Abstract
We examined the differentiation potential of an adult liver stem cell line (WB F344) in a cardiac microenvironment, ex vivo. WB F344 cells were established from a single cloned nonparenchymal epithelial cell isolated from a normal male adult rat liver. Genetically modified, WB F344 cells that express β-galactosidase and green fluorescent protein or only β-galactosidase were co-cultured with dissociated rat or mouse neonatal cardiac cells. After 4 to 14 days, WB F344-derived cardiomyocytes expressed cardiac-specific proteins and exhibited myofibrils, sarcomeres, and a nascent sarcoplasmic reticulum. Further, rhythmically beating WB F344-derived cardiomyocytes displayed calcium transients. Fluorescent recovery after photobleaching demonstrated that WB F344-derived cardiomyocytes were coupled with adjacent neonatal cardiomyocytes and other WB F344-derived cardiomyocytes. Fluorescence in situ hybridization experiments suggested that fusion between WB F344 cells and neonatal mouse cardiomyocytes did not take place. Collectively, these results support the conclusion that these adult-derived liver stem cells respond to signals generated in a cardiac microenvironment ex vivo acquiring a cardiomyocyte phenotype and function. The identification ex vivo of microenvironmental signals that appear to cross germ layer and species specificities should prove valuable in understanding the molecular basis of adult stem cell differentiation and phenotypic plasticity.
Stem cells in the liver in vivo are quiescent until the liver is injured and the proliferative capacity of its differentiated hepatocytes and biliary duct cells is inhibited. Then, a facultative stem cell compartment is believed to become activated. Stem cell lines have been derived from normal and diseased livers and established in propagable cultures from experimental animals by several investigators.1–8
We have previously used such a stem cell line (WB F344) cloned from a single nonparenchymal epithelial stem cell of a normal adult male rat liver to investigate the differentiation potential of these stem cells in the heart, in vivo.9 This cell line is diploid, nontumorigenic, and demonstrates phenotypic properties of oval cells.10 WB F344 cells engraft in the livers of rats that are deficient for dipeptidyl peptidase IV activity, integrate into the hepatic plates, and differentiate into morphologically and functionally mature hepatocytes that express dipeptidyl peptidase IV.1,2 When transplanted in the myocardium of nude mice,9 WB F344 cells acquire a phenotype characteristic of mature cardiomyocytes.
It has been suggested that the plasticity of adult nonhematopoietic stem cells, in vivo, may be a reflection of multipotent hematopoietic stem cells circulating in the blood11–13 or residing quiescent in the tissues outside the bone marrow.14 Moreover, cell fusion of embryonic and some adult stem cells has been suggested, in some cases, to explain the apparent plasticity of adult stem cells.15–17
Here, we demonstrate that WB F344 cells acquire a cardiomyocyte phenotype and function in a neonatal cardiac environment generated ex vivo. Taking advantage of xenogeneic differences between the mouse and rat, our results suggest that these rat WB F344-derived cardiomyocytes are not the product of fusion with differentiated mouse cardiomyocytes. Furthermore, the cloning of the WB F344 cell line from a single liver epithelial cell argues against a contaminating hematopoietic stem cell giving rise to our finding.
Materials and Methods
A myocardial microenvironment was generated by dissociating neonatal rat or mouse hearts and establishing cardiomyocyte primary cultures. Neonatal cardiomyocytes were isolated from the hearts of 1- to 2-day-old Sprague-Dawley rats or CD1 mice in accordance with accepted guidelines for the care and treatment of experimental animals at the University of North Carolina School of Medicine and the National Institutes of Health. Neonatal cardiomyocytes were isolated using a Worthington Neonatal Cardiomyocyte Isolation System (Worthington Biochemical Corp., Lakewood, NJ). They were plated on laminin-coated cover slides at 1 × 106 cells per 22-mm cover slide and grown in Richter (Irvine Scientific, Santa Ana, CA) medium supplemented with 10% fetal calf serum. They were maintained for 48 hours before WB F344 cells were added.
WB F344 Cells and Genetic Modification of WB F344 Cells
WB F344 cells were genetically modified to express the Escherichia coli lac Z gene (β-galactosidase) by infection with the CRE BAG2 retrovirus.18 WB F344 cells intended for transplantation were harvested by trypsinization from subconfluent cultures, washed, and resuspended in cell culture medium. They were counted and plated at a 1/2000 to 1/3000 ratio (300 to 500 cells per 22-mm cover slide) with neonatal cardiac cells in the same medium for 1 to 4 days, then the serum was replaced with 2% horse serum. The co-cultures were grown in a 5% CO2, 95% air environment at 37°C and interrupted at 4 to 15 days. In control experiments, WB F344 cells were grown in conditioned medium collected from 4- to 6-day-old cultures of neonatal cells. Furthermore, neonatal cells were cultured on 0.02-μm Anopore membrane inserts (Nunc, Naperville, IL) that physically separated them from WB F344 cells in the same culture.
For functional studies, a subset of WB F344 cells was transfected with a plasmid containing a green fluorescent protein (GFP) gene that enables the identification of living WB F344 cells. An enhanced GFP gene was obtained from Clontech (Palo Alto, CA) and inserted between the promoter and polyadenylation signal of the murine phosphoglycerate kinase gene (pgk-1). The pgk-1 sequences were derived from the vector pJNS2.19 The vector pPGKpropA, which contains the pgk-1 promoter and polyadenylation sequences separated by a unique Pst1 site, was constructed by standard cloning. The GFP gene was amplified by polymerase chain reaction (PCR) and inserted into the Pst1 site of pPGKpropA to generate the final pPGKBSGFPpropA (BlueScript). WB F344 cells were transfected with this plasmid using 50 μl of lipofectamine (Life Technologies, Inc., Grand Island, NY) to 10 μg of DNA. The cells were plated and screened for GFP expression 3 days later. Fluorescent cell nests were trypsinized using cloning rings and expanded to enrich for GFP-expressing cells. Fluorescent cells were fluorescence-activated cell sorted, amplified in culture, and subsequently cloned at limiting dilutions. Clones were screened, and selected clones were cloned again after which a single clone was selected and amplified.
Histochemistry, Immunocytochemistry, and Electron Microscopy
Cells grown on cover slides were rinsed in 0.1 mol/L sodium phosphate buffer, pH 7.3, and fixed for 5 minutes in 2% formaldehyde in 0.1 mol/L sodium phosphate buffer, pH7.3. After three washes with phosphate buffer, the β-galactosidase reaction was performed by incubating the fixed cells in the X-gal substrate for 2 to 4 hours.20 Expression of cardiac-specific proteins was demonstrated using antibodies against cardiac myosin heavy chain (Chemicon International, Temecula, CA), cardiac troponin I (cTnI),21 and cTnT.22 The anti-cTnT monoclonal antibody (mAb) (designated mAb 13-11) recognizes a cardiac-specific TnT epitope22 conserved across phyla. mAb 13-11 does not immunoreact with skeletal muscle because this cardiac-specific epitope is absent from skeletal muscle TnT. The expression of connexin 43 was demonstrated using a polyclonal antibody against connexin 43 (Zymed, San Francisco, CA). An anti-myogenin antibody (PharMingen, San Diego, CA) was used to rule out a skeletal muscle phenotype. After the β-galactosidase reaction, the cells were permeabilized with 0.4% Triton, washed, and incubated with the respective antibodies, as previously described.9 Rhodamine-, fluorescein isothiocyanate-, or Texas Red-labeled goat anti-mouse and anti-rabbit antibodies were used as secondary antibodies (Rockland Immunochemicals, Gilbertsville, PA). Normal mouse or rabbit sera were substituted for the primary antibodies in control conditions. After performing the β-galactosidase reaction, the cells were prepared for electron microscopy examinations, as previously described.9 β-galactosidase-positive cardiomyocytes were selected, sectioned at 70 nm, and examined with a Leo EM 910 transmission electron microscope at 80 kV.
Fluorescence in Situ Hybridization (FISH) on Neonatal Mouse Cell/Rat WB F344 Cell Co-Cultures
Taking advantage of xenogeneic differences between mouse and rat DNA, dissociated cardiac cells from neonatal mice were co-cultured with WB F344 cells to investigate whether WB F344 cells fused with differentiated neonatal mouse cardiac cells. After the β-galactosidase reaction, the cells were permeabilized and incubated with mAb 13-11, the cardiac-specific anti-cTnT monoclonal antibody. Texas Red-labeled anti-mouse antibody was used as a secondary antibody.
FISH experiments were conducted using two probes simultaneously on the same co-culture. A rat-specific tandemly repetitive 200-bp DNA sequence, predominant in the rat Y-chromosome, was used to identify the rat-derived WB F344 cardiomyocyte nuclei.9,23 This rat-specific probe was labeled with biotin (Biotin-Nick translation mix; Roche, Indianapolis, IN) and used at 50 ng in the probe mix. Fluorescein-labeled avidin (Roche) was used as the secondary label. In control experiments, we investigated by PCR whether this rat-specific DNA is present in the mouse genome.
A second probe, specific for the mouse L1 Md-A2 DNA element (L1 DNA),24 was used to identify mouse cell nuclei. This repetitive L1 DNA sequence is dispersed throughout the mouse genome and has been estimated to be repeated 10,000 times. In control experiments, we investigated by PCR whether this L1 DNA sequence is present in the rat genome. The mouse L1 DNA probe was amplified from mouse genomic DNA using the following primers: 5′-AATCGCACGGAACTTGAGACTGCA-3′ tandemly repeated in the 5′ end of the mouse L1 and 5′-CGTTTGCCTTTCGCCATCTGGTA-3′ at position 1756. A 400-bp PCR product was cloned into a TA cloning vector (Invitrogen, Carlsbad, CA). The resulting clones were screened for the mouse L1 DNA sequence by PCR, and positive clones were subjected to automated DNA sequencing to confirm that the sequence was identical to the published mouse L1 sequence.24 The L1 DNA-containing plasmid was labeled with digoxigenin (Dig-Nick translation mix, Roche) and used at a concentration of 100 ng in the double probe mix.
The FISH experiments were performed on WB F344 cell/mouse cell co-cultures after performing the β-galactosidase reaction and immunocytochemistry to demonstrate in the same WB F344-derived cardiomyocytes the expression of cTnT. After fixation of the immune complexes in 10% formaldehyde, the cells were washed in sodium phosphate buffer. This was followed by proteinase K digestion at 37°C for 20 minutes in 0.01 μg/ml of proteinase K and 200 mmol/L Tris-HCl, 300 mmol/L sodium acetate, 50 mmol/L ethylenediaminetetraacetic acid, 1% sodium dodecyl sulfate, pH 9.0. After three washes in 2× standard saline citrate (SSC, 0.3 mol/L NaCl, 30 mmol/L sodium citrate), the cells were incubated in a prehybridization solution at 40°C (50% formamide, 50% 2× SSC, and 0.1% Tween). After adding both probes into the same hybridization mix, the DNA was simultaneously targeted and denatured at 90°C for 5 minutes. Hybridization was performed overnight at 37°C in a humid chamber, after which the cells were washed three times for 2 minutes each in 50% formamide in 2× SSC at 40°C, and two washes in 2× SSC. After incubation in preblocking solution (3% bovine serum albumin in 0.1 mol/L phosphate buffer) for 30 minutes at 37°C, a sheep anti-digoxigenin rhodamine-labeled antibody (Roche) that recognizes the digoxigenin-labeled mouse L1 DNA and avidin-fluorescein (Roche) that binds to the biotin-labeled rat-specific DNA were applied simultaneously for 30 minutes at 37°C in a humid chamber. After this incubation, the cells were washed in phosphate-buffered saline in 0.1% Tween three times for 5 minutes each and covered using a mounting medium containing 4,6-diamidine-2-phenylindole counter stain (Vectashield; Vector Laboratories, Inc, Burlingame, CA). The cells were examined using a Zeiss LSM 5 Pascal confocal microscope equipped with a rhodamine long-pass filter.
Fluorescence Recovery after Photobleaching (FRAP)
Functional gap junction mediated cell-to-cell communication was measured in the neonatal cardiomyocyte/WB F344 cell co-cultures using a FRAP analysis at room temperature. A 1-mmol/L stock solution of red SNARF calcein acetoxymethylester (SNARF calcein AM; Molecular Probes, Eugene, OR) was prepared in dimethyl sulfoxide. Using fluorescence microscopy this cytosolic fluoroprobe is visible at wavelengths greater than 590 nm when excited at 560 nm. SNARF calcein has a molecular weight less than 1 kd, allowing it to pass through the connexons of neighboring cells. The dissolved SNARF calcein AM was added to a modified Tyrode’s composed of 124 mmol/L NaCl, 4.5 mmol/L KCl, 0.5 mmol/L MgCl2, 0.4 mmol/L NaH2PO4, 1.8 mmol/L CaCl2, 5 mmol/L sodium pyruvate, 10 mmol/L HEPES, 20 mmol/L glucose, insulin 1 U/L, pH 7.2. The cytosol of the co-cultures was labeled by incubation in 1 μmol/L of SNARF calcein AM Tyrode’s for 30 minutes at 37°C. De-esterification inside the cell renders SNARF calcein membrane impermeant. The cell cultures were washed several times with dye-free modified Tyrode’s before measurements were recorded. Cell cultures were bathed in a modified Tyrode’s, pH 7.2, and placed on the stage of a Zeiss LSM410 laser-scanning confocal microscope. High-resolution optics (Zeiss ×63/1.4 NA oil immersion objective) were focused on the co-cultures, identifying native cardiomyocytes and WB F344-derived cardiomyocytes. The latter were identified, as binucleated cells expressing GFP (Figure 1D) and were located on top or integrated within a group of neonatal cardiomyocytes. A 488-nm excitation wavelength and 540 ± 25-nm band pass emission filter were used to locate the WB F344-expressing GFP. Using Zeiss software, a rectangular region of interest encompassing 90% of a WB F344 stem cell, WB F344-derived cardiomyocyte, or a neonatal cardiomyocyte was selected. A high-intensity laser pulse (5 to 10 seconds) excitation wavelength of 568 nm bleached the SNARF calcein molecules within the region of interest cell. This caused an immediate loss of SNARF calcein fluorescence emission recorded through a 590-nm long-pass filter. SNARF calcein redistribution from adjacent unbleached cells through connexin pores into the bleached region of interest was recorded in subsequent confocal images acquired at 30- to 120-second intervals. Monitoring the fluorescence emission intensity of the SNARF calcein in the region of interest cell as a function of time resulted in a single exponential recovery curve, yielding a rate constant for the fluoroprobe transport (k = min−1), a measure of gap junction permeability.25
Figure 1.
WB F344 cells co-cultured with neonatal cardiac cells. A: Proliferating WB F344 cells that attached away from cardiac cells formed nests of undifferentiated WB F344 cells (black triangle). B and C: Differentiating WB F344-derived cardiomyocytes (blue cell) adjacent to neonatal cardiomyocytes. C: Binucleated, WB F344-derived cardiomyocyte. D: WB F344-derived cardiomyocyte expressing GFP. Original magnifications, ×40.
Inhibition of cell-to-cell gap junction coupling was measured in the co-cultures with FRAP analysis in the presence of the gap junction uncoupler 3β-hydroxy-11-oxoolean-12-en-30-oic acid 3-hemisuccinate (carbenoxolone, Sigma-Aldrich).26 For this series of measurements, a WB F344-derived cardiomyocyte in modified Tyrode’s was identified and photobleached, and the fluorescence recovery recorded. A modified Tyrode’s with 100 μmol/L of carbenoxolone was added to the co-cultures, and a second photobleach and recovery sequence was repeated in the same WB F344-derived cardiomyocyte.
Measurement of Intracellular [Ca2+] Transients
A multisite ratiometric fiber optic recording and imaging system provided high temporal and spatial resolution recordings of whole-cell [Ca2+]i transients in the co-cultures.27 After indo-1 labeling, the cell cultures were washed with fluoroprobe-free modified Tyrode’s and placed on the stage of an inverted Olympus IMT-2 microscope. Temperature was maintained at 37°C. The co-cultures were electrically stimulated with a platinum bipolar electrode. Rectangular impulses, 1 ms in duration, 2× threshold intensity, were delivered continuously to the co-cultures at a cycle length of 1 to 2 Hz (Grass Instruments). High-resolution images of cell alignment and geometry were acquired with a Cohu CCD camera mounted to the trinocular of the microscope. Whole-cell [Ca2+]i transients were recorded with an Olympus ×40/1.4 NA UV oil immersion objective. An excitation wavelength of 350 nm was focused on the co-culture, and indo-1 emission fluorescence was collected for up to 2 seconds through a 406-nm BP and 460-nm LP filter to two identical, precisely aligned fiber optic arrays. The ratio of the emission fluorescence (406 nm/460 nm) provided a measure of [Ca2+]i. Recorded signals were normalized for individual fiber optics. Fluorescent signals were collected simultaneously from eight sites (16 channels) at a sampling rate of 70 kHz.
Results
WB F344-Derived Cardiomyocytes
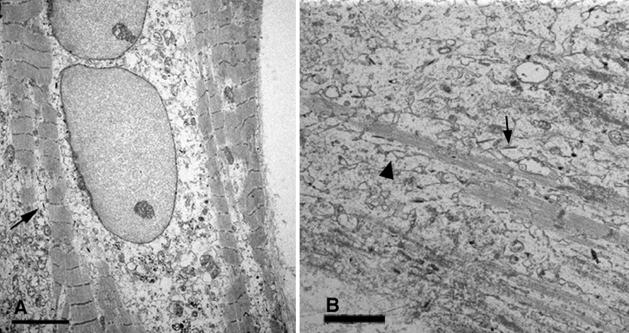
WB F344 cells that attached to the coverslips away from the neonatal cardiac cells behaved as WB F344 cells normally would when plated in the absence of neonatal cardiac cells. They multiplied to form nests of confluent, small polygonal cells (8.5 to 10 μm in diameter) (Figure 1A), that as shown previously were coupled via connexons made up of connexin 43.2,3,28,29 Undifferentiated WB F344 cells continued to proliferate in the co-cultures where they became the predominant cells in older (>15 days) co-cultures occupying the majority of the culture field. In the periphery of younger WB F344 nests, WB F344 cells that were bordered by neonatal cardiac cells acquired various shapes consisting of larger elongated and fusiform cells (Figure 1A). Because the neonatal heart cells are enriched with cardiomyocytes but also contain other types of cells, we suspect that the fusiform and elongated WB F344 cells surrounding the nests, such as seen in Figure 1A, may be responding to signals from cell types other than cardiomyocytes. WB F344-derived cardiomyocytes measuring up to 90 to 120 μm in length were most frequently present among neonatal cardiomyocytes. These WB F344-derived cardiomyocytes were identified by their blue color, the product of the β-galactosidase reaction in the cytoplasm (Figure 1, B and C). Neonatal cardiac cells alone did not demonstrate any β-galactosidase expression. Like the neonatal cardiomyocytes, the majority of which were multinucleated, the WB F344-derived cardiomyocytes exhibited double nuclei (Figure 1C), a characteristic of developing cardiomyocytes in culture,30 and occasionally demonstrated cross-striations by light microscopy. The number of WB F344-derived cardiomyocytes was examined at 6 to 7 days in the co-culture and was 99.8 ± 8.1 (n = 6, mean ± SEM) per coverslip. The number of WB F344 cardiomyocytes increased with time but so did the undifferentiated WB F344 cells. When examined by electron microscopy, the WB F344-derived myocytes exhibited nascent myofibrils (Figure 2A) and sarcoplasmic reticulum (Figure 2B). The WB F344-derived cardiomyocytes were identified through the electron dense crystalline deposits in the cytoplasm, the product of the β-galactosidase reaction (Figure 2, arrows).
Figure 2.
Myofibrillogenesis of WB F344-derived myocytes. Nascent myofibrils in A and B and sarcoplasmic reticulum (black triangle) in a WB F344-derived myocyte. Electron dense crystalline deposits (arrows in A and B) are product of β-galactosidase reaction. Scale bars: 10 μm (A); 20 μm (B).
The WB F344-derived cardiomyocytes expressed cardiac proteins, namely, myosin, cTnI, cTnT, and connexin 43 (Figure 3). They did not express skeletal muscle myogenin. The striated pattern obtained with the immunolocalization of cardiac myosin, cTnI, and cTnT is consistent with localization of these proteins in the cardiac sarcomeres (Figure 3; a, b, and c). By confocal microscopy, neonatal cardiomyocytes were not found to underlie or overlap the WB F344-derived cardiomyocytes. The diffuse distribution of connexin 43 along the WB F344-derived cardiomyocyte cell membrane reflects the relative immaturity of these cardiomyocytes (Figure 3d).
Figure 3.
WB F344 myocytes express cardiac-specific protein. Undifferentiated WB F344 cells and WB F344-derived myocytes express β-galactosidase (blue cells in a–d). WB F344-derived cardiomyocytes co-express cardiac myosin, striated red fluorescence in a, 2 and 3 (13 days co-culture); cardiac-specific cTnI, red fluorescence in b (13 days co-culture); cardiac-specific cTnT, striated red fluorescence in c (7 days co-culture); connexin 43, green fluorescence in d (13 days co-culture). Select confocal microscopic stacks in a and b demonstrate that neonatal myocytes do not overlie or underlie the WB F344-derived myocytes. Expression of myosin and TnI is inherent to the WB F344-derived myocytes that co-express β-galactosidase. Undifferentiated WB F344 cells are predominant in the 13-day co-cultures (a and b). Arrow in a3 depicts the nucleus of a neonatal cardiomyocyte. Arrow in a7 depicts the body of the same neonatal cardiomyocyte in contact with the WB F344-derived myocyte. The distribution of connexin 43 throughout the WB F344-derived myocyte cell membranes (d) is consistent with the immaturity of the myocyte. The arrow at the top in d represents connexin 43 expression in an adjacent neonatal cardiac myocyte that does not express β-galactosidase. Original magnifications, ×40.
Contact between the WB F344 cells and neonatal cells or their extracellular matrix in the neonatal cardiac microenvironment seems to be necessary for WB F344 cells to acquire the cardiomyocyte phenotype. In conditioned media from neonatal cardiac cell cultures, or when WB F344 cells were grown on 0.02-μm Anopore membrane inserts (Nunc) that prevented their direct contact with the neonatal cardiac microenvironment, WB F344 cells did not acquire the cardiomyocyte phenotype.
FISH in Mouse/Rat Co-Cultures
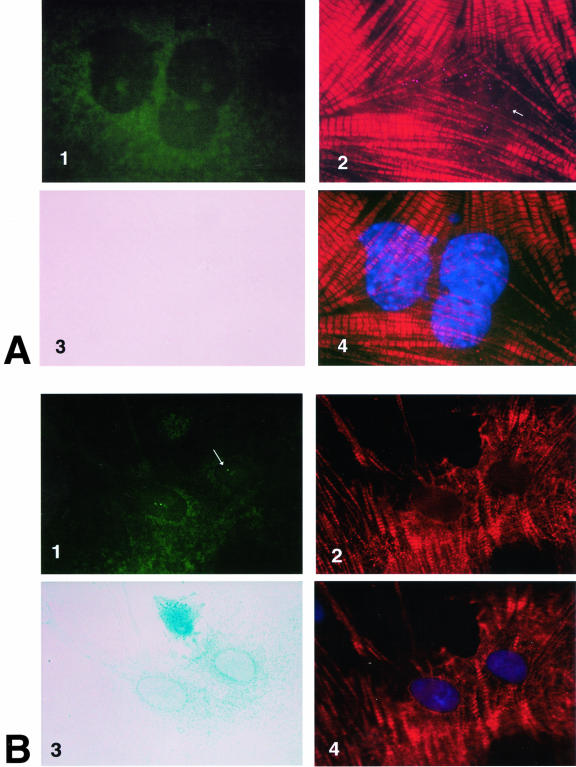
Control experiments on WB F344 cells demonstrated that FISH analysis did not identify 100% of the sites in the WB F344 cell nuclei. Despite this and other technical difficulties encountered in the double FISH experiments, we found in one experiment six cardiomyocytes and in another experiment four cardiomyocytes that fulfilled criteria that allowed us to identify them as WB F344-derived cardiomyocytes (Figure 4B), namely, 1) β-gal expression, 2) cardiac troponin T expression, 3) positive in situ hybridization with the rat-specific probe, and 4) negative in situ hybridization with the mouse-specific line 1 probe. By the same token, we examined more than 100 neonatal mouse cardiomyocytes in each mouse/rat co-culture (Figure 4A). All of these demonstrated 1) cardiac troponin T expression, 2) positive in situ hybridization with the mouse line 1-specific probe, 3) no β-gal expression, and 4) no hybridization with the rat-specific probe. It is important to point out that co-hybridization of the rat-specific probe and the mouse-specific line 1 probe was never observed in the same cardiomyocytes in the mouse/rat co-cultures.
Figure 4.
Double FISH on mouse neonatal cell/WB F344 cell co-cultures. The FISH was conducted after performing the β-gal reaction and the immunochemistry to demonstrate β-galactosidase and cardiac-specific protein expression in WB F344-derived cardiomyocytes. The mouse line 1 DNA and the rat-specific DNA probes were used simultaneously in the same probe mix on the same mouse/rat co-culture. A: The mouse L1 DNA probe hybridized with a mouse myocyte nuclear DNA. Punctate rhodamine-positive speckles in the nuclei in A2 (arrow) represent clusters of mouse L1 DNA. Some are artificially enhanced to differentiate their signal from the Texas Red striations in the sarcomeres. The nuclei of the same cell are shown with the 4,6-diamidino-2-phenylindole stain in A4. This mouse-derived myocyte does not express β-galactosidase (A3), nor does it hybridize with the rat DNA probe (A1). B: The rat-specific DNA probe hybridizes with the rat WB F344-derived myocyte nuclei. The two green fluorescent (fluorescein isothiocyanate-positive) dots in the nuclei of a WB F344-derived myocyte (B1, arrow) reflect high-affinity hybridization sites of the rat DNA probe with the Y-chromosome of the male WB F344-derived myocyte and with less affinity with alleles on chromosome 3 and/or 12.23 This WB F344-derived myocyte does not hybridize with mouse L1 DNA (B2). The nuclei of this WB F344-derived myocyte are shown with the 4,6-diamidino-2-phenylindole stain in B4. Unlike the mouse-derived myocyte, this WB F344-derived myocyte expresses β-galactosidase. Both the mouse neonatal myocyte and the WB F344-derived myocyte express cTnT (striations), probed with mAb 13-11. mAb 13-11 recognizes a cTnT epitope conserved across species and does not recognize skeletal muscle TnT. A Texas Red-labeled anti-mouse antibody was used as a secondary antibody. Using a long-pass filter, the Texas Red emission bled through the rhodamine range, and sarcomeric cTnT striations are evident in the myocytes (A2 and A4, B2 and B4). Only the WB F344-derived myocyte (B3) expresses β-galactosidase. Original magnifications, ×100.
Neonatal Mouse Cardiomyocytes
Neonatal mouse cardiomyocyte nuclei hybridized with the mouse L1 DNA probe labeled with rhodamine (Figure 4A2). cTnT expression was demonstrated with a Texas Red-labeled secondary antibody in the same cell (Figure 4A, 2 and 4). Using a long-pass emission filter, the Texas Red emission maximum of 635 nm bled through the rhodamine range used to probe the mouse L1 DNA. Taking advantage of this technical overlap, the presence of rhodamine-positive punctate clusters (Figure 4A2) of mouse L1 DNA in the nuclei (Figure 4A4) and Texas Red-labeled cTnT striations in the cytoplasm were clearly demonstrated in the same mouse cardiomyocytes. These mouse neonatal cardiomyocytes did not express β-galactosidase (Figure 4A3) nor did their nuclear DNA hybridize with the rat-specific fluorescein-labeled DNA probe (Figure 4A1). In control PCR experiments, the mouse L1 DNA was not present in the rat genome.
WB F344-Derived Cardiomyocytes
In contrast, the WB F344-derived cardiomyocytes identified with β-galactosidase (Figure 4B3) and cTnT expression (Figure 4B2) hybridized with the fluorescein-labeled rat Y-chromosome-specific DNA probe (Figure 4B1). That some cardiomyocytes had more than one labeled nuclear DNA site is consistent with the findings of Essers et al23 that the rat-specific DNA, used in the FISH experiment as a probe for rat DNA, is specific for a repetitive sequence in the rat Y-chromosome, but hybridization with alleles on rat chromosomes 3 and 12 may occur in less stringent conditions. None of the WB F344-derived cardiomyocytes hybridized with the rhodamine-labeled mouse L1 DNA probe (Figure 4B2). In control PCR experiments, the rat-specific DNA was not present in the mouse genome.
Functional Cell-to-Cell Communication
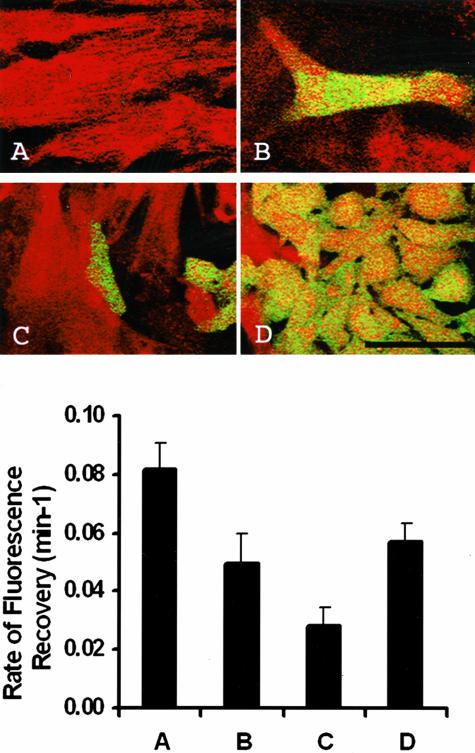
Gap junction-mediated cell-to-cell communication between the neonatal cardiomyocytes and WB F344-derived cardiomyocytes was measured with FRAP (n = 15). WB F344-derived cardiomyocytes were identified by their expression of GFP (Figure 1D) and characterized by their elongated shape and double nuclei. The confocal images (optical thickness, 3 μm) shown in Figure 5 (top) distinguish the four types of cell-to-cell interactions examined with FRAP (Figure 5, bottom) in cultures: A) neonatal rat cardiomyocyte to cardiomyocyte; B) WB F344-derived cardiomyocyte to rat neonatal cardiomyocytes; C) undifferentiated WB F344 cell to rat neonatal cardiomyocytes; and D) WB F344 cell nests. In Figure 5, B and C, only portions of the surrounding cardiomyocytes are visible as the optics were focused on the WB F344 cells. Also shown in Figure 5 (bottom) is the magnitude of gap junction mediated cell-to-cell communication for each of the classes of cell-to-cell interaction, as assessed by the rate of fluorescence recovery. The highest rate of fluorescence recovery was measured in the neonatal cardiomyocytes surrounded by neonatal cardiomyocytes (Figure 5A, top and bottom; k = 0.082 ± 0.009), whereas the lowest rate of fluorescence recovery was measured between neonatal cardiomyocytes and undifferentiated WB F344 cells (Figure 5C, top and bottom; k = 0.029 ± 0.006). The rate of fluorescence recovery measured between the neonatal cardiomyocyte and WB F344-derived cardiomyocyte (Figure 5B, top and bottom) is ∼60% of that measured between adjacent neonatal cardiomyocytes (k = 0.049 ± 0.006, k = 0.082 ± 0.009, P < 0.01). In addition the rate of fluorescence recovery between the neonatal cardiomyocyte and WB F344-derived cardiomyocyte (Figure 5B, top and bottom; k = 0.049 ± 0.006) was approximately equal to that measured in the confluent WB F344 nests (Figure 5D, top and bottom; k = 0.057 ± 0.010). Repeated photobleach and recovery sequences on WB F344-derived cardiomyocytes demonstrated reproducible measures of FRAP with no photochemical damage or cell injury (data not shown). After the first photobleach and recovery sequence, WB F344-derived cardiomyocytes recovered to 70% of the initial fluorescence with similar rates of fluorescence recovery (k = 0.09 minutes−1). In modified Tyrode’s, after the WB F344-derived cardiomyocyte was bleached a second time, fluorescence recovered to 60% of the initial fluorescence within 15 minutes with a similar rate of fluorescence recovery as measured in the first photobleach and recovery sequence. However, when the co-cultures were exposed to 100 μm carbenoxolone in Tyrode’s, the fluorescence recovery rate in the photobleached WB F344-derived cardiomyocyte decreased (k = 0.03 minutes−1) with minimal change in fluorescence emission. These results demonstrate decreased gap junction permeability in the presence of carbenoxolone.
Figure 5.
Top: Cellular coupling in co-cultures measured with FRAP. Neonatal myocyte (red) adjacent to another neonatal myocyte (A), WB F344-derived myocyte (green) adjacent to a neonatal myocyte (B), undifferentiated WB F344 cell adjacent to neonatal myocyte (C), and WB F344 nests (D) are shown. Confocal images of co-cultures (optical thickness, 3 μm) were obtained with a ×63/1.4 NA oil immersion objective (scale bar, 50 μm). Bottom: The rate of fluorescence recovery for the cell relations described at the top are provided here. Column A, neonatal cardiomyocyte adjacent to another neonatal cardiomyocyte; Column B, WB F344-derived cardiomyocyte adjacent to a neonatal cardiomyocyte; Column C, undifferentiated WB F344 cell adjacent to neonatal cardiomyocyte; and Column D, WB F344 cells within a nest of WB F344 cells.
Intracellular [Ca2+]i Transients
The change in [Ca2+]i in response to an electrical stimulus in co-cultured cardiomyocytes and WB F344-derived cardiomyocytes is shown in Figure 6. The image in the top panel depicts cell orientation and fiber optic placement. The highlighted optical fibers labeled A and B overlay a cardiomyocyte and WB F344-derived cardiomyocyte (GFP-positive), respectively, separated by 120 μm and labeled with indo-1. [Ca2+]i transients recorded from optical fibers A and B are shown in Figure 6 (panel 2). When the co-cultures were stimulated at 1 Hz, the recorded [Ca2+]i transients from the neonatal cardiomyocytes (A) and WB F344-derived cardiomyocytes (B) were of similar amplitude and duration. In contrast, in the same co-culture, [Ca2+]i transients were not recorded from WB F344 cell nests that had not acquired a cardiac phenotype (Figure 5D). Increasing the frequency of the pacing stimulus to 2 Hz resulted in a corresponding increase in the frequency of [Ca2+]i transients in the neonatal cardiomyocytes. However, [Ca2+]i transients in the WB F344-derived cardiomyocytes evidenced 2:1 or higher degrees of conduction block.
Figure 6.
Intracellular [Ca2+]i transients. 1: Highlighted fiber optics overlayed on image of co-culture with neonatal cardiac myocyte (A) and WB F344-derived myocyte expressing GFP (B), intracellularly labeled with indo-1 (scale bar, 100 μm). 2: [Ca2+]i transients recorded at sites A and B. Measured change in [Ca2+]i recorded at sites A and B demonstrated conduction block when the pacing stimulus increased from 1 Hz to 2 Hz. Recorded signals were normalized for fiber optic A and B and offset for display.
Discussion
Our findings suggest that stem cells cloned from a single epithelial cell of a normal adult rat liver acquire a cardiac phenotype in a cardiac microenvironment generated, ex vivo, from mouse or rat neonatal cardiac cells, as WB F344 cells did in vivo9 in the mouse. We furthermore demonstrate that these ex vivo WB F344-derived cardiomyocytes function as cardiomyocytes.
WB F344 cells that engrafted near differentiated neonatal cardiomyocytes, acquired a cardiomyocyte phenotype. In contrast, when WB F344 cells were engrafted away from neonatal cardiac cells, were separated from the neonatal cardiac cells through a barrier membrane, or were grown in a neonatal cardiac cell preconditioned medium, the WB F344 cells multiplied and formed confluent nests of undifferentiated WB F344 cells as they do when grown alone. This suggested that some of the inductive signals present in the neonatal cardiac microenvironment acted on the WB F344 cells through either cell-cell or cell-extracellular matrix contact, rather than as soluble factors alone. It has previously been demonstrated that the WB F344 cells in culture are joined by gap junctions and express liver and cardiac connexin isoforms, namely connexin 43 and 26.2,3,28,29 It is tempting to speculate that connexin 43, being the predominant isoform expressed in ventricular cardiomyocytes, might have facilitated the transfer of cardiogenic signals from the neonatal cardiomyocytes to adjacent undifferentiated WB F344 cells through shared connexons. Interestingly, intercellular signaling across connexons have been reported in WB F344 cells28,29 and other types of cells31 and propagation of Ca2+ waves in C6 glioma cells, which have low endogenous connexin levels, have been shown after their transfection with connexin 43.31,32
The spatial requirement necessary for the acquisition by the WB F344 cells of a cardiomyocyte phenotype within the neonatal cardiac microenvironment raises the hypothetical possibility that fusion between WB F344 cells and neonatal cardiomyocytes might have occurred, allowing β-galactosidase-expressing WB F344 cells to adopt the phenotype of neonatal cardiomyocytes. We examined this possibility in mouse cell/WB F344 cell co-cultures using FISH experiments with DNA probes that differentiated between rat and mouse. We demonstrate that WB F344-derived cardiomyocytes were not a product of fusion between WB F344 cells and mouse neonatal cardiac cells as the mouse-specific and rat-specific DNA probes were never co-localized in the same cardiomyocyte nuclei. Furthermore, mouse cardiomyocytes that hybridized with the mouse-specific DNA probe were never found to express β-galactosidase. In addition, our finding of functional disparity between the immature WB F344-derived cardiomyocytes and the relatively more mature neonatal cardiomyocytes argues against fusion between these two types of cells. It is conceivable that fusion between some WB F344 cells and differentiated neonatal cardiac cells took place in these cultures but was not seen in the cells we examined because fusion between somatic stem cells and ES cells was reported to occur as a rare event in culture.15,16
An important aspect of this study is the confirmation that the WB F344-derived cardiomyocytes that acquire a cardiomyocyte phenotype also function as cardiomyocytes. Functional integration was verified by observing the presence of Ca2+ transients in response to propagated electrical wavefronts and the diffusion of a fluorescent probe between WB F344-derived cardiomyocytes and adjacent neonatal cardiomyocytes. These studies show that WB F344-derived cardiomyocytes cycle intracellular Ca2+ during contraction, a characteristic of cardiomyocytes. In general, the amplitude and contour of the Ca2+ transient measured in the cardiomyocytes and WB F344-derived cardiomyocytes were similar. Two observations, however, indicate that the electrical properties of the WB F344-derived cardiomyocytes are not as well developed as the neonatal cardiomyocytes. First, cell-to-cell coupling, as measured by FRAP, was decreased between WB F344-derived cardiomyocytes and neonatal cardiomyocytes when compared to neonatal cardiomyocytes to neonatal cardiomyocytes. And, secondly electrical coupling or excitability of the WB F344-derived cardiomyocytes, as measured by the effect of increasing pacing rate, appeared to be decreased compared to neonatal cardiomyocytes. The reduced cell-to-cell coupling measured between WB F344-derived cardiomyocytes and neonatal cardiomyocytes observed in FRAP experiments may contribute to the decreased excitability. Other possibilities such as differences in inward and repolarizing currents cannot be excluded.
Oval cells have been isolated from diseased liver and reported to repopulate the injured liver of FAH −/− mice.33 In analogy, progenitor/stem cells from the heart have also been described to reconstitute the myocardium in vivo after experimental infarction.34 Because of their clonal nature, which eliminates the contribution from other cell types, WB F344 cells provide an ideal experimental system in understanding the plasticity of adult stem cells and to investigate, ex vivo, the signals that induce lineage commitment and phenotypic differentiation.35 These signals are likely to be of broad importance in that they cross species specificities.
Footnotes
Address reprint requests to Nadia N. Malouf, Dept. of Pathology and Laboratory Medicine, Room 614, Brinkhous Bullitt Building CB #7525, University of North Carolina at Chapel Hill, Chapel Hill, North Carolina 27599. E-mail: malouf@med.unc.edu.
Supported by the National Institutes of Health (grants R01 HL42250 and R01 HL67358).
References
- Grisham JW, Thorgeirsson SS. Liver stem cells. Potten CS, editor. London: Academic Press; Stem Cells. 1997:pp 233–282. [Google Scholar]
- Coleman WB, Grisham JW. Epithelial stem-like cells of the rodent liver. Strain A, Diehl AM, editors. New York: Chapman and Hall; Liver Growth and Repair. 1998:pp 50–99. [Google Scholar]
- Grompe M, Finegold M. Liver stem cells. Gottlieb D, editor. New York: Cold Spring Harbor Laboratory Press; Stem Cell Biology. 2001:pp 455–498. [Google Scholar]
- Fausto N. Liver regeneration. J Hepatol. 2000;32:19–31. doi: 10.1016/s0168-8278(00)80412-2. [DOI] [PubMed] [Google Scholar]
- Fausto N, Lemire JM, Shiojiri N. Cell lineages in hepatic development and the identification of progenitor cells in normal and injured liver. Proc Soc Exp Biol Med. 1993;204:237–241. doi: 10.3181/00379727-204-43659. [DOI] [PubMed] [Google Scholar]
- Koch KS, Leffert HL. Normal liver progenitor cells in culture. Sell S, editor. Totowa: Humana Press Inc.; Stem Cells Handbook. 2004:pp 367–384. [Google Scholar]
- Strick-Marchand H, Weis MC. Permanent lines of stem cells from the liver. Sell S, editor. Totowa: Humana Press Inc.; Stem Cells Handbook. 2004:pp 385–395. [Google Scholar]
- Strain AJ, Nijjar SS, Crosby HA. Biology of human liver stem cells. Sell S, editor. Totowa: Humana Press, Inc.; Stem Cells Handbook. 2004:pp 397–407. [Google Scholar]
- Malouf NN, Coleman WB, Grisham JW, Lininger RA, Madden VJ, Sproul M, Anderson PA. Adult-derived stem cells from the liver become myocytes in the heart in vivo. Am J Pathol. 2001;158:1929–1935. doi: 10.1016/S0002-9440(10)64661-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao MS, Smith JD, Nelson KG, Grisham JW. A diploid epithelial cell line from normal adult rat liver with phenotypic properties of ‘oval’ cells. Exp Cell Res. 1984;154:38–52. doi: 10.1016/0014-4827(84)90666-9. [DOI] [PubMed] [Google Scholar]
- McKinney-Freeman SL, Jackson KA, Camargo FD, Ferrari G, Mavilio F, Goodell MA. Muscle-derived hematopoietic stem cells are hematopoietic in origin. Proc Natl Acad Sci USA. 2002;99:1341–1346. doi: 10.1073/pnas.032438799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman IL, Anderson DJ, Gage F. Stem and progenitor cells: origins, phenotypes, lineage commitments, and transdifferentiations. Annu Rev Cell Dev Biol. 2001;17:387–403. doi: 10.1146/annurev.cellbio.17.1.387. [DOI] [PubMed] [Google Scholar]
- Wright DE, Wagers AJ, Gulati AP, Johnson FL, Weissman IL. Physiological migration of hematopoietic stem and progenitor cells. Science. 2001;294:1933–1936. doi: 10.1126/science.1064081. [DOI] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis and stem cells: plasticity versus developmental heterogeneity. Nat Immunol. 2002;3:323–328. doi: 10.1038/ni0402-323. [DOI] [PubMed] [Google Scholar]
- Terada N, Hamazaki T, Oka M, Hoki M, Mastalerz DM, Nakano Y, Meyer EM, Morel L, Petersen BE, Scott EW. Bone marrow cells adopt the phenotype of other cells by spontaneous cell fusion. Nature. 2002;416:542–545. doi: 10.1038/nature730. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Evans EP, Smith AG. Changing potency by spontaneous fusion. Nature. 2002;416:545–548. doi: 10.1038/nature729. [DOI] [PubMed] [Google Scholar]
- Alvarez-Dolado M, Pardal R, Garcia-Verdugo JM, Fike JR, Lee HO, Pfeffer K, Lois C, Morrison SJ, Alvarez-Buylla A. Fusion of bone-marrow-derived cells with Purkinje neurons, cardiomyocytes and hepatocytes. Nature. 2003;425:968–973. doi: 10.1038/nature02069. [DOI] [PubMed] [Google Scholar]
- Coleman WB, Wennerberg AE, Smith GJ, Grisham JW. Regulation of the differentiation of diploid and some aneuploid rat liver epithelial (stemlike) cells by the hepatic microenvironment. Am J Pathol. 1993;142:1373–1382. [PMC free article] [PubMed] [Google Scholar]
- Dombrowicz D, Flamand V, Brigman KK, Koller BH, Kinet JP. Abolition of anaphylaxis by targeted disruption of the high affinity immunoglobulin E receptor alpha chain gene. Cell. 1993;75:969–976. doi: 10.1016/0092-8674(93)90540-7. [DOI] [PubMed] [Google Scholar]
- Ausubel FM. New York: J Wiley; Current Protocols in Molecular Biology. 2001 [Google Scholar]
- Bodor GS, Oakeley AE, Allen PD, Crimmins DL, Ladenson JH, Anderson PA. Troponin I phosphorylation in the normal and failing adult human heart. Circulation. 1997;96:1495–1500. doi: 10.1161/01.cir.96.5.1495. [DOI] [PubMed] [Google Scholar]
- Malouf NN, McMahon D, Oakeley AE, Anderson PA. A cardiac troponin T epitope conserved across phyla. J Biol Chem. 1992;267:9269–9274. [PubMed] [Google Scholar]
- Essers J, de Stoppelaar JM, Hoebee B. A new rat repetitive DNA family shows preferential localization on chromosome 3, 12 and Y after fluorescence in situ hybridization and contains a subfamily which is Y chromosome specific. Cytogenet Cell Genet. 1995;69:246–252. doi: 10.1159/000133974. [DOI] [PubMed] [Google Scholar]
- Loeb DD, Padgett RW, Hardies SC, Shehee WR, Comer MB, Edgell MH, Hutchison CA., III The sequence of a large L1Md element reveals a tandemly repeated 5′ end and several features found in retrotransposons. Mol Cell Biol. 1986;6:168–182. doi: 10.1128/mcb.6.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade MH, Trosko JE, Schindler M. A fluorescence photobleaching assay of gap junction-mediated communication between human cells. Science. 1986;232:525–528. doi: 10.1126/science.3961495. [DOI] [PubMed] [Google Scholar]
- Goldberg GS, Bechberger JF, Naus CC. A pre-loading method of evaluating gap junctional communication by fluorescent dye transfer. Biotechniques. 1995;18:490–497. [PubMed] [Google Scholar]
- Hyatt CJ. Chapel Hill: University of North Carolina at Chapel Hill; Multisite ratiometric intracellular Ca2+ imaging with temporal resolution in cultured neonatal rat cardiomyocytes. A novel superfusion device and optical imaging system to study border zone physiology in cultured heart cell monolayers. Biomedical Engineering. 2000 [Google Scholar]
- Frame MK, de Feijter AW. Propagation of mechanically induced intercellular calcium waves via gap junctions and ATP receptors in rat liver epithelial cells. Exp Cell Res. 1997;230:197–207. doi: 10.1006/excr.1996.3409. [DOI] [PubMed] [Google Scholar]
- Upham BL, Suzuki J, Chen G, Wang Y, McCabe LR, Chang CC, Krutovskikh VA, Yamasaki H, Trosko JE. Reduced gap junctional intercellular communication and altered biological effects in mouse osteoblast and rat liver oval cell lines transfected with dominant-negative connexin 43. Mol Carcinog. 2003;37:192–201. doi: 10.1002/mc.10137. [DOI] [PubMed] [Google Scholar]
- Claycomb WC, Bradshaw HD., Jr Acquisition of multiple nuclei and the activity of DNA polymerase alpha and reinitiation of DNA replication in terminally differentiated adult cardiac muscle cells in culture. Dev Biol. 1983;99:331–337. doi: 10.1016/0012-1606(83)90283-x. [DOI] [PubMed] [Google Scholar]
- Goodenough DA, Paul DL. Beyond the gap: functions of unpaired connexon channels. Nat Rev Mol Cell Biol. 2003;4:285–294. doi: 10.1038/nrm1072. [DOI] [PubMed] [Google Scholar]
- Charles AC, Naus CC, Zhu D, Kidder GM, Dirksen ER, Sanderson MJ. Intercellular calcium signaling via gap junctions in glioma cells. J Cell Biol. 1992;118:195–201. doi: 10.1083/jcb.118.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Foster M, Al-Dhalimy M, Lagasse E, Finegold M, Grompe M. The origin and liver repopulating capacity of murine oval cells. Proc Natl Acad Sci USA. 2003;100(Suppl 1):11881–11888. doi: 10.1073/pnas.1734199100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, Kasahara H, Rota M, Musso E, Urbanek K, Leri A, Kajstura J, Nadal-Ginard B, Anversa P. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114:763–776. doi: 10.1016/s0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- Doyonnas R, Blau HM. What is the future for stem cell research? Sell S, editor. Totowa: Humana Press Inc.; Stem Cells Handbook. 2004:pp 491–499. [Google Scholar]