Methods section. Original magnifications, ×1000 (C and D).
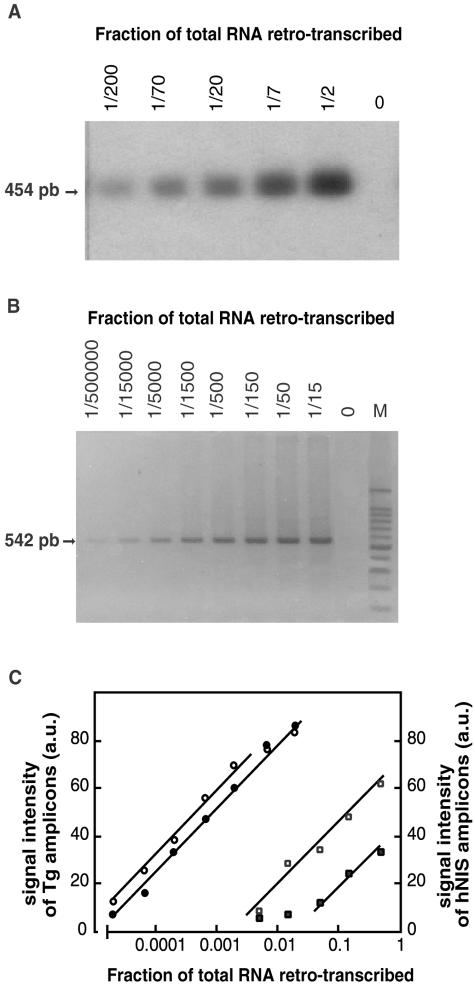
Transcripts from the hNIS gene were amplified by RT-PCR and hNIS amplicons were detected by Southern blot. Data of Figure 2 show that the amount of hNIS amplicons obtained after 35 cycles (Figure 2A) was proportional to the logarithm of the hNIS cDNA template concentration expressed as the fraction of total RNA retro-transcribed. Similar results were obtained for Tg amplicons after 30 PCR cycles (Figure 2B). Results of the amplification of hNIS and Tg transcripts from a tumor and its paired NT are reported in Figure 2C. The two samples had comparable Tg transcript levels but the hNIS transcript content of the tumor was ∼10 times lower than that of the NT. The fractions of total RNA retro-transcribed that were chosen for subsequent semiquantitative assays of hNIS and Tg transcripts were 1/2 and 1/750, respectively. These conditions were chosen to have the highest sensitivity in the hNIS transcript assay and a good accuracy in the Tg transcript determination.
Figure 2.
Semiquantitative measurements of hNIS transcripts. Validation of the RT-PCR assay conditions. Total thyroid RNA from a NT sample was reverse-transcribed using specific hNIS and thyroglobulin (Tg) primers; hNIS and Tg cDNAs were amplified from serial dilutions (1/2 to 1/200 and 1/15 to 1/500,000, respectively) of the reverse transcription reaction mixtures. After amplification, equal volumes of the PCR solutions were analyzed by electrophoresis on 2% nondenaturing agarose gels. PCR-amplified products were detected by hybridization with a specific probe in the case of hNIS (A) or by ethidium bromide staining in the case of Tg (B). In A and B, arrows indicate the size of the expected amplicons: 454 bp for hNIS and 542 bp for Tg. M, 100-bp ladder. The intensity of the fluorescent signal of Tg amplicons and the radioactive signal of hNIS amplicons was quantified using Adobe Photoshop software and expressed in arbitrary units (a.u.). Results from two tissue samples: FA5 (filled symbols) and its paired NT (open symbols) are reported in C. Signals obtained for Tg (circles) and hNIS (squares) are plotted as a function of the logarithm of the fraction of total RNA retro-transcribed.
Compared Levels of hNIS Protein and hNIS Transcripts in the Same Hypofunctioning Thyroid Tumors
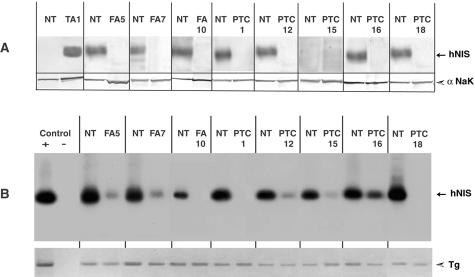
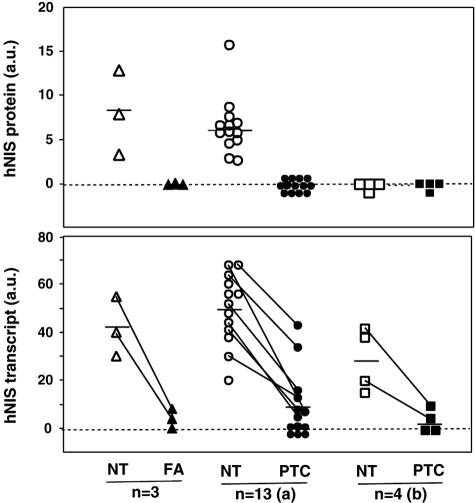
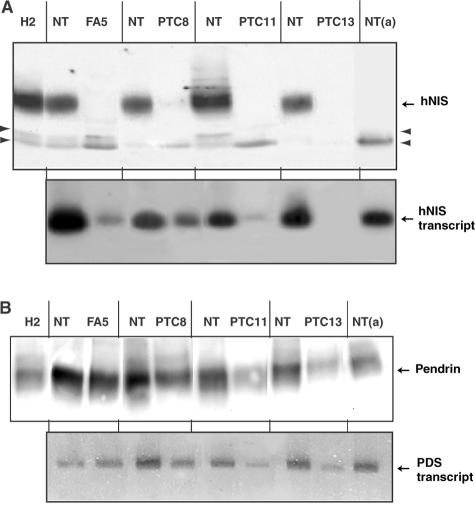
hNIS protein and hNIS transcript were assayed on duplicate samples of 20 hypofunctioning tumors and paired NT (3 FAs and 17 TCs). Western blot signals obtained from 7 representative paired samples of the 20 and from 1 TA are shown on Figure 3A. RT-PCR/Southern blot signals obtained from the same tumor/NT paired samples are shown in Figure 3B. Values obtained by quantification of hNIS protein and hNIS transcript signals corresponding to the 20 hypofunctioning tumors and paired NT, are reported in Figure 4.
Figure 3.
Analyses of the hNIS protein and hNIS transcript contents of thyroid tumors and paired NT samples. A: Membrane fractions (30 μg of protein) from paired samples (tumor and NT) were subjected to Western blot analysis using pAb 795 IgG and the enhanced chemiluminescence procedure. The α-subunit of Na+K+-ATPase (αNaK) was then detected on the same transfer membranes using a mAb and the alkaline phosphatase-based colorimetric detection. Data from eight representative hypofunctioning tumor/NT-paired samples: three FAs and five PTCs, and from one hyperfunctioning tumor (TA1) and paired NT. B: Total thyroid RNA was reverse-transcribed using specific hNIS and Tg primers. PCR was performed from cDNA template solution representing 1/2 and 1/1500 of total RNA retro-transcribed for hNIS and Tg, respectively. Data from the eight hypofunctioning tumor/NT-paired samples analyzed in A and positive (+) and negative (−) PCR controls. Top: hNIS amplicons detected by Southern blot. Bottom: Ethidium bromide-labeled Tg amplicons.
Figure 4.
Comparative analyses of the hNIS protein and transcript levels in 20 tumor/NT-paired samples. Twenty tumors (3 FA and 17 PTC) and their paired NT were assayed for hNIS protein by Western blot and hNIS transcript by RT-PCR/Southern blot as presented in Figure 3. Western blot signals (top) and Southern blot signals (bottom) were quantified as mentioned in the Materials and Methods section; values are expressed in arbitrary units (a.u.). Data obtained for the 17 PTC are presented in two subgroups: (a) and (b); the (b) subgroup corresponds to PTC from patients under T4 treatment, the NT of which was hNIS protein-negative. Horizontal bars represent mean values. Bottom: Lines were drawn between NT and tumor symbols to identify paired samples with a hNIS transcript-positive tumor.
The 75- to 80-kd hNIS was detected in all NT samples paired to hypofunctioning tumors except four (one case, PTC15, is shown in Figure 3). The four hNIS-negative NT samples were from patients under T4 therapy in the presurgical period. Unexpectedly, the 75- to 80-kd hNIS was not detected in any of the 20 hypofunctioning tumors. NT samples paired to TA were hNIS-negative (see NT paired to TA1 in Figure 3A) and the hNIS content of TA was three to five times higher than the average hNIS content of NT from euthyroid patients (data not shown).
hNIS transcripts were detected in 20 of 20 NT samples, including the 4 NT samples that were hNIS protein-negative, and in 11 of 20 tumors (2 of 3 FAs and 9 of 17 TCs). In the 11 tumors, in which hNIS transcripts were detected, the average signal intensity (expressed in arbitrary units) was 14 ± 4 versus 47 ± 4 (mean ± SEM) for the paired NT. By reference to the standard plot of Figure 2C, this decrease in signal intensity corresponded to a 10-fold reduction in hNIS transcript content.
Data of Figures 3 and 4 show that there was a dissociation between hNIS protein and hNIS transcript contents in 15 of the 40 samples: 11 tumors and the 4 NT samples from T4-treated patients. As the lack of detection of hNIS protein in samples containing hNIS transcript could simply be because of a lack of sensitivity of the protein detection method, samples giving discordant results were subjected to Western blot assay at high membrane protein input, ie, 180 μg (instead of 30 μg). In no case could the 75- to 80-kd hNIS protein be detected. This is illustrated for three tumors (FA5, PTC8, and PTC11) and one NT (NTa) in Figure 5A; these samples contained hNIS transcripts but no 75- to 80-kd protein. To determine whether the dissociation between transcript and protein levels was a general feature of hypofunctioning tumors, transcripts of the PDS gene and the corresponding protein, pendrin (another membrane protein with an iodide transport activity), were assayed in the same tumors and NT samples, as previously described.16 Transcripts from the PDS gene and pendrin were detected in all samples. Data from four paired samples are reported in Figure 5B. As compared to NT, PDS transcripts and pendrin were decreased by 30 to 50% in hypofunctioning thyroid tumors. There was a relationship between transcript and protein levels (Figure 5B); tumors exhibiting a decreased PDS transcript level had a decreased pendrin content. The pendrin to PDS transcript ratio estimated from signal quantification values corresponding to the 13 PTC/NT paired samples (analyzed for their hNIS transcript and protein contents in Figure 4) was not statistically different in tumors and NT (0.90 ± 0.18 versus 1.28 ± 0.25).
Figure 5.
Parallel investigations of transcript and protein from hNIS (A) and PDS (B) genes in the same tumor/NT-paired samples. A: Attempt to detect hNIS at high membrane protein input. Identification of low-molecular mass hNIS-immunoreactive species. Top: Tumor and NT samples that were hNIS-negative in the assay from 30 μg of membrane protein were subjected to new Western blot analyses starting from 180 μg of protein. NT samples that were hNIS-positive and the H2 reference membrane preparation were analyzed from 90 μg of membrane protein. Data from four paired samples (one FA and three PTC) and a NT (NTa) paired to a PTC. Arrowheads identify the immunoreactive species evidenced at high membrane protein input. Bottom: Results of the hNIS transcript assay (RT-PCR/Southern blot) performed on duplicate specimens of the same tumors and NT. B: Relationship between PDS transcript and pendrin levels in hypofunctioning tumors and paired NT. Top: Western blot analyses of pendrin from 30 μg of membrane protein of the samples analyzed for hNIS protein in A. The anti-pendrin antibodies used in these experiments (IgG fraction of the pAb 827 immune serum) have been previously characterized.16 Bottom: Analyses of PDS transcripts in the samples analyzed for hNIS transcripts in A. RT-PCR was performed as previously reported.16 Amplicons were detected by ethidium bromide staining.
Hypofunctioning Benign and Malignant Tumors Only Express Low Levels of Nonglycosylated hNIS
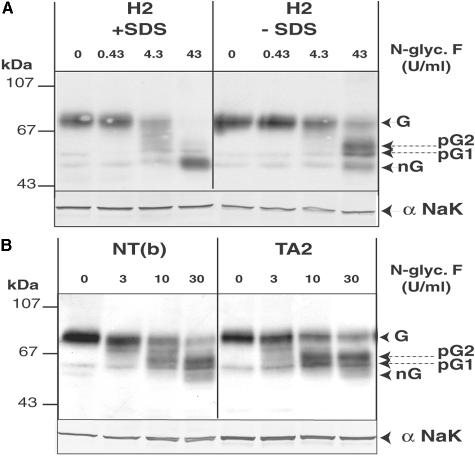
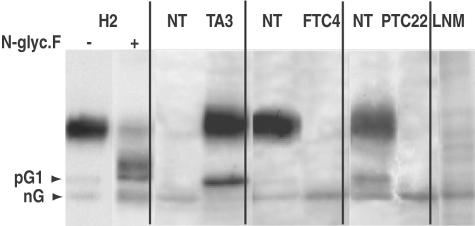
Western blot analyses performed at high membrane protein input reported in Figure 5A revealed that tumors (as well as NT from T4-treated patients) containing hNIS transcripts contained hNIS-immunoreactive species with an apparent molecular mass ranging from 50 to 55 kd. These immunoreactive species were not detected in hNIS transcript-negative samples such as PTC13 (Figure 5A). Given their size, we thought they could correspond to nonmature form of hNIS. To test this hypothesis, we generated hNIS maturation intermediates by deglycosylation using N-glycosidase F (Figure 6) and we compared the electrophoretic properties of the deglycosylated forms of hNIS with those of the low-molecular mass immunoreactive species identified in tumors and unstimulated NT (Figure 7). Treatment of thyroid membrane fractions by N-glycosidase F in the presence of SDS led to the transformation of the 75- to 80-kd hNIS protein into a main component of ∼50 kd, likely representing the nonglycosylated (nG) hNIS polypeptide chain as found for rat NIS.18,19 In the absence of SDS, the enzyme had a lower efficiency and generated two main intermediate forms and a low amount of the 50-kd species. The hNIS deglycosylation pattern (Figure 6) composed of two partially glycosylated forms named pG1, pG2, and nG was reproducibly obtained. The electrophoretic mobility of another membrane protein, the α-subunit of Na+/K+-ATPase, a nonglycosylated protein, was not modified by the enzyme treatment. The low-molecular mass hNIS-immunoreactive species, detected in hypofunctioning tumors and unstimulated NT, exactly co-migrated with the nG form of hNIS; this is shown for an unstimulated NT (paired to a TA) and two tumors (FTC4 and PTC22) in Figure 7. The nG and/or the pG1 forms of hNIS were present in low amounts in NT (from euthyroid patients) and hyperfunctioning tissue (TA) containing mature hNIS. The nG form of hNIS was also detected in a lymph node metastasis (LNM) of a PTC, devoid of the 75- to 80-kd mature hNIS.
Figure 6.
Identification of the nonglycosylated form and deglycosylation intermediates of hNIS. Thyroid membrane fractions (40 μg of protein) were incubated with increasing concentrations of N-glycosidase (N-glyc.F) in PBS supplemented with protease inhibitors for 3 hours at 37°C and analyzed by Western blot using pAb 795 IgG and then the mAb directed against the Na+K+-ATPase α-subunit (αNaK). A: Treatment of the H2 reference preparation in the absence or in the presence of SDS. B: Treatment of a NT paired to FA5 (NTb) and TA2 in the absence of SDS. G and nG, glycosylated and nonglycosylated hNIS; pG1 and pG2, partially deglycosylated forms of hNIS.
Figure 7.
Relationship between the molecular forms and the cellular distribution of hNIS in normal and tumoral thyroid tissue. Western blot analysis of hNIS-immunoreactive species. Membrane fractions (180 μg of protein) from thyroid tumors (one hyperfunctioning tumor, TA3, and two hypofunctioning tumors, FTC4 and PTC26) and paired NT samples and from a lymph node metastasis (LNM) were subjected to Western blot analysis using pAb 795 IgG. The H2 reference preparation either untreated or treated with N-glycosidase F (43 U/ml) was run in parallel. Arrows identify nG and pG1 hNIS-immunoreactive forms. Duplicate specimens of these tissue samples were processed for immunohistochemical detection of hNIS (see Figure 8).
Relationships between the Presence of Immature Forms of hNIS and the Subcellular Distribution of hNIS Immunoreactivity
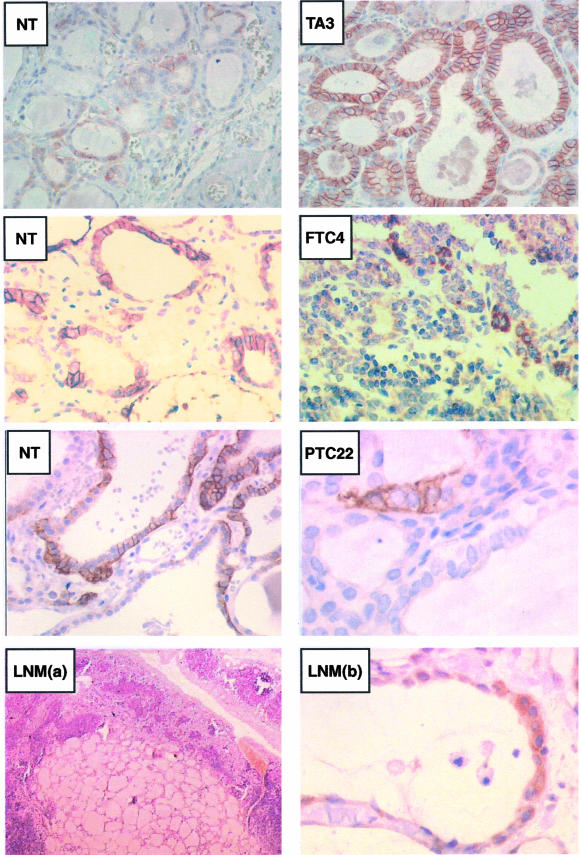
Tumor/NT-paired samples analyzed by Western blot in Figure 7 were processed for immunohistochemical detection of hNIS immunoreactivity (Figure 8). NT samples paired to either FTC4 or PTC22 that contained mature hNIS (and in addition a low amount of pG1 and/or nG forms), showed an intense labeling (although nonuniform among cells and follicular structures) of thyroid cell plasma membrane. Corresponding tumors that did not contain the 75- to 80-kd hNIS protein but only a low amount of nG hNIS, exhibited scarce groups of immunolabeled cells and, in the positive cells, the labeling did not segregate with the plasma membrane but was patchy and covered the entire cell surface. The hyperfunctioning tissue (TA3) with a high mature hNIS content presented the classical basolateral plasma membrane immunolabeling that was intense and homogeneous between cells and follicles; as compared to nonepithelial cells located between follicles, the inside of thyrocytes was clearly immunostained. By contrast, the NT paired to TA, which only contained the nG form of hNIS on Western blot, was faintly immunolabeled and the weakly positive cells mainly exhibited a diffuse intracellular labeling. The lymph node metastasis, containing the nG form of hNIS (Figure 7), exhibited a clear intracellular immunoreactive staining of the metastatic thyroid cells showing the typical follicle organization.
Figure 8.
Relationship between the molecular forms and the cellular distribution of hNIS in normal and tumoral thyroid tissue. Immunohistochemical location of hNIS. Paraffin-embedded tissue sections were prepared from duplicate specimens of the samples analyzed by Western blot in Figure 7. Tissue sections were processed as indicated in the Materials and Methods section. Images on the right correspond to the immunostaining of tumors, and those on the left to the immunostaining of paired NT, except the bottom left image LNM(a), which corresponds to a low-magnification image of the lymph node metastasis after H&E staining. Immunostaining appeared as a brown color. Original magnifications: ×40 [LNM(a)]; ×250 (TA3 and paired NT, FTC4 and paired NT, NT paired to PTC22); ×1000 [PTC22 and LNM(b)].
Discussion
By performing semiquantitative measurements of hNIS transcript and hNIS protein in the same tumors and paired NT samples, we confirm that hNIS is markedly underexpressed in all hypofunctioning benign and malignant thyroid tumors, and we show that the alterations of hNIS gene expression probably takes place at both transcriptional and posttranscriptional levels and the only form of hNIS present in hypofunctioning thyroid tumors corresponds to nonglycosylated hNIS exhibiting a predominant intracellular location.
The use of nonsaturated PCR conditions and a Southern blot procedure to identify hNIS amplicons allowed us to measure changes of hNIS transcript levels of a rather large amplitude. In approximately half of the tumors, there was an average 10-fold decrease in hNIS transcript as compared to paired NT, and in the other half, hNIS transcripts were not detected, indicating a 100-fold reduction. These quantitative changes, which are in full agreement with those obtained by real-time PCR,4,5,8 give evidence for an impairment of transcription of the hNIS gene in all hypofunctioning tumors.
The Western blot assay, that reproducibly identified the 75- to 80-kd glycosylated hNIS from 3 to 10 μg of membrane protein of NT, failed to detect the protein in all of the hypofunctioning thyroid tumors using up to 180 μg of membrane protein. This finding indicates that the mature hNIS protein content of the tumors was at least 20 times lower than that of the surrounding NT. In approximately half of the tumors, the lack of hNIS transcripts coincided with the lack of protein. In the other tumors, hNIS transcripts were present (sometimes at levels comparable to those found in NT), and we only detected low amounts of nonglycosylated hNIS. This dissociation between transcript and protein levels (not observed in the same tumors for the products of another gene, the PDS gene) indicates that the expression of hNIS gene could also be altered at posttranscriptional step(s), ie, synthesis and/or maturation of hNIS. The striking association between the presence of nonglycosylated hNIS and the intracellular location of hNIS immunoreactivity in hypofunctioning tissue, whether normal or tumoral, suggests that, when synthesized in low amounts, hNIS would not undergo glycosylation and would remain in intracellular compartments. Because nonglycosylated NIS expressed in nonthyroid cells was reported to be transported at the plasma membrane and to be active,20 alteration of hNIS glycosylation in tumors would not be per se a limitation of its transport at the cell surface. In tissues expressing mature hNIS and showing the classical basolateral plasma membrane staining, intracellular hNIS immunoreactivity was also found; it would correspond to hNIS in the course of synthesis and trafficking toward and from the plasma membrane.21,22
It is noteworthy that the observations made on hypofunctioning tumors applied in all respects to nonstimulated NT. Indeed, nonstimulated NT samples from patients under T4 treatment contained hNIS transcripts and only low amounts of nonglycosylated hNIS. Similarly, NT samples paired to TA, devoid of mature hNIS, contained nonglycosylated hNIS. Thus, the impairment of hNIS gene expression occurring in hypofunctioning thyroid tumors could simply result from a switching off of the pathway controlling the expression of hNIS gene. In vitro studies on thyroid cell models and in vivo studies in rodents indicate that the main regulator of NIS gene expression is TSH and that the hormone primarily acts at the level of transcription,23–27 but also exerts posttranscriptional regulatory actions.21 Because patients with a cold nodule, be it an adenoma or a carcinoma, are generally euthyroid with a plasma TSH concentration within the normal range, and as TSH receptor expression is maintained in these tumors,28 it is reasonable to think that the impairment of hNIS expression occurring in hypofunctioning thyroid tumors could result from alterations of regulatory elements along the TSH-activated pathway.
The difference in the hNIS protein content of tumors (either adenomas or carcinomas) versus paired NT accounts for the difference of iodide uptake activity between tumors appearing as cold nodules at scintigraphy and the surrounding functional normal thyroid tissue. Similarly, the absence of mature hNIS in NT from patients with TA fits with the loss of iodide uptake activity and shows that a lowering or a suppression of TSH secretion dramatically impairs hNIS expression. Changes in hNIS expression level in normal human thyroid tissue according to the activation and/or functional state of the tissue were expected, but had not yet been demonstrated. Interestingly, in the NT from the four T4-treated patients, hNIS protein content was dramatically diminished, whereas hNIS transcript levels were only slightly modified. This strongly suggests that the down-regulation of hNIS expression in response to a lowering of TSH could primarily involve posttranscriptional (possibly translational) regulatory mechanisms.
Although there is no simple explanation for the divergent data in the literature regarding the level of expression of hNIS in hypofunctioning thyroid tumors, one might think that some of the previous published results showing an overexpression of hNIS could have been influenced by the reference used to establish the normal hNIS expression level. This might be the case in the work of Saito and colleagues;13 these authors only detected trace amounts of hNIS transcript and hNIS protein in the NT samples they used as reference. A lowering of hNIS content of NT could be related to a T4-treatment (as we showed) or to an elevated iodide supply of patients. Indeed, iodide intake in Japan is known to be high and it has been shown that iodide exerts down-regulatory actions on NIS expression.29,30 This remark might also be applied to the immunohistochemical study of Dohan and colleagues14 who reported an overexpression of hNIS in 70% of thyroid cancer (as compared to adjacent NT) and no expression in the 30% remaining cases. The authors did not give information on the thyroid status of the patients. In addition, the interpretation of the findings of Dohan and colleagues14 is obscured by the fact that tumors expected to be hNIS-negative such as anaplastic carcinomas corresponding to fully dedifferentiated tumors no longer expressing thyroid-specific genes31,32 and medullary carcinomas that develop from C cells of neuronal crest origin, were also found highly hNIS-positive. Identification of the molecular species responsible for the immunolabeling of these tumors by Western blot would have been of a major interest to credit these findings. Using a similar immunohistochemical approach, Tonacchera and colleagues15 observed that ∼50% of benign thyroid tumors were hNIS-negative, whereas 50% exhibited a high intracellular immunolabeling as compared to surrounding NT. The intensity of tumor labeling, assessed by the percentage of positive cells, was however very variable (from 15 to 80%) and the intensity of labeling of NT was most often very low (from 1 to 3%). A comparative analysis of the hNIS protein content of the tumors and their paired NT by Western blot would have represented a useful complement of immunohistochemistry to conclude on the hNIS expression status of hypo- or nonfunctioning benign thyroid tumors. Indeed, contrary to Western blot measurements, immunohistochemical observations hardly give quantitative information and furthermore do not allow comparison between samples often processed separately. Dohan and colleagues14 and Tonacchera and colleagues15 both detected hNIS immunoreactivity inside the cells of benign or malignant tumors. We have made comparable observations, but in no case was this related to an overexpression of hNIS. In our study, intracellular hNIS immunoreactivity was restricted to tumor or nonstimulated NT samples that contained minute amounts of hNIS in a nonmature form.
In conclusion, our findings further document the relationship between the iodide uptake deficit of hypofunctioning benign or malignant thyroid tumors and the alterations of hNIS gene expression. In these tumors as in NT weakly or no longer stimulated by TSH, 1) transcription of the hNIS gene is reduced and frequently to a large extent, 2) translation of hNIS transcript might be less efficient (considering that minute amounts of protein were found in samples with definite hNIS transcript levels), and 3) maturation of residual hNIS—particularly with regard to glycosylation—and hNIS cell surface targeting are impaired.
Acknowledgments
We thank the organizing and supervision committees of the Lyon Thyroid Tumor Bank for their contributions to this study, Rachida Rabilloud for her technical assistance, and Catherine Limoge for her involvement in the preparation of the manuscript.
Footnotes
Address reprint requests to Professor Bernard Rousset, INSERM Unit 369, Faculté de Médecine Lyon-RTH Laennec, Rue Guillaume Paradin, 69372 Lyon Cedex 08, France. E-mail: u369@laennec.univ-lyon1.fr.
Supported by a grant from the Programme Hospitalier de Recherche Clinique “Cancer Différencié de la Thyroïde.”
S.T.-M. and S.S.-R. are first co-authors.
References
- Dai G, Levy O, Carrasco N. Cloning and characterization of the thyroid iodide transporter. Nature. 1996;379:458–460. doi: 10.1038/379458a0. [DOI] [PubMed] [Google Scholar]
- Smanik P, Liu Q, Furminger T, Ryu K, Xing S, Mazzeferra E, Jhiang S. Cloning of the human sodium iodide symporter. Biochem Biophys Res Commun. 1996;226:339–345. doi: 10.1006/bbrc.1996.1358. [DOI] [PubMed] [Google Scholar]
- Arturi F, Russo D, Schlumberger M, duVillard JA, Caillou B, Vigneri P, Wicker R, Chiefari E, Suarez HG, Filetti S. Iodide symporter gene expression in human thyroid tumors. J Clin Endocrinol Metab. 1998;83:2493–2496. doi: 10.1210/jcem.83.7.4974. [DOI] [PubMed] [Google Scholar]
- Lazar V, Bidart JM, Caillou B, Mahe C, Lacroix L, Filetti S, Schlumberger M. Expression of the Na+/I− symporter gene in human thyroid tumors: a comparison study with other thyroid-specific genes. J Clin Endocrinol Metab. 1999;84:3228–3234. doi: 10.1210/jcem.84.9.5996. [DOI] [PubMed] [Google Scholar]
- Ryu KY, Senokozlieff ME, Smanik PA, Wong MG, Siperstein AE, Duh QY, Clark OH, Mazzaferri EL, Jhiang SM. Development of reverse transcription competitive polymerase chain reaction method to quantitate the expression levels of human sodium iodide symporter. Thyroid. 1999;9:405–409. doi: 10.1089/thy.1999.9.405. [DOI] [PubMed] [Google Scholar]
- Park HJ, Kim JY, Park KY, Gong G, Hong SJ, Ahn IM. Expressions of human sodium iodide symporter mRNA in primary and metastatic papillary thyroid carcinomas. Thyroid. 2000;10:211–217. doi: 10.1089/thy.2000.10.211. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Otsuki T, Sonoo H, Yamamoto Y, Udagawa K, Kunisue H, Arime I, Yamamoto S, Kurebayashi J, Shimozuma K. Semi-quantitative comparison of the differentiation markers and sodium iodide symporter messenger ribonucleic acids in papillary thyroid carcinomas using RT-PCR. Eur J Endocrinol. 2000;142:340–346. doi: 10.1530/eje.0.1420340. [DOI] [PubMed] [Google Scholar]
- Ringel MD, Anderson J, Souza SL, Burch HB, Tambascia M, Shriver CD, Tuttle RM. Expression of the sodium iodide symporter and thyroglobulin genes are reduced in papillary thyroid cancer. Mod Pathol. 2001;14:289–296. doi: 10.1038/modpathol.3880305. [DOI] [PubMed] [Google Scholar]
- Caillou B, Troalen F, Baudin E, Talbot M, Filetti S, Schlumberger M, Bidart JM. Na+/I− symporter distribution in human thyroid tissues: an immunohistochemical study. J Clin Endocrinol Metab. 1998;83:4102–4106. doi: 10.1210/jcem.83.11.5262. [DOI] [PubMed] [Google Scholar]
- Jhiang SM, Cho JY, Ryu KY, DeYoung BR, Smanik PA, McGaughy VR, Fischer AH, Mazzaferri EL. An immunohistochemical study of Na+/I− symporter in human thyroid tissues and salivary gland tissues. Endocrinology. 1998;139:4416–4419. doi: 10.1210/endo.139.10.6329. [DOI] [PubMed] [Google Scholar]
- Castro MR, Bergert ER, Beito TG, McIver B, Goellner JR, Morris JC. Development of monoclonal antibodies against the human sodium iodide symporter: immunohistochemical characterization of this protein in thyroid cells. J Clin Endocrinol Metab. 1999;84:2957–2962. doi: 10.1210/jcem.84.8.5871. [DOI] [PubMed] [Google Scholar]
- Castro MR, Bergert ER, Beito TG, Roche PC, Ziesmer SC, Jhiang SM, Goellner JR, Morris JC. Monoclonal antibodies against the human sodium iodide symporter: utility for immunocytochemistry of thyroid cancer. J Endocrinol. 1999;163:495–504. doi: 10.1677/joe.0.1630495. [DOI] [PubMed] [Google Scholar]
- Saito T, Endo T, Kawaguchi A, Ikeda M, Katoh R, Kawaoi A, Muramatsu A, Onaya T. Increased expression of the sodium/iodide symporter in papillary thyroid carcinomas. J Clin Invest. 1998;101:1296–1300. doi: 10.1172/JCI1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohan O, Baloch Z, Banrevi Z, Livolsi V, Carrasco N. Rapid communication: predominant intracellular overexpression of the Na(+)/I(−) symporter (NIS) in a large sampling of thyroid cancer cases. J Clin Endocrinol Metab. 2001;86:2697–2700. doi: 10.1210/jcem.86.6.7746. [DOI] [PubMed] [Google Scholar]
- Tonacchera M, Viacava P, Agretti P, de Marco G, Perri A, di Cosmo C, de Servi M, Miccoli P, Lippi F, Naccarato AG, Pinchera A, Chiovato L, Vitti P. Benign nonfunctioning thyroid adenomas are characterized by a defective targeting to cell membrane or a reduced expression of the sodium iodide symporter protein. J Clin Endocrinol Metab. 2002;87:352–357. doi: 10.1210/jcem.87.1.8173. [DOI] [PubMed] [Google Scholar]
- Porra V, Bernier-Valentin F, Trouttet-Masson S, Berger-Dutrieux N, Peix JL, Perrin A, Selmi-Ruby S, Rousset B. Characterization and semiquantitative analyses of pendrin expressed in normal and tumoral human thyroid tissues. J Clin Endocrinol Metab. 2002;87:1700–1707. doi: 10.1210/jcem.87.4.8372. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Levy O, Dai G, Riedel C, Ginter CS, Paul EM, Lebowitz AN, Carrasco N. Characterization of the thyroid Na+/I− symporter with an anti-COOH terminus antibody. Proc Natl Acad Sci USA. 1997;94:5568–5573. doi: 10.1073/pnas.94.11.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paire A, Bernier-Valentin F, Selmi-Ruby S, Rousset B. Characterization of the rat thyroid iodide transporter using anti-peptide antibodies. Relationship between its expression and activity. J Biol Chem. 1997;272:18245–18249. doi: 10.1074/jbc.272.29.18245. [DOI] [PubMed] [Google Scholar]
- Levy O, DelaVieja A, Ginter CS, Riedel C, Dai G, Carrasco N. N-linked glycosylation of the thyroid Na+/I− symporter (NIS)—implications for its secondary structure model. J Biol Chem. 1998;273:22657–22663. doi: 10.1074/jbc.273.35.22657. [DOI] [PubMed] [Google Scholar]
- Riedel C, Levy O, Carrasco N. Post-transcriptional regulation of the sodium/iodide symporter by thyrotropin. J Biol Chem. 2001;276:21458–21463. doi: 10.1074/jbc.M100561200. [DOI] [PubMed] [Google Scholar]
- Kaminsky SM, Levy O, Salvador C, Dai G, Carrasco N. Na(+)-I−symport activity is present in membrane vesicles from thyrotropin-deprived non-I(−)-transporting cultured thyroid cells. Proc Natl Acad Sci USA. 1994;91:3789–3793. doi: 10.1073/pnas.91.9.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Endo T, Kawaguchi A, Ikeda M, Nakazato M, Kogai T, Onaya T. Increased expression of the Na+/I− symporter in cultured human thyroid cells exposed to thyrotropin and in Graves’ thyroid tissue. J Clin Endocrinol Metab. 1997;82:3331–3336. doi: 10.1210/jcem.82.10.4269. [DOI] [PubMed] [Google Scholar]
- Kogai T, Endo T, Saito T, Miyazaki A, Kawaguchi A, Onaya T. Regulation by thyroid-stimulating hormone of sodium/iodide symporter gene expression and protein levels in FRTL-5 cells. Endocrinology. 1997;138:2227–2232. doi: 10.1210/endo.138.6.5189. [DOI] [PubMed] [Google Scholar]
- Ohno M, Zannini M, Levy O, Carrasco N, di Lauro R. The paired-domain transcription factor Pax8 binds to the upstream enhancer of the rat sodium/iodide symporter gene and participates in both thyroid-specific and cyclic-AMP-dependent transcription. Mol Cell Biol. 1999;19:2051–2060. doi: 10.1128/mcb.19.3.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmi-Ruby S, Watrin C, Trouttet-Masson S, Bernier-Valentin F, Flachon V, Munari-Silem Y, Rousset B. The porcine sodium/iodide symporter gene exhibits an uncommon expression pattern related to the use of alternative splice sites not present in the human or murine species. Endocrinology. 2003;144:1074–1085. doi: 10.1210/en.2002-220971. [DOI] [PubMed] [Google Scholar]
- Marians RC, Ng L, Blair HC, Unger P, Graves PN, Davies TF. Defining thyrotropin-dependent and -independent steps of thyroid hormone synthesis by using thyrotropin receptor-null mice. Proc Natl Acad Sci USA. 2002;99:15776–15781. doi: 10.1073/pnas.242322099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabant G, Maenhaut C, Kohrle J, Scheumann G, Dralle G, Hoang-Vu C, Hesch RD, von zur Muhlen A, Vassart G, Dumont JE. Human thyrotropin receptor gene: expression in thyroid tumors and correlation to markers of thyroid differentiation and dedifferentiation. Mol Cell Endocrinol. 1991;82:R7–R12. doi: 10.1016/0303-7207(91)90018-n. [DOI] [PubMed] [Google Scholar]
- Uyttersprot N, Pelgrims N, Carrasco N, Gervy C, Maenhaut C, Dumont JE, Miot F. Moderate doses of iodide in vivo inhibit cell proliferation and the expression of thyroperoxidase and Na+/I− symporter mRNAs in dog thyroid. Mol Cell Endocrinol. 1997;131:195–203. doi: 10.1016/s0303-7207(97)00108-1. [DOI] [PubMed] [Google Scholar]
- Eng PHK, Cardona GR, Fang SL, Previti M, Alex S, Carrasco N, Chin WW, Braverman LE. Escape from the acute Wolff-Chaikoff effects is associated with a decrease in thyroid sodium/iodide symporter messenger ribonucleic acid and protein. Endocrinology. 1999;140:3404–3410. doi: 10.1210/endo.140.8.6893. [DOI] [PubMed] [Google Scholar]
- Hoang-Vu C, Dralle H, Scheumann G, Maenhaut C, Horn R, von zur Muhlen A, Brabant G. Gene expression of differentiation- and dedifferentiation markers in normal and malignant human thyroid tissues. Exp Clin Endocrinol. 1992;100:51–56. doi: 10.1055/s-0029-1211176. [DOI] [PubMed] [Google Scholar]
- Ain KB. Management of undifferentiated thyroid cancer. Baillieres Best Pract Res Clin Endocrinol Metab. 2000;14:615–629. doi: 10.1053/beem.2000.0106. [DOI] [PubMed] [Google Scholar]