Abstract
We recently characterized gene expression patterns in gastrointestinal stromal tumors (GISTs) using cDNA microarrays, and found that the gene FLJ10261 (DOG1, discovered on GIST-1), encoding a hypothetical protein, was specifically expressed in GISTs. The immunoreactivity of a rabbit antiserum to synthetic DOG1 peptides was assessed on two soft tissue tumor microarrays. The tissue microarrays included 587 soft tissue tumors, with 149 GISTs, including 127 GIST cases for which the KIT and PDGFRA mutation status was known. Immunoreactivity for DOG1 was found in 136 of 139 (97.8%) of scorable GISTs. All seven GIST cases with a PDGFRA mutation were DOG1-positive, while most of these failed to react for KIT. The immunohistochemical findings were confirmed with in situ hybridization probes for DOG1, KIT, and PDGFRA. Other neoplasms in the differential diagnosis of GIST, including desmoid fibromatosis (0 of 17) and Schwannoma (0 of 3), were immunonegative for DOG1. Only 4 of 438 non-GIST cases were immunoreactive for DOG1. DOG1, a protein of unknown function, is expressed strongly on the cell surface of GISTs and is rarely expressed in other soft tissue tumors. Reactivity for DOG1 may aid in the diagnosis of GISTs, including PDGFRA mutants that fail to express KIT antigen, and lead to appropriate treatment with imatinib mesylate, an inhibitor of the KIT tyrosine kinase.
Gastrointestinal stromal tumors occur in the wall of the bowel and have been proposed to arise from the interstitial cells of Cajal. The differential diagnosis of these tumors includes desmoid fibromatosis, Schwannoma, leiomyosarcoma, and, in some cases, high-grade sarcomas.1 Accurate diagnosis of gastrointestinal stromal tumor (GIST) is important, because imatinib mesylate has been shown to significantly inhibit these tumors presumably through inhibition of the KIT tyrosine kinase receptor, which is highly expressed in these tumors.2–5 As a result, the diagnosis of GIST relies heavily on KIT immunoreactivity. Current recommendations in the literature emphasize a diffuse, strong KIT immunoreactivity for the diagnosis of GIST.6 CD34 immunostaining can also aid in the diagnosis, but a subset of cases is immunonegative while many other types of sarcomas are immunoreactive for this marker.7–10 In the vast majority of GISTs, high levels of KIT expression are accompanied by a KIT gene mutation in exons 9, 11, 13, or 17.11,12
Recently, a subset of GISTs have been found to have PDGFRA mutations rather than KIT mutations.13,14 Patients with GISTs containing mutations in PDGFRA may still benefit from imatinib therapy, but these tumors often fail to react with antibodies against KIT and hence may remain undiagnosed as GIST.2 In addition, some GISTs with KIT mutations may have low KIT expression by immunohistochemistry yet will still respond to imatinib therapy.15
Although much work has been done on the biology of GISTs and KIT, additional insight has recently been gained through gene microarray studies.16–18 These studies have identified a number of genes whose expression is relatively increased compared to other soft tissue tumors. This includes genes known to be involved with GISTs, such as KIT and CD34, but also includes a number of genes that have not been well characterized. We have generated an antiserum against one GIST-specific gene, encoding for the hypothetical protein FLJ10261, which we have named “Discovered on GIST 1” (DOG1). Using immunohistochemistry with this antiserum and in situ hybridization with DOG1-specific probes, we show that DOG1 is highly expressed not only in typical GISTs but also in KIT-mutation-negative GISTs.
Materials and Methods
Tissue Microarray (TMA)
The studies described here were performed with the approval of the Institutional Review Board at Stanford University Hospital. Two TMAs were used for this study. The first TMA contained 460 different soft tissue tumors from 421 patients, with each tumor represented by two cores. The samples were distributed over two array blocks that were constructed using a technique previously described19 with a tissue arrayer from Beecher Instruments, Silver Spring, MD. Cores (0.6 mm) were taken from paraffin-embedded soft tissue tumors archived from the Stanford University Medical Center between 1995 and 2001. This array has also been used for characterization of apolipoprotein D expression.20 The second TMA used GISTs that were obtained from the pathology archives of Oregon Health and Science University Hospital, the Portland VA Medical Center, and the Kaiser Permanente Northwest Regional Laboratory. This single-block array consisted of 0.6-mm cores from formalin-fixed, paraffin-embedded tumor assembled using a semiautomated tissue arrayer.21 There was one core for each tumor, and all of the GISTs on this TMA were analyzed for mutations in exons 9, 11, 13, and 17 of the KIT gene using a combination of denaturing high pressure liquid chromatography and direct sequencing, as previously described.13,22 KIT wild-type tumors included on the array were also screened for mutations in exons 12 and 18 of the PDGFRA gene.13
Antibody Generation
The cDNA-derived protein sequence of DOG1 showed no significant homology with other genes, including the KIT gene. A rabbit polyclonal antibody was raised by injecting three peptides derived from the gene sequence (Applied Genomics Inc., Hunstville, AL). These peptides have no sequence homology to KIT. The peptides were synthesized by standard FMOC chemistry: peptide 1, EEAVKDHPRAEYEARVLEKSLK; peptide 2, DHEECVKR-KQRYEVDYNLE; peptide 3, KEKVLMVELFMREEQDK. The peptides were conjugated to keyhole limpet hemocyanin and injected into two out-bred rabbits. The serum (S284) was harvested after the rabbits demonstrated a significant anti-peptide titer. Affinity-purified antibodies were obtained by passing the antiserum over an affinity column conjugated with the three peptides; bound antibodies were eluted with a pH gradient.
Immunohistochemistry
Primary antibodies were directed toward DOG1 (S284, rabbit polyclonal, 1:50; Applied Genomics Inc.) and KIT (rabbit polyclonal, 1:50; DAKO, Carpinteria, CA). Serial sections of 4 μm were cut from the tissue array blocks, deparaffinized in xylene, and hydrated in a graded series of alcohol. Staining was then performed using the EnVision+ anti-rabbit system (DAKO).
In Situ Hybridization
In situ hybridization of TMA sections was performed based on a protocol published previously.23,24 Briefly, digoxigenin-labeled sense and anti-sense RNA probes are generated by polymerase chain reaction amplification of 400- to 600-bp products with the T7 promoter incorporated into the primers. In vitro transcription was performed with a digoxigenin RNA-labeling kit and T7 polymerase according to the manufacturer’s protocol (Roche Diagnostics, Indianapolis, IN). Sections (5 μm thick) cut from the paraffin blocks, deparaffinized in xylene, were hydrated in graded concentrations of ethanol for 5 minutes each. Sections were then incubated with 1% hydrogen peroxide, followed by digestion in 10 μg/ml of proteinase K at 37°C for 30 minutes. Sections were hybridized overnight at 55°C with either sense or anti-sense riboprobes at 200 ng/ml dilution in mRNA hybridization buffer (DAKO). The following day, sections were washed in 2× standard saline citrate and incubated with 1:35 dilution of RNase A cocktail (Ambion, Austin, TX) in 2× standard saline citrate for 30 minutes at 37°C. Next, sections were stringently washed in 2× standard saline citrate/50% formamide twice, followed by one wash at 0.08× standard saline citrate at 50°C. Biotin-blocking reagents (DAKO) were applied to the section to block the endogenous biotin. For signal amplification, a horseradish peroxidase-conjugated rabbit anti-digoxigenin antibody (DAKO) was used to catalyze the deposition of biotinyl tyramide, followed by secondary streptavidin complex (GenPoint kit, DAKO). The final signal was developed with diaminobenzidine (GenPoint kit, DAKO), and the tissues were counterstained in hematoxylin for 15 seconds.
Scoring of Immunohistochemistry and in Situ Hybridization
Cores were scored as follows. A score of 0 was given for absent or insignificant staining: less than 5% tumor cells with light brown staining. A score of 1 was given for unscorable cores. A score of 2 was given for light brown stain in greater than 5% of tumor cells or dark brown stain in less than 50% of tumor cells. A score of 3 was given for dark brown staining in greater that 50% tumor cells. Nontumor cells and cells of unknown origin were not scored. The cores were independently reviewed by two pathologists (RBW and MvdR) and disagreements were reviewed together to achieve a consensus score.
Digital Image Collection and Data Analysis
To aid in the analysis of numerous tissue cores stained by immunohistochemistry and in situ hybridization, digital images were collected using the BLISS instrument (Bacuslabs, Lombard IL; http://bacuslabs.com). Scoring results were combined using Deconvoluter and represented in Treeview,25 as shown on the accompanying website (http://microarray-pubs.stanford.edu/tma_portal/dog1/), where more than 4000 digital images are available.
Results
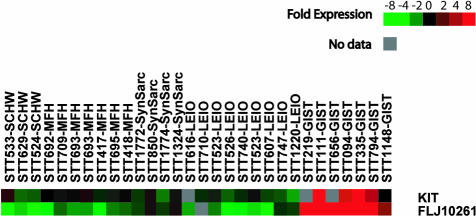
Previously, we examined the gene expression profile of GISTs using cDNA microarrays and identified a number of genes, in addition to the KIT gene, that demonstrated a specific pattern of elevated mRNA expression in GISTs.18 Figure 1 shows the relative level of mRNA expression for one of these genes, DOG1 (FLJ10261), compared with KIT in a variety of soft tissue tumors, including those in the differential diagnosis of GIST. Searches failed to show any sequence similarity between the genes on either the DNA or protein level.
Figure 1.
Gene array measurement of KIT and DOG1 mRNA expression in 30 soft tissue tumors. Red indicates a relatively high level of expression whereas green denotes a low level of expression. Gene array data for STTs 524, 629, 417, 418, 219, 111, 656, 94, 335, 794, 1148, 850, 616, 710, 523, 526, 740, 607, and 1220 have been previously reported.18
A rabbit antiserum was generated against synthetic peptides derived from the putative coding sequence of DOG1. Antiserum immunoreactivity was characterized on two separate TMAs containing soft tissue tumors. The first TMA contained 460 different soft tissue tumor samples representing more than 50 different diagnostic entities.20 This array included 22 KIT-immunoreactive GISTs. The second TMA included 127 GIST cases for which the KIT and PDGFRA mutation status was previously determined. On this TMA there were 102 cases with an activating mutation in KIT, 8 cases with a mutation in PDGFRA, and 17 cases that were wild type for both kinases but nevertheless had clinical, histological, and immunophenotypic features typical for GIST.
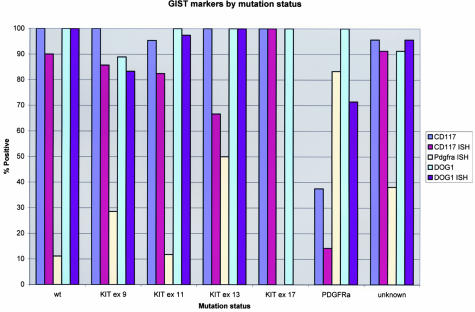
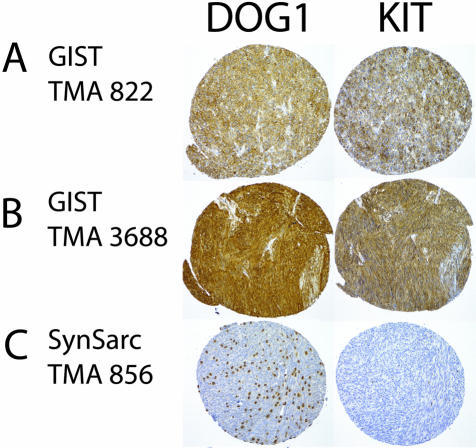
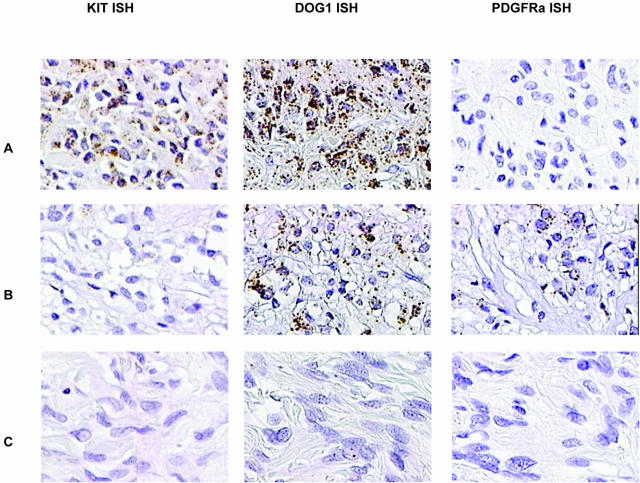
In these two TMAs, 136 of 139 scorable GISTs (97.8%) demonstrated immunoreactivity with DOG1 antiserum (Figures 2 and 3, Table 1). The staining observed with DOG1 antisera appeared predominately localized to the plasma membrane (Figure 4A). In some very strongly immunoreactive samples, the subcellular distribution of the staining could not be evaluated (Figure 4B). Mast cells present in some of the samples, for example synovial sarcoma, were strongly immunoreactive as well (Figure 4C), whereas the same samples showed only weak staining in the mast cells with KIT antibodies. We confirmed these results with in situ hybridization studies (Figures 5 and 6). Interestingly, DOG1 antisera stained all eight scorable PDGFRA-mutant GISTs (one case from first TMA and seven cases from second TMA), whereas the KIT antibody staining was weak in three of these cases and negative in the remaining five. These findings were further extended by in situ hybridization with PDGFRA (Figure 6). PDGFRA expression was predominately, but not exclusively, present in the PDGFRA-mutant GISTs. Five of six (83%) scorable PDGFRA-mutant GISTs were positive for PDGFRA in situ hybridization (Figures 2 and 3, Table 1). In contrast, only 10 of 70 (14%) KIT-mutant and KIT-wild-type GISTs were positive for PDGFRA in situ hybridization. Correlation of KIT in situ hybridization with KIT immunohistochemistry was good, with the in situ hybridization signal detectable in almost all immunopositive cases (Figure 2). However, a difference was seen in the PDGFRA-mutant GISTs with regard to KIT expression. Three cases were immunopositive for KIT, but only one case was positive by KIT in situ hybridization. Hierarchical clustering analysis of immunohistochemistry and in situ hybridization data were performed as previously described.25 Among these parameters—KIT immunohistochemistry, KIT in situ hybridization, DOG1 immunohistochemistry, DOG1 in situ hybridization, and PDGFRA in situ hybridization—the most distinguishing feature was PDGFRA in situ hybridization positivity (Figure 2), with overexpression of PDGFRA by PDFGRA in situ hybridization seen in only in a small subset of GISTs. Images of all cores from both TMAs were digitally captured and are available at the accompanying website (http://microarray-pubs.stanford.edu/tma_portal/dog1/).
Figure 2.

Hierarchical clustering of CD117 (KIT) immunohistochemistry, CD117 in situ hybridization, PDGFRA in situ hybridization, DOG1 immunohistochemistry, and DOG1 in situ hybridization. The results for GISTs on the two TMAs have been combined. Antisera or hybridization probes are in columns, tumors in rows. Bright red denotes strong reactivity, whereas dark red and green indicate low and absent reactivity, respectively. White means missing data.
Figure 3.
Staining results on GISTs for CD117 (KIT) immunohistochemistry, CD117 in situ hybridization, PDGFRA in situ hybridization, DOG1 immunohistochemistry, and DOG1 in situ hybridization in graphic form (see also Table 1).
Table 1.
Staining Results for CD117 IHC, CD117 ISH, PDGFRA ISH, DOG1 IHC, and DOG1 ISH in Tabular Form (see also Figure 3)
| Mutation status | CD117 | CD117 ISH | PDGFRA ISH | DOG1 | DOG1 ISH | |
|---|---|---|---|---|---|---|
| wt | 14 | 10 | 9 | 14 | 3 | Total scorables |
| 14 | 9 | 1 | 14 | 3 | Total positive | |
| 100 | 90 | 11 | 100 | 100 | % positive | |
| KIT ex 9 | 9 | 7 | 7 | 9 | 6 | Total scorables |
| 9 | 6 | 2 | 8 | 5 | Total positive | |
| 100 | 86 | 29 | 89 | 83 | % positive | |
| KIT ex 11 | 86 | 57 | 51 | 81 | 39 | Total scorables |
| 82 | 47 | 6 | 81 | 38 | Total positive | |
| 95 | 82 | 12 | 100 | 97 | % positive | |
| KIT ex 13 | 3 | 3 | 2 | 3 | 2 | Total scorables |
| 3 | 2 | 1 | 3 | 2 | Total positive | |
| 100 | 67 | 50 | 100 | 100 | % positive | |
| KIT ex 17 | 1 | 1 | 1 | 1 | 0 | Total scorables |
| 1 | 1 | 0 | 1 | 0 | Total positive | |
| 100 | 100 | 0 | 100 | NA | % positive | |
| PDGFRA | 8 | 7 | 6 | 8 | 7 | Total scorables |
| 3 | 1 | 5 | 8 | 5 | Total positive | |
| 37.5 | 14 | 83 | 100 | 71 | % positive | |
| Unknown | 23 | 23 | 21 | 23 | 23 | Total scorables |
| 22 | 21 | 8 | 21 | 22 | Total positive | |
| 96 | 91 | 38 | 91 | 96 | % positive |
IHC, immunohistochemistry; ISH, in situ hybridization.
Figure 4.
Immunohistochemical staining with anti-DOG1 serum (S284) and KIT on two GISTs [TMA 822 (A) and 3688 (B)] and a synovial sarcoma [TMA 856 (C)].
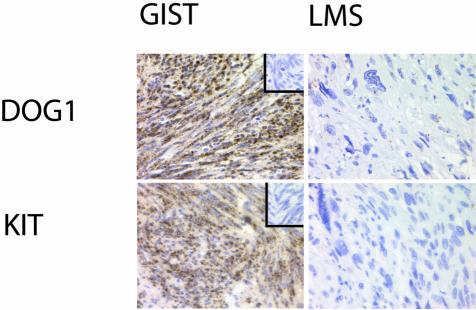
Figure 5.
In situ hybridization of a GIST and leiomyosarcoma with anti-sense probes to DOG1 and KIT on a GIST and a leiomyosarcoma (LMS). The corresponding negative control sense probes are included in the inset in the top right corner of the GIST sample.
Figure 6.
In situ hybridization of KIT, DOG1, and PDGFRA with GISTs. A: GIST with mutation in KIT shows positive in situ hybridization for KIT and DOG1 but not PDGFRA. B: GIST with mutation in PDGFRA shows positive in situ hybridization for DOG1 and PDGFRA but not for KIT. C: Negative control leiomyosarcoma.
From the 460 tumor samples that were not classified as GIST in the first TMA, only four cases that were not histologically and immunophenotypically consistent with GIST were immunoreactive with DOG1 antiserum: one synovial sarcoma (1 of 20 = 5%), one (1 of 40 = 2.5%) leiomyosarcoma, one (1 of 4 = 25%) fibrosarcoma, and (1 of 9 = 11%) one Ewing’s sarcoma/PNET. Of the 40 leiomyosarcomas, 17 originated in the abdomen and none of these were DOG1 immunoreactive. Other tumors in the GIST differential diagnosis failed to stain with the DOG1 antisera. These include desmoid fibromatosis (17 cases) and Schwannoma (3 cases). Parenthetically, under the staining conditions used, none of the fibromatosis cases were positive for KIT by immunohistochemistry or in situ hybridization. One leiomyosarcoma was positive for KIT immunohistochemistry only (TMA 3725). Interestingly, the staining was exclusively in a diffuse nuclear pattern. This tumor was negative for DOG1 by both immunohistochemistry and in situ hybridization and for KIT in situ hybridization.
Seven cases in the first TMA, not counted among the 22 unequivocal GISTs, showed histological features indeterminate between GIST and smooth muscle tumor. All of these tumors were located in the wall of the stomach or intestine, with four tumors from the stomach, one from the duodenum, one from the gastro-esophageal junction, and one from the rectum. All seven cases were negative for KIT by immunohistochemistry and thus might not be considered GISTs according to current recommendations.6 However, four of the seven cases were positive by KIT in situ hybridization, while DOG1 immunoreactivity was seen in two cases, and all seven cases were positive for DOG1 by in situ hybridization. Furthermore, two cases (TMAs 863 and 3696) were positive for PDGFRA in situ hybridization. Subsequent sequence analysis of cases 863 and 3696 revealed a point mutation and a deletion in exon 18 of PDGFRA, respectively. To date, such mutations have only been described in GISTs. We conclude that the seven KIT immunonegative cases with morphological features between GIST and smooth muscle tumor actually represent GISTs.
We also stained a TMA containing a spectrum of normal tissues with the DOG1 antiserum (data not shown). We observed staining in the epithelium of breast, prostate, salivary gland, liver, stomach, testis, pancreas, and gallbladder. The pattern of DOG1 immunostaining of the interstitial cells of Cajal was similar to KIT. In addition, DOG1 antiserum reacted with a number of tumor cores in a carcinoma array, including some that did not stain with KIT antiserum (data not shown).
Discussion
GISTs have a high rate of local recurrence.1 Imatinib, a small molecule inhibitor of several type III receptor tyrosine kinases, including KIT and PDGFRA, has demonstrated promise in controlling GIST growth.3–5 The majority of GISTs (80 to 85%) harbor oncogenic mutations of KIT, and for this reason KIT has been regarded as the primary target for imatinib therapy. Indeed, initial trials of imatinib were limited to KIT-immunoreactive GISTs. Recently it was discovered that a subset of GISTs (5 to 7%) has activating mutations of PDGFRA.13,14 Most of these tumors are weak or negative in immunostaining for KIT, which may lead to underdiagnosis and possible withholding of imatinib therapy. Furthermore, identification of PDGFRA-mutant GISTs requires molecular analysis, a laborious process that is not ideal for application in a routine clinical setting.
In this article, we demonstrate that a novel gene, DOG1, identified in a DNA microarray analysis of gene expression patterns as associated with GIST, is highly expressed in both KIT- and PDGFRA-mutant GISTs. Expression of DOG1 in GISTs was demonstrated both by immunodetection of the protein and by in situ hybridization. DOG1 immunoreactivity was assessed on two soft tissue tumor microarrays representing 587 soft tissue tumors, including 149 GISTs. Of scorable GISTs 97.8% demonstrated immunoreactivity with DOG1 antisera. Only four KIT-negative, non-GIST soft tissue tumors were DOG1 immunoreactive. Several GISTs with mutations in the PDGRFA gene were found to react only by in situ hybridization for DOG1 and to be negative for DOG1 by immunohistochemistry. Future studies are necessary to determine whether monoclonal antibodies against purified DOG1 might yield tools with sensitivity similar to that seen with in situ hybridization probes. We also confirm PDGFRA expression in a subset of GISTs using in situ hybridization. PDGRFA expression and KIT expression are not mutually exclusive. A subset of KIT-mutated GISTs expresses PDGRFA in addition to KIT while a subset of PDGRFA-mutated tumors also expresses KIT. These data were seen with both immunohistochemical and in situ hybridization techniques.
In addition to the marked similarity in reactivity for DOG1 protein on non-GIST sarcomas, DOG1 protein can also be seen in a subset of melanomas and germ cell tumors as has been described for KIT (West et al, in preparation). Furthermore just as seen with the KIT molecule, a variety of carcinomas also express DOG1. These tumors mostly overlap with the KIT-positive tumors. Although within the field of soft tissue tumors DOG1 expression seems quite specific for GIST, in a differential diagnostic setting DOG1 reactivity does not exclude carcinomas. Therefore additional markers such as keratin stains should be performed when the differential diagnosis includes carcinoma.
We also demonstrated the feasibility of assessing GIST markers by in situ hybridization on paraffin-embedded tissue. Correlation between immunohistochemistry and in situ hybridization for DOG1 on GISTs was excellent. In the case of KIT, the correlation was not as strong because of relatively weak or absent in situ hybridization signals in some CD117-positive GISTs. It is likely that this reflects lower sensitivity of the KIT in situ hybridization assay, although cross-reactivity of the CD117 antibody to another epitope on GISTs has not been excluded. In situ hybridization for PDGFRA proved to be valuable in identifying KIT-negative GISTs, although DOG1 immunohistochemistry was equally sensitive for these cases. Overall, we have found that in situ hybridization techniques are complementary to immunohistochemistry tests in the evaluation of GISTs.
DOG1 has been recently identified as a gene in the CCND1-EMS1 locus on human chromosome 11q13, which is amplified in esophageal cancer, bladder tumors, and breast cancer.26 Human DOG1 protein showed 89.8% total amino acid identity with mouse DOG1 protein, and also 58.4%, 38.3%, and 38.6% identity with human C12orf3, C11orf25, and FLJ34272/BAC03704 proteins, respectively. Sequence analysis predicts the presence of eight transmembrane-spanning segments. This correlates with our observations of the immunohistochemical localization to the cell membrane. DOG1 may be part of an as yet unclassified ion transporter family.
Because the biological function is unknown, it is unclear why DOG1 is so widely expressed in GISTs. Two broad possibilities exist. It may be that the protein has a role in receptor kinase type III signal transduction pathways. On the other hand, DOG1 may be a fortuitous marker of the GIST phenotype, with no direct connection to the KIT and PDGFRA signaling pathways. The finding that mast cells are also immunoreactive for DOG1 tends to favor the former possibility.
In summary, we demonstrate that detection of a novel gene, DOG1, identifies the vast majority of both KIT- and PDGFRA-mutated GISTs. This may be of clinical value in identifying candidates for Gleevec therapy. As a cell membrane-associated protein, with markedly elevated expression in GISTs, DOG1 may also be a potential therapeutic target.
Footnotes
Address reprint requests to Matt van de Rijn: Department of Pathology, Stanford University Medical Center, 300 Pasteur Dr., Stanford, CA, 94305. E-mail: mrijn@stanford.edu.
Supported by the National Institutes of Health (grants CA85129 and CA84967) and the Howard Hughes Medical Institute.
P.O.B. is an Associate Investigator of the Howard Hughes Medical Institute.
References
- Weiss SW, Goldblum JR. St Louis: Mosby; Soft Tissue Tumors. 2001 [Google Scholar]
- Heinrich MC, Corless C, Demetri GD, Blanke C, von Mehren M, Joensuu H, McGreevey L, Chen CJ, Van den Abbeele A, Druker B, Kiese B, Eisenberg B, Roberts P, Singer S, Fletcher CD, Silberman S, Dimitrijevic S, Fletcher JA. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol. 2003;21:4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- van Oosterom AT, Judson I, Verweij J, Stroobants S, Donato di Paola E, Dimitrijevic S, Martens M, Webb A, Sciot R, Van Glabbeke M, Silberman S, Nielsen OS, European Organization for Research and Treatment of Cancer Soft Tissue and Bone Sarcoma Group Safety and efficacy of imatinib (STI571) in metastatic gastrointestinal stromal tumours: a phase I study. Lancet. 2001;358:1421–1423. doi: 10.1016/s0140-6736(01)06535-7. [DOI] [PubMed] [Google Scholar]
- Demetri G, von Mehren M, Blanke C, Van den Abbeele A, Eisenberg B, Roberts P, Heinrich M, Tuveson D, Singer S, Janicek M, Fletcher J, Silverman S, Silberman S, Capdeville R, Kiese B, Peng B, Dimitrijevic S, Druker B, Corless C, Fletcher C, Joensuu H. Efficacy and safety of imatinib mesylate in advanced gastrointestinal stromal tumors. N Engl J Med. 2002;347:472–480. doi: 10.1056/NEJMoa020461. [DOI] [PubMed] [Google Scholar]
- Joensuu H, Roberts P, Sarlomo-Rikala M, Andersson L, Tervahartiala P, Tuveson D, Silberman S, Capdeville R, Dimitrijevic S, Druker B, Demetri G. Effect of the tyrosine kinase inhibitor STI571 in a patient with a metastatic gastrointestinal stromal tumor. N Engl J Med. 2001;344:1052–1056. doi: 10.1056/NEJM200104053441404. [DOI] [PubMed] [Google Scholar]
- Fletcher CD, Berman JJ, Corless C, Gorstein F, Lasota J, Longley BJ, Miettinen M, O’Leary TJ, Remotti H, Rubin BP, Shmookler B, Sobin LH, Weiss SW. Diagnosis of gastrointestinal stromal tumors: a consensus approach. Hum Pathol. 2002;33:459–465. doi: 10.1053/hupa.2002.123545. [DOI] [PubMed] [Google Scholar]
- van de Rijn M, Rouse RV. CD34: a review. Appl Immunohistochem. 1994;2:71–80. [Google Scholar]
- van de Rijn M, Hendrickson MR, Rouse RV. CD34 expression by gastrointestinal tract stromal tumors. Hum Pathol. 1994;25:766–771. doi: 10.1016/0046-8177(94)90245-3. [DOI] [PubMed] [Google Scholar]
- Yantiss RK, Spiro IJ, Compton CC, Rosenberg AE. Gastrointestinal stromal tumor versus intra-abdominal fibromatosis of the bowel wall: a clinically important differential diagnosis. Am J Surg Pathol. 2000;24:947–957. doi: 10.1097/00000478-200007000-00006. [DOI] [PubMed] [Google Scholar]
- Smithey BE, Pappo AS, Hill DA. C-kit expression in pediatric solid tumors: a comparative immunohistochemical study. Am J Surg Pathol. 2002;26:486–492. doi: 10.1097/00000478-200204000-00011. [DOI] [PubMed] [Google Scholar]
- Hirota S, Nishida T, Isozaki K, Taniguchi M, Nakamura J, Okazaki T, Kitamura Y. Gain-of-function mutation at the extracellular domain of KIT in gastrointestinal stromal tumours. J Pathol. 2001;193:505–510. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH818>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Rubin B, Singer S, Tsao C, Duensing A, Lux M, Ruiz R, Hibbard M, Chen C, Xiao S, Tuveson D, Demetri G, Fletcher C, Fletcher J. KIT activation is a ubiquitous feature of gastrointestinal stromal tumors. Cancer Res. 2001;61:8118–8121. [PubMed] [Google Scholar]
- Heinrich MC, Corless CL, Duensing A, McGreevey L, Chen CJ, Joseph N, Singer S, Griffith DJ, Haley A, Town A, Demetri GD, Fletcher CD, Fletcher JA. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299:708–710. doi: 10.1126/science.1079666. [DOI] [PubMed] [Google Scholar]
- Hirota S, Ohashi A, Nishida T, Isozaki K, Kinoshita K, Shinomura Y, Kitamura Y. Gain-of-function mutations of platelet-derived growth factor receptor alpha gene in gastrointestinal stromal tumors. Gastroenterology. 2003;125:660–667. doi: 10.1016/s0016-5085(03)01046-1. [DOI] [PubMed] [Google Scholar]
- Bauer S, Corless C, Heinrich M, Dirsch O, Antoch G, Kanja J, Seeber S, Schutte J. Response to imatinib mesylate of a gastrointestinal stromal tumor with very low expression of KIT. Cancer Chemother Pharmacol. 2003;51:261–265. doi: 10.1007/s00280-002-0564-x. [DOI] [PubMed] [Google Scholar]
- Allander SV, Nupponen NN, Ringner M, Hostetter G, Maher GW, Goldberger N, Chen Y, Carpten J, Elkahloun AG, Meltzer PS. Gastrointestinal stromal tumors with KIT mutations exhibit a remarkably homogeneous gene expression profile. Cancer Res. 2001;61:8624–8628. [PubMed] [Google Scholar]
- Khan J, Wei JS, Ringner M, Saal LH, Ladanyi M, Westermann F, Berthold F, Schwab M, Antonescu CR, Peterson C, Meltzer PS. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nat Med. 2001;7:673–679. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen TO, West RB, Linn SC, Alter O, Knowling MA, O’Connell JX, Zhu S, Fero M, Sherlock G, Pollack JR, Brown PO, Botstein D, van de Rijn M. Molecular characterisation of soft tissue tumours: a gene expression study. Lancet. 2002;359:1301–1307. doi: 10.1016/S0140-6736(02)08270-3. [DOI] [PubMed] [Google Scholar]
- Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844–847. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- West RB, Harvell J, Linn S, Lui C, Prapong W, Hernandez-Boussard T, Montgomery K, Nielsen TO, Rubin BP, Patel R, Goldblum JR, Brown P, van de Rijn M: Apo D in soft tissue tumors: a novel marker for dermatofibrosarcoma protuberans. Am J Surg Pathol (In press) [DOI] [PubMed] [Google Scholar]
- Torhorst J, Bucher C, Kononen J, Haas P, Zuber M, Kochli O, Mross F, Dieterich H, Moch H, Mihatsch M, Kallioniemi O, Sauter G. Tissue microarrays for rapid linking of molecular changes to clinical endpoints. Am J Pathol. 2001;159:2249–2256. doi: 10.1016/S0002-9440(10)63075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corless C, McGreevey L, Haley A, Town A, Heinrich M. KIT mutations are common in incidental gastrointestinal stromal tumors one centimeter or less in size. Am J Pathol. 2002;160:1567–1572. doi: 10.1016/S0002-9440(10)61103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Croix B, Rago C, Velculescu V, Traverso G, Romans K, Montgomery E, Lal A, Riggins G, Lengauer C, Vogelstein B, Kinzler K. Genes expressed in human tumor endothelium. Science. 2000;289:1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- Iacobuzio-Donahue CA, Ryu B, Hruban RH, Kern SE. Exploring the host desmoplastic response to pancreatic carcinoma: gene expression of stromal and neoplastic cells at the site of primary invasion. Am J Pathol. 2002;160:91–99. doi: 10.1016/S0002-9440(10)64353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CL, Prapong W, Natkunam Y, Alizadeh A, Montgomery K, Gilks CB, van de Rijn M. Software tools for high-throughput analysis and archiving of immunohistochemistry staining data obtained with tissue microarrays. Am J Pathol. 2002;161:1557–1565. doi: 10.1016/S0002-9440(10)64434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M, Katoh M. FLJ10261 gene, located within the CCND1-EMS1 locus on human chromosome 11q13, encodes the eight-transmembrane protein homologous to C12orf3, C11orf25 and FLJ34272 gene products. Int J Oncol. 2003;22:1375–1381. [PubMed] [Google Scholar]