Abstract
The M184V substitution in human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT), encoding high-level resistance to lamivudine (3TC), results in decreased HIV-1 replicative capacity, diminished RT processivity, and increased RT fidelity in biochemical assays. We assessed the effect of M184V on the development of resistance to the nonnucleoside RT inhibitors efavirenz (EFV) and nevirapine, and to the protease inhibitor amprenavir (APV) in tissue culture. Genotypic analysis revealed differences in EFV resistance-conferring mutations in subtype B (K103N) versus subtype C (V106 M), and the appearance of both was significantly delayed in the M184V-containing variants compared with the wild type (WT). Similarly, there was a marked delay in the emergence of mutations associated with APV resistance (I54 M/L/V) in subtype B viruses harboring M184V compared with paired WT viral isolates.
The YMDD catalytic domain of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) is highly conserved among polymerase enzymes, and mutations within this region have a significant effect on RT catalytic activity (23). The M184V mutation results in high-level resistance to the (−) enantiomer of 2′,3′-dideoxy-3′-thiacytidine (3TC) as well as low-level resistance to other nucleoside RT inhibitors (NRTIs), including 2′,3′-dideoxycytidine, 2′,3′-dideoxyinosine (ddI), and abacavir (2, 6, 8, 20). M184V is also associated with diminished RT processivity and viral replication fitness in primary cells (1, 3, 12, 13, 21) as well as increased RT fidelity, as assessed by deoxynucleotide misincorporation and misinsertion experiments (9, 16, 21).
Interestingly, in vivo studies have revealed that 3TC can continue to exert virological benefit in spite of the M184V mutation (5). The presence of M184V can also result in increased susceptibility to zidovudine (ZDV) and other drugs in vitro (13, 22). In addition, a residual antiviral activity of 3TC may sometimes still be detected in spite of M184V (17), and M184V RT can impair the rescue of ZDV-terminated primers during synthesis of viral DNA (7).
However, other work has shown that exposure of HIV-IIIB-containing M184V to nine different non-NRTIs (NNRTIs) resulted in the rapid emergence of NNRTI-resistant virus and that this occurred at a rate similar to that seen with wild-type (WT) HIV-IIIB (10, 13). We have monitored the evolutionary potential and resistance patterns of M184V-containing versus parental WT variants when grown in the presence of efavirenz (EFV) for both subtypes B and C and amprenavir (APV) for subtype B viruses and found that the variants containing M184V required a longer period of selection (i.e., delays of 10 and 29 weeks, respectively) to develop phenotypic resistance to EFV and APV under conditions in which the genotypic patterns of the WT and M184V-containing variants were similar.
Selection of resistance to EFV by using subtype B and C clinical isolates.
HIV-infected subtype B clinical isolates were obtained with informed consent from drug-naïve individuals at our clinics in Montreal, Canada, and subtype C clinical isolates were obtained from treatment-naïve subjects from Ethiopia and Botswana (14). Viral strains were amplified as described by coculture of peripheral blood mononuclear cells from infected patients with uninfected cord blood mononuclear cells (11). Cord blood mononuclear cells were also used to generate 3TC-resistant variants of both of the subtype HIV-1 isolates as described by using a multiplicity of infection of 0.01 (6). Drug susceptibility assays and 50% inhibitory concentrations (IC50) of 3TC were determined by RT assay as described (6, 14); at each passage, cells were collected and kept at −80°C for sequencing. Insofar as drug selections with EFV and APV required 30 to 52 weeks for development of resistance, viruses were maintained in the presence of low concentrations of 3TC (i.e., 0.1 μM) to prevent back-mutation of M184V and overgrowth with WT variants. In the absence of 3TC, M184V-containing viruses reverted to WT within 20 weeks (data not shown). Samples were genotyped by using the Open Gene automated DNA system (Visible Genetics Inc., Toronto, Ontario, Canada). We received 3TC, EFV, and APV as gifts from Shire Biochem. Inc. (Montreal, Canada), Bristol-Myers-Squibb Pharmaceuticals (Montreal, Quebec, Canada), and GlaxoSmithKline (Research Triangle Park, N.C.), respectively.
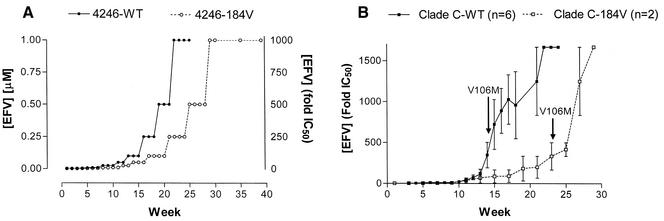
For all EFV selections, the starting and endpoint concentrations were 0.001 μM and 1 μM, respectively (Fig. 1A). EFV concentrations had to be increased slowly due to marked decreases in RT activity of treated cultures compared with parallel cultures grown in the absence of drug (Fig. 1A), and a WT virus required 30 weeks to become fully resistant to EFV through the acquisition of the K103N mutation compared with 40 weeks in the case of a matched virus containing the M184V substitution (Table 1). In contrast, resistance to NVP (K103N) arose within 10 weeks for both M184V-containing and WT viruses, and the appearance of K103N was necessary for high-level phenotypic resistance to EFV to occur (Table 1). There was a significant delay in time to the first appearance of a mixture of K103N and WT viruses in studies performed with M184V-containing viruses compared with WT (Tables 1 and 2).
FIG. 1.
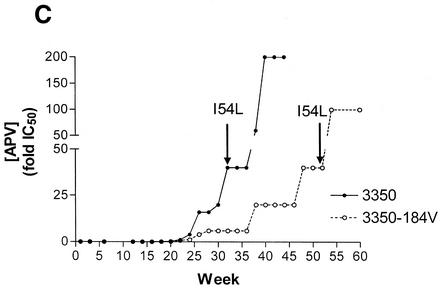
Selection of resistance to drugs in tissue culture. (A) Stepwise progression of EFV concentrations and development of resistance in a paired experiment involving the WT and M184V-containing 4246 clinical isolates of subtype B origin. (B) Selection of resistance to EFV in WT and M184V-containing clinical isolates of subtype C origin. Concentrations of EFV are expressed as changes in mean IC50 values (n-fold). Arrows indicate time to first appearance of V106 M. Values represent the mean ± standard error of the mean (n = 6 and 2 for WT and M184V isolates, respectively). (C) Selection of resistance to APV in WT and M184V-containing clinical isolates of subtype C origin. Concentrations of APV are expressed as changes in mean IC50 values (n-fold). Arrows indicate time to first appearance of the I54L mutation.
TABLE 1.
Time to development of major and minor mutations associated with resistance to EFV in WT and M184V subtype B isolatesa
| EFV concn (μM) | Wk | B-4246-WT
|
B-4246-184V
|
5346-WT
|
3350-184V
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Major | Minor | Major | Minor | Major | Minor | Major | Minor | ||
| 0 | 1 | WT | WT | M184V | WT | WT | WT | M184V | WT |
| 0.004 | 10 | WT | V108I/V, V179D/V | M184V | WT | WT | L100I/L | M184V | L100I/L, K101K/E, K101E |
| 0.1 | 20 | WT | L100I/L, V108I/V, V179D/V | M184V | L100I/L | K103K/Nb | L100I/L | M184V | |
| 1 | 24 | K103K/Nb | L100I/L, V108I/V, V179D/V | M184V | L100I | K103N | L100I | M184V | K101E |
| 1 | 30 | K103N | A98G, L100I, V108I/V, V179D | K103K/N,b M184V | L100I | K103N | L100I | M184V | K101E |
| 1 | 35 | K103N | A98G, L100I, V108I/V, V179D | K103N, M184V | L100I | K103N | L100I | K103K/N,b M184V | L100I/L |
| 1 | 40 | NDc | ND | ND | ND | K103N | L100I | K103N, M184V | L100I |
Genotypic analysis was performed at the designated weeks based on RT activity to determine the time to first appearance of resistance mutations. Baseline genotype is also presented.
Time to first appearance of the major/primary EFV resistance mutation in the quasispecies, i.e. K103N. The remaining mutations are accessory mutations that contribute to the evolution of high-level phenotypic resistance and/or increased viral fitness.
ND, not done.
TABLE 2.
Time to selection of phenotypic resistance to APV and EFV using WT versus M184V subtype B and C isolatesa
| Virus | Drug | Primary/major resistance mutation | Time to first appearance of primary resistance mutation (weeks) |
|---|---|---|---|
| B-WT (n = 5) | EFV | K103N | 22.0 ± 2.0 |
| B-M184V (n = 2) | EFV | K103N, M184V | 32.5 ± 2.5b |
| C-WT (n = 5) | EFV | V106M | 13.6 ± 1.6 |
| C-M184V (n = 2) | EFV | V106M, M184V | 24 ± 1.0b |
| B-WT (n = 2) | APV | I54M/L | 24.5 ± 1.5 |
| B-M184V (n = 2) | APV | I54L/V, M184V | 53.5 ± 5.2b |
APV and EFV concentrations were increased in stepwise progression based on RT activities. Genotypic analysis was performed to monitor the time to first appearance of major/primary resistance mutations associated with high-level phenotypic resistance (see Tables 1 and 3). Values represent the mean ± standard error of the mean of experiments performed on different clinical isolates.
P < 0.05 for t tests of WT versus M184V isolates.
Subtype C clinical isolates.
Resistance to EFV in subtype C viruses arises through the acquisition of a V106 M mutation that also yields high-level cross-resistance to all currently approved NNRTIs (7). EFV resistance through the acquisition of V106 M arose more rapidly in subtype C viruses than in subtype B viruses that acquired the K103N substitution. In addition, the time to appearance of V106 M was delayed by approximately 10 weeks in viruses that also contained M184V compared to WT (Fig. 1B, Table 2). Phenotypic assays showed >50-fold cross-resistance to NNRTIs upon acquisition of V106 M or K103N (4).
Selection of resistance to APV by using subtype B clinical isolates.
The B-3350-WT virus and its M184V-containing counterpart were selected for resistance to APV as described above, with RT assays performed on a weekly basis (Fig. 1C). APV concentrations were increased slowly, and no increases in drug concentrations were applied when levels of RT activity were <10% of those of control cultures grown in the absence of drug, since a too-rapid increase in protease inhibitor concentration caused complete viral suppression. A period of >6 months was required for the acquisition of mutations in the viral protease (PR) associated with high-level phenotypic resistance to APV, i.e., I54L/M/V (Fig. 1C, Table 3).
TABLE 3.
Development of resistance to APV in WT and M184V clade B isolatesa
| APV concn (μM) | Wk | 4246-WT | 4246-M184V | 3350-WT | 3350-M184V |
|---|---|---|---|---|---|
| 0 | 1 | WT | WT (M184V) | WT (L63P, V77I, I93L) | WT (L63P, V77I, I93L; M184V) |
| 0.1 | 23 | V32I, I54I/Mb | WT | WT | WT |
| 0.4 | 30 | V32I, I54M | WT | V32I, M46I, I54Lb | WT |
| 0.4 | 34-35 | V32I, I54M | V32I/V, N37S | V32I, M46I, I54L | M46I/L |
| 10 | 46-47 | V32I, M46I, I54M | V32I, M46I/M | V32I, M46I, I54L | M46L |
| 10 | 52 | V32I, M46I, I54M | V32I/V, M46I, I54I/Vb | ND | M46I, I54I/Lb |
| 10 | 55 | NDc | ND | ND | M46I, I54Lb |
| 10 | 58 | V32I, M46I, I54M | V32I/V, M46I, I54V | ND | ND |
Genotypic analysis was performed at designated weeks based on RT activity to determine the time to appearance of resistance mutations. The baseline genotype included mutations shown in parentheses that were sustained at subsequent time points.
Time to first appearance of the major resistance mutations, I54M/V/L, that confer high-level phenotypic resistance to protease inhibitors.
ND, not done.
Genotypic results comparing paired WT and M184V viruses show that 24 weeks were required for an initial emergence of phenotypic resistance to APV in WT viruses (Tables 2 and 3). This was delayed by >19 weeks for the M184V variants compared to WT, under conditions in which only very low concentrations of APV, i.e., <0.1 μM, were present for over 22 weeks (Fig. 1C, Table 3).
There was also a lag in regard to appearance of the major I54 M/V/L mutations associated with resistance to APV (Table 3) in the M184V variants versus WT. Phenotypic analysis revealed that acquisition of the I54V/L/M mutations conferred >50-fold resistance to APV as well as cross-resistance to indinavir and nelfinavir (data not shown). In addition to I54 M/V/L, other accessory mutations, i.e., V32I and M46I, were also present. Selections of subtype C viruses with APV have been in progress for over 1 year. We have not yet succeeded in documenting resistance to APV using either WT subtype C viruses or subtype C viruses containing the M184V substitution in RT.
Our study was designed to determine the impact of the M184V mutation in RT on the development of resistance to EFV and APV as drugs representative of NNRTIs and protease inhibitors, respectively. Overall, our data are in agreement with previously published data on the evolutionary potential of viruses harboring the M184V mutation (12), i.e., M184V attenuated the emergence of resistance to EFV in subtype B and C infections as well as resistance to APV in subtype B viruses. While these data appear to conflict with earlier findings that M184V viruses were not delayed in the development of resistance to NNRTIs (10), it is noteworthy that EFV resistance develops more slowly than that to NVP and delavirdine (DLV), and it was only NVP and not EFV that had previously been assessed (18, 19). Moreover, our study employed clinical isolates and primary cells compared with recombinant viruses and cell lines. In this regard, it should be noted that the lower fitness of M184V-containing viruses is more pronounced in primary cells than in cell lines (1, 13). Similar results were obtained in regard to the slower appearance of the I54L/M/V mutations in M184V variants of subtype B viruses that were selected with APV.
It has been demonstrated that a single-cycle recombinant assay can yield increased mutational rates of RTs resistant to ZDV in the presence of each of ZDV and 3TC (15). However, recombinant single-cycle assays do not take into account the cumulative effects of M184V on HIV-1 replicative rates, the ability of HIV-1 to initiate new rounds of infection, or the accumulation of mutations that may require months or even years to develop either in vivo or in cell culture.
Overall, our results demonstrate that the low replicative fitness of HIV-1 variants that contain M184V can delay the occurrence of resistance to at least some antiviral drugs, notably APV and EFV, in primary cells.
Nucleotide accession numbers.
Accession numbers for the clinical isolates used in this study are as follows: 3350 (PR-AY236362; RT AY236363), 5346 (PR-AY236360; RT-AY236361), 4246 (PR-AY213138; RT-AY213139). Accession numbers for the subtype C isolates have already been designated (21).
(This work was performed by Karidia Diallo in partial fulfillment of the requirements for a Ph.D. degree, faculty of Graduate Studies and Research, McGill University, Montreal, Quebec, Canada.)
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research and by a donation from Aldo and Diane Bensadoun.
Footnotes
Dedicated to the memory of James-Paul Marois.
REFERENCES
- 1.Back, N. K. T., M. Nijhuis, W. Keulen, C. A. B. Boucher, B. B. O. Essnik, A. B. P. van Kuilenburg, A. H. van Gennip, and B. Berkhout. 1996. Reduced replication of 3TC-resistant HIV-l variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 15:4040-4049. [PMC free article] [PubMed] [Google Scholar]
- 2.Boucher, C. A. B., N. Cammack, P. Schipper, R. Schuurman, P. L. Rouse, and J. M. Cameron. 1993. High level resistance to (−) enantiomeric 2′-deoxy-3′-thiacytidine (3TC) in vitro is due to one amino acid substitution in the catalytic site of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 37:2231-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenner, B. G., J. P. Routy, M. Petrella, D. Moisi, M. Oliveira, M. Deterio, B. Spira, V. Essabag, B. Conway, R. Lalonde, R. P. Sekaly, and M. A. Wainberg. 2002. Persistence and fitness of multidrug-resistant human immunodeficiency virus type 1 acquired in primary infection. J. Virol. 76:1753-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brenner, B. G., D. Turner, M. Oliveira., D. Moisi, M. Detorio, M. Carobene, R. G. Marlink, J. Schapiro, M. Roger, and M. A. Wainberg. 2003. A V106M mutation in HIV-1 clade C viruses exposed to efavirenz confers cross-resistance to non-nucleoside reverse transcriptase inhibitors. AIDS 17:F1-F5. [DOI] [PubMed] [Google Scholar]
- 5.Eron, J. J., S. L. Benoit, J. Jemsek, R. D. MacArthur, J. Santana, J. B. Quinn, D. R. Kuritzkes, M. A. Fallon, and M. Rubin. 1995. Treatment with lamivudine, zidovudine, or both in HIV-positive patients with 200 to 500 CD4+ cells per cubic millimeter. North American HIV Working Party. N. Engl. J. Med. 333:1662-1669. [DOI] [PubMed] [Google Scholar]
- 6.Gao, Q., Z. Gu, M. A. Paniak, J. Cameron, N. Cammack, C. Boucher, and M. A. Wainberg. 1993. The same mutation that encodes low-level human immunodeficiency virus type 1 resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine confers high-level resistance to the (−) enantiomer of 2′,3′-dideoxy-3′-thiacytidine. Antimicrob. Agents Chemother. 37:1390-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Götte, M., D. Arion, M. A. Parniak, and M. A. Wainberg. 2000. The M184V mutation in the reverse transcriptase of human immunodeficiency virus type 1 impairs rescue of chain-terminated DNA synthesis J. Virol. 74:3579-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu, Z., Q. Gao, X. Li, M. A. Parniak, and M. A. Wainberg. 1992. Novel mutation in the human immunodeficiency virus type 1 reverse transcriptase gene that encodes cross-resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine. J. Virol. 66:7128-7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu, M., P. Inouye, L. Rezende, N. Richard, Z. Li, V. R. Prasad, and M. A. Wainberg. 1997. Higher fidelity of RNA-dependent DNA mispair extension by M184V drug-resistant than wild-type reverse transcriptase of human immunodeficiency virus type 1. Nucleic Acids Res. 25:4532-4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jonckheere, H., M. Witvrouw, E. De Clercq, and J. Anné. 1998. Lamivudine resistance of HIV type 1 does not delay development of resistance to nonnucleoside HIV type 1-specific reverse transcriptase inhibitors as compared with wild-type HIV type 1. AIDS Res. Hum. Retrovir. 14:249-253. [DOI] [PubMed] [Google Scholar]
- 11.Keulen, W., N. K. Back, A. van Wijk, C. A. Boucher, and B. Berkhout. 1997. Initial appearance of the 184Ile variant in lamivudine-treated patients is caused by the mutational bias of human immunodeficiency virus type l reverse transcriptase J. Virol. 71:3346-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keulen, W., A. van Wijk, R. Schuurman, B. Berkhout, and C. A. B. Boucher. 1999. Increased polymerase fidelity of lamivudine-resistant HIV-1 variants does not limit their evolutionary potential. AIDS 13:1343-1349. [DOI] [PubMed] [Google Scholar]
- 13.Larder, B. A., S. D. Kemp, and P. R. Harrigan. 1995. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science 269:696-699. [DOI] [PubMed] [Google Scholar]
- 14.Loemba, H., B. Brenner, M. A. Parniak, S. Ma'ayan, S. B. Spira, D. Moisi, M. Oliveira, M. Deterio, and M. A. Wainberg. 2002. Genetic divergence of human immunodeficiency virus type 1 Ethiopian clade C reverse transcriptase (RT) and rapid development of resistance against nonnucleoside inhibitors of RT. Antimicrob. Agents Chemother. 46:2087-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mansky, L. M., D. K. Pearl, and L. C. Gajary. 2002. Combination of drugs and drug-resistant reverse transcriptase results in a multiplicative increase of human immunodeficiency virus type 1 mutant frequencies. J. Virol. 76:9253-9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pandey, V., N. Kaushik, N. Rege, S. G. Sarafianos, P. N. S. Yadav, and M. J. Modal. 1996. Role of methionine 184 of human immunodeficiency virus type-1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Biochemistry 35:2168-2179. [DOI] [PubMed] [Google Scholar]
- 17.Quan, Y., B. G. Brenner, M. Oliveira, and M. A. Wainberg 2003. Lamivudine can exert a modest antiviral effect against human immunodeficiency virus type 1 containing the M184V mutation. Antimicrobial Agents Chemother. 47:747-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richman, D. D., C. K. Shih, I. Lowy, J. Rose, P. Prodanovich, S. Goff, and J. Griffin. 1991. Human immunodeficiency virus type 1 mutants resistant to non-nucleoside inhibitors of reverse transcriptase arise in cell culture. Proc. Natl. Acad. Sci. USA 88:11241-11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shulman, N., A. R. Zolopa, D. Passaro, R. W. Shafer, W. Huang, D. Katzenstein, D. M. Israelski, N. Hellman, C. Petropoulos, and J. Witcomb. 2001. Phenotypic hypersusceptibility to non-nucleoside reverse transcriptase inhibitors in treatment-experienced HIV-infected patients: impact on virological response to efavirenz-based therapy. AIDS 15:1125-1132. [DOI] [PubMed] [Google Scholar]
- 20.Tisdale, M., S. D. Kemp, N. R. Parry, and B. A. Larder. 1993. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc. Natl. Acad. Sci. USA 90:5653-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wainberg, M. A., W. C. Drosopoulos, H. Salomon, M. Hsu, G. Borkow, M. A. Parniak, Z. Gu., Q. Song, J. Manne, S. Islam, G. Castriota, and V. R. Prasad. 1996. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science 217:1282-1285. [DOI] [PubMed] [Google Scholar]
- 22.Wainberg, M. A., D. M. Miller, Y. Quan, H. Salomon, A. S. Mulato, P. D. Lamy, N. A. Margot, K. E. Anton, and J. M. Cherrington. 1999. In vitro selection and characterization of HIV-1 with reduced susceptibility to PMPA. Antiviral Ther. 4:87-94. [DOI] [PubMed] [Google Scholar]
- 23.Wakefield, J. K., S. A. Jablonski, and C. D. Morrow. 1992. In vitro enzymatic activity of human immunodeficiency virus type 1 reverse transcriptase mutants in the highly conserved YMDD amino acid motif correlates with the infectious potential of the proviral genome. J. Virol. 66:6806-6812. [DOI] [PMC free article] [PubMed] [Google Scholar]