Abstract
Four clinical Helicobacter pylori isolates with high-level resistance to β-lactams exhibited low- to moderate-level resistance to the structurally and functionally unrelated antibiotics ciprofloxacin, chloramphenicol, metronidazole, rifampin, and tetracycline. This pattern of multidrug resistance was transferable to susceptible H. pylori by natural transformation using naked genomic DNA from a clinical multidrug-resistant isolate. Acquisition of the multidrug resistance was also associated with a change in the genotype of the transformed multidrug-resistant H. pylori. DNA sequence analyses of the gene encoding penicillin binding protein 1A (PBP 1A) showed 36 nucleotide substitutions resulting in 10 amino acid changes in the C-terminal portion (the putative penicillin binding domain). Acquisition of β-lactam resistance was consistently associated with transfer of a mosaic block containing the C-terminal portion of PBP 1A. No changes of genes gyrA, rpoB, rrn16S, rdxA, and frxA, and nine other genes (ftsI, hcpA, llm, lytB, mreB, mreC, pbp2, pbp4, and rodA1) encoding putative PBPs or involved in cell wall synthesis were found among the transformed resistant H. pylori. Antibiotic accumulations of chloramphenicol, penicillin, and tetracycline were all significantly decreased in the natural and transformed resistant H. pylori compared to what was seen with susceptible H. pylori. Natural transformation also resulted in the outer membrane protein profiles of the transformed resistant H. pylori becoming similar to that of the clinical resistant H. pylori isolates. Overall, these results demonstrate that high-level β-lactam resistance associated with acquired multidrug resistance in clinical H. pylori is mediated by combination strategies including alterations of PBP 1A and decreased membrane permeability.
Helicobacter pylori is a spiral-shaped gram-negative bacterium and an important human gastric pathogen. H. pylori infections have proven to be difficult to cure (15, 16). The most successful treatment regimens use combinations of two or more antibiotics, such as amoxicillin, clarithromycin, metronidazole, or tetracycline, along with an antisecretory agent or bismuth. Current regimens produce only 60 to 85% cure rates in clinical practice (16). The presence in clinical isolates of antibiotic resistance to amoxicillin, clarithromycin, metronidazole, and tetracycline has been reported, and antibiotic resistance is one of the major causes of treatment failure (16). The prevalence of antibiotic-resistant H. pylori varies among different geographical areas but generally has been increasing worldwide. For example, clarithromycin was first used to treat H. pylori infection in 1992 (R. P. Logan, P. A. Gummett, B. T. Hegarty, M. M. Walker, J. H. Baron, and J. J. Misiewicz, Letter, Lancet 340:239, 1992) but by 1994 it was evident that clarithromycin resistance was going to become an impediment to therapy with that drug (29). Currently, rates of clarithromycin-resistant clinical isolates are reported to be approximately 12% in North America (37), 15% in Europe (4, 30), and up to 13% in East Asia (23, 24).
Mechanisms of resistance to clarithromycin and metronidazole have been extensively studied in H. pylori. Resistance to clarithromycin is generally caused by mutational alterations of specific nucleotides on the macrolide binding sites of the 23S rRNA which prevent the macrolide from binding (45). Metronidazole is a prodrug activated by nitroreductases in the bacterial cell, and metronidazole resistance is caused by either the absence or the inactivation of these nitroreductases (8). Tetracycline binds to the 30S subunit of ribosomes, which blocks the binding of aminoacyl-tRNA, resulting in stalled synthesis of nascent peptide chains. It has recently been reported that mutations in the 16S rRNA are related to tetracycline resistance in H. pylori (12, 44). Resistance to β-lactams in bacteria is most often due to the production of β-lactamase. Other resistance mechanisms to β-lactams include alterations in penicillin binding proteins (PBPs), decreased membrane permeability of antibiotics into the bacterial cell, or combinations of these resistance strategies. Active efflux pumps that excrete drugs can also confer resistance to β-lactams (22, 28, 34). Low-level resistance to amoxicillin (MICs, <8 μg/ml) has been linked to a point mutation on PBP 1A of H. pylori (5, 35, 39).
Our group has previously reported the presence of stable high-level amoxicillin-resistant clinical H. pylori isolates that also exhibited high-level resistance to ampicillin, cephalothin, and penicillin (M. P. Dore, D. H. Kwon, A. R. Sepulveda, D. Y. Graham, and G. Realdi, Letter, Helicobacter 6:79, 2001). We now report evidence that the high-level resistance to β-lactams in the clinical resistant H. pylori isolates is associated with acquired multidrug resistance and that the resistance is mediated by alterations in a mosaic block of pbp-1A in combination with decreased membrane permeability.
MATERIALS AND METHODS
H. pylori isolates and culture conditions.
Amoxicillin-resistant H. pylori isolated in Sardinia, Italy, as clinical isolates (Dore et al., letter) and reference H. pylori strains ATCC 700392, ATCC 43629, and ATCC 43504 obtained from the American Type Culture Collection (Rockville, Md.) were used in this study. H. pylori was routinely cultured on brain heart infusion (BHI; Difco, Detroit, Mich.) agar plates containing 5% horse blood under a microaerobic atmosphere (10% CO2 and 5% O2) at 37°C for 2 to 3 days.
Antibiotics.
The antibiotics used in this study included amoxicillin, ampicillin, aztreonam, cephalothin, chloramphenicol, ciprofloxacin, imipenem, metronidazole, penicillin, rifampin, and tetracycline. With the exceptions of rifampin (Boehringer Mannheim, Indianapolis, Ind.) and imipenem (Merck & Co., Inc., West Point, Pa.), the antibiotics were purchased from Sigma Co. (St. Louis, Mo.). Antibiotics were dissolved in appropriate solvent or distilled water as suggested by the manufacturers.
Determination of MIC.
MICs were determined by twofold agar dilution basically as recommended by the National Committee for Clinical Laboratory Standards (32, 36). Agar dilution plates were prepared by using Mueller-Hinton agar as the base medium. Aged sheep blood (2 weeks old) was added at a concentration of 5%. The range for the antibiotic dilutions was 0.125 to 64 μg/ml. Fresh H. pylori isolates (2- to 3-day culture) were prepared in sterile saline and adjusted to an optical density at 625 nm of 0.38 to 0.40. Using a Steers-type replicating device, 2 to 5 μl of the adjusted inoculum was delivered to agar plates. All plates were incubated under CampyPak Plus conditions (Becton Dickinson BBL, Cockeysville, Md.) at 37°C for up to 4 days. A quality control organism (H. pylori ATCC 43504) was included in every MIC determination. The accepted quality control MIC of metronidazole, as established by the NCCLS, was ≥64 μg/ml (32, 36). Any test in which the quality control MICs fell outside this range was repeated. The MIC was defined as the lowest concentration of the antibiotics at which the growth of the inoculum was completely inhibited.
Resistance stability testing.
To assess the stability of resistance during storage at low temperature, H. pylori strains were stored at −80°C for 1 month and recultured on nonselective 5% horse blood BHI agar plates at 37°C in microaerobic conditions prior to determination of the MICs. To assess the stability of resistance during repeated subculture, cells grown on nonselective media were also repeatedly subcultured by transfer to fresh media for 10 passages (1 month). MICs were assessed using the cells from the last passage.
Natural transformation.
Natural transformation for H. pylori was performed using the methods described by Haas et al. (18). Briefly, approximately 3 × 107 viable cells (optical density at 550 nm = 0.1) of H. pylori were preincubated for 5 h and mixed with 1 to 3 μg of H. pylori genomic DNA purified as previously described (25). The transformed H. pylori colonies were selected on BHI agar plates supplemented with 5% horse blood and amoxicillin (2 μg/ml) or ampicillin (2 μg/ml).
REP-PCR amplification.
H. pylori genomic DNA was purified from individual clinical isolates and from H. pylori ATCC 700392 for PCR templates as previously described (25). One hundred nanograms of template DNA was used for each repetitive extragenic palindromic DNA sequence (REP)-PCR. The REP-PCR conditions were essentially the same as those described previously (25). Each 25-μl REP-PCR volume contained the following: 0.625 mM concentrations of each of the four deoxynucleoside triphosphates (Pharmacia LKB Biotechnology, Piscataway, N.J.), 16.6 mM ammonium acetate, 67 mM Tris-HCl, 6.7 mM magnesium chloride, 6.7 μM EDTA, 30 mM β-mercaptoethanol, 170 μg of bovine serum albumin/ml [pH 8.8], 5 U of Taq polymerase (Promega, Madison, Wis.), and 50 pmol of primer R1 (5′-ICGICTTATCIGGCCTAC-3′). REP-PCR amplification was performed in a thermal cycler (MJ Research, New York, N.Y.) with an initial step of denaturation of target DNA at 95°C for 7 min, followed by 30 cycles of denaturation at 90°C for 30 s, annealing at 40°C for 1 min, and extension at 65°C for 8 min. An additional extension step at 65°C for 16 min completed the PCR. Aliquots of the PCR-amplified products (10 to 20 μl) were resolved in 1% agarose gels containing 0.5× TAE (0.04 M Tris-acetate-0.001 M EDTA) and stained with ethidium bromide. The REP-PCR amplifications were performed twice to confirm the reproducibility of the results.
β-Lactamase assay.
The chromogenic cephalosporin method was used to test the production of β-lactamase. Cefinase disks (Becton Dickinson Microbiology Systems, Cockeysville, Md.) impregnated with nitrocefin, a chromogenic cephalosporin, were moistened with a drop of sterile distilled water, and several well-isolated fresh colonies from the BHI agar plates containing 8 μg of amoxicillin or ampicillin/ml were selected and smeared on the disk surface. β-Lactamase activity was read as positive by the change in color of the chromogenic cephalosporin after 6 to 12 h of incubation at room temperature. Escherichia coli harboring pUC19 was used as a positive control.
DNA sequence analyses.
To determine DNA sequences in H. pylori genes encoding GyrA (resistance to ciprofloxacin [46]), RpoB (resistance to rifampin [46]), 16S rRNA (resistance to tetracycline [44]), and RdxA/FrxA (resistance to metronidazole [14, 26]), PCR amplification of the genes was performed by employing the specific PCR primers used previously (26, 44, 46). DNA sequence analysis of the 10 genes encoding putative PBPs and involved in cell wall synthesis (i.e., PBP 4 [pbp4], cysteine-rich protein A [hcpA], lysis tolerance protein [lytB], PBP 1A [pbp-1A], three rod shape-determining proteins [rodA1, mreC, and mreB], cell division protein [ftsI], PBP 2 [pbp2], and methicillin resistance protein [llm]) (11) was performed using genomic DNA from the transformed resistant H. pylori. PCR amplification for these genes was carried out to amplify DNA fragments of less than 400 bps, and the resulting fragments were used for DNA sequence analysis. DNA sequences of the PCR fragments (both directions for each fragment) were determined at a commercial DNA sequencing facility (SEQWRITE, Houston, Tex.). In parallel, PCR amplification and determination of the known DNA sequences of gyrA, rpoB, rrn16S, rdxA, frxA, and the 10 genes noted above from H. pylori ATCC 700392 (43) was also performed to ensure PCR amplification fidelity. The resulting DNA sequences from the PCR fragments were analyzed by comparison to the known DNA sequences.
Genomic library construction and isolation of a gene encoding PBP 1A (pbp-1A) from amoxicillin-resistant H. pylori IH-1.
Genomic DNA was extracted from amoxicillin-resistant H. pylori IH-1 and fractionated to ∼5- to 10-kb fragments after digestion with Sau3AI on 0.7% agarose gels. The agarose gel-purified fragments (1 μg) were ligated with a ZAP Express BamHI predigested vector (Stratagene, La Jolla, Calif.) containing phagemid pBK-CMV in lambda phage arms. The ligation mixture was packaged by using a commercial packaging extract (Stratagene) and infected into E. coli XL1-Blue with blue-white plaque selection. To isolate a gene encoding PBP 1A, a PCR fragment containing the C terminus of PBP 1A (1,048 bp), which was amplified by primer pairs BP2F (5′GCGTCTAATGAAGATGAAGA3′) and SP3R (5′TTAAAGTCCCTATAGCCATG3′) from H. pylori ATCC 700392, was labeled using digoxigenin. Plaque hybridization using the digoxigenin-labeled probe was performed to isolate pbp-1A as described by the manufacturer (Roche Molecular Biochemicals, Indianapolis, Ind.). DNA sequencing of the inserts was performed at a commercial DNA sequencing facility (SEQWRITE) by using appropriate synthetic primers, including T3/T7 promoter primers.
Antibiotic accumulation assay.
Antibiotic accumulation assays for chloramphenicol, penicillin, and tetracycline were performed essentially as described by Perreten et al. (40) and George et al. (10). H. pylori cells grown for 2 to 3 days on BHI agar plates supplemented with 5% horse blood were harvested in assay buffer containing 50 mM KPO4 and 1 mM MgSO4 (pH 6.6). The cells (5 × 109 per ml) were centrifuged and resuspended in the same assay buffer. Antibiotic accumulation assays were commenced by the addition of d-threo-[dichloroacetyl-1-14C]chloramphenicol (50 to 62 mCi mmol−; 1.85 to 2.29 GBq mmol−; Amersham Pharmacia Biotech, Piscataway, N.J.), [penyl-4(n)-3H]benzylpenicillin (10 to 30 Ci mmol−; 0.37 to 1.1 TBq mmol−; Amersham Pharmacia Biotech), or [7-3H]tetracycline (0.6 Ci mmol−; 22.2 GBq mmol−; Dupont/NEN Research Products, Boston, Mass.) to 5 μM. One-milliliter aliquots were taken every 10 min. After 20 min, each cell suspension was divided in half, and 100 μM CCCP (carbonyl cyanide m-chlorophenylhydrazone) was added to one half to de-energize the cells. Then, 1-ml aliquots were taken every 10 min from each half. Each aliquot was immediately centrifuged and washed three times in phosphate-buffered saline (PBS). The resulting pellets were then diluted in scintillation fluid (CytoScint; Fisher Biotech) and analyzed for radioactivity in an LS 6500 scintillation counter (Beckman Instruments, Palo Alto, Calif.). Antibiotic accumulation assays for starved H. pylori cells were also performed using the same cells suspended in sterilized saline and incubated at 37°C in microaerobic conditions for 12 h. Then, the radioactively labeled chloramphenicol, penicillin, or tetracycline was added to commence the accumulation assays. After 20 min, each cell suspension was divided into halves as described above and 0.5% (vol/vol) glucose was added to one half to energize the cells. Subsequently, 1-ml aliquots were taken at 10-min intervals and immediately washed three times before analysis for radioactivity as described above.
Preparation and analysis of OMPs.
H. pylori cells grown for 2 to 3 days (two plates) were harvested in 5 ml (approximately 5 × 109 cells/ml) of PBS (pH 7.4). The cells were washed once with PBS. Crude extracts of the cells were prepared by disruption using a French pressure cell (600 lb/in2; Aminco, Urbana, Ill.) and then centrifuged at 12,000 × g for 10 min. Outer membrane insoluble fractions were isolated by incubation of the supernatant in 3% N-lauroyl sarcosyl for 45 min at room temperature as described previously (2, 9). The outer membranes were precipitated by centrifugation at 130,000 × g for 45 min at 15°C and resuspended in the PBS. Protein concentrations in outer membrane preparations were determined by the Bradford procedure (Bio-Rad Laboratories, Richmond, Calif.) with bovine serum albumin as the standard. OMP profiles (10 μg per sample) were examined on a 7% polyacrylamide gel containing 8 M urea and 0.2% sodium dodecyl sulfate as described previously (3). OMP bands were stained with 0.1% Coomassie brilliant blue R250 (Bio-Rad Laboratories) following destaining with 10% methanol-7% acetic acid.
Nucleotide sequence accession numbers.
The nucleotide sequences for the pbp-1A genes from amoxicillin-resistant H. pylori IH-1 and susceptible H. pylori ATCC 43629 were deposited in GenBank under accession numbers AY241260 and AY241259, respectively.
RESULTS
High-level β-lactam-resistant H. pylori isolates associated with acquired multidrug resistance.
Patient information relating to the clinical H. pylori isolates is shown in Table 1. The high-level MICs of amoxicillin, ampicillin, cephalothin, and penicillin were similar to those previously reported (Dore et al., letter). The MICs of other β-lactams were also significantly increased (e.g., MIC of aztreonam, 64 μg/ml; MIC of imipenem, 1 μg/ml) compared with those for the susceptible reference H. pylori ATCC 700392 strain (Table 2). The MICs of ciprofloxacin (2 μg/ml), chloramphenicol (4 μg/ml), metronidazole (32 μg/ml), rifampin (4 μg/ml), and tetracycline (8 μg/ml) (Table 2) for these isolates were also increased compared with those for the susceptible H. pylori ATCC 700392 strain, implying that the isolates had acquired multidrug resistance.
TABLE 1.
Patient information relating to the high-level β-lactam-resistant H. pylori isolates
| Isolate | Patient informationa
|
|||
|---|---|---|---|---|
| Disease | Histology | Gender | Age (yr) | |
| IH-1 | Erosive gastropathy | Chronic active gastritis | Male | 36 |
| IH-2 | Bile reflux and gastritis | Mild chronic active gastritis | Female | 62 |
| IH-3 | Gastropathy | Mild chronic active gastritis | Female | 60 |
| IH-4 | Gastropathy | Chronic active gastritis | Female | 66 |
For all patients, the antrum was the biopsy site. None of the patients had previous anti-H. pylori treatment.
TABLE 2.
MICs for H. pylori isolates
| H. pylori strain | MIC (μg/ml)a
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AMX | AMC | ATM | CEF | IPM | PEN | CIP | CHL | MTZ | RIF | TET | |
| ATCC 700392 | ≤0.125 | 0.5 | 4 | 0.5 | ≤0.125 | ≤0.125 | 0.25 | 1 | 2 | 0.5 | 0.5 |
| Clinical isolates | |||||||||||
| IH-1 | 64 | 64 | 64 | 64 | 1 | 16 | 2 | 4 | 32 | 4 | 8 |
| IH-2 | 64 | 64 | 64 | 64 | 1 | 16 | 2 | 4 | 32 | 4 | 8 |
| IH-3 | 64 | 64 | 64 | 64 | 1 | 16 | 2 | 4 | 32 | 4 | 8 |
| IH-4 | 64 | 64 | 64 | 64 | 1 | 16 | 2 | 4 | 32 | 4 | 8 |
| Transformed colonies (with naked genomic DNA)b | |||||||||||
| 1 | 32 | 64 | 32 | 64 | 0.5 | 8 | 4 | 8 | 16 | 8 | 4 |
| 2 | 64 | 64 | 32 | 16 | 0.25 | 8 | 2 | 4 | 16 | 8 | 4 |
| 3 | 16 | 32 | 32 | 64 | 0.25 | 8 | 2 | 8 | 16 | 8 | 4 |
| 4 | 64 | 64 | 32 | 32 | 0.25 | 8 | 4 | 8 | 16 | 4 | 4 |
| 5 | 32 | 64 | 32 | 64 | 1 | 8 | 2 | 8 | 16 | 8 | 4 |
| 6 | 32 | 64 | 32 | 64 | 0.25 | 16 | 4 | 8 | 16 | 8 | 8 |
| 7 | 16 | 64 | 64 | 32 | 0.5 | 8 | 4 | 4 | 16 | 4 | 4 |
| 8 | 16 | 32 | 64 | 64 | 0.25 | 8 | 4 | 8 | 16 | 8 | 8 |
| 9 | 16 | 32 | 32 | 64 | 0.5 | 8 | 2 | 8 | 8 | 8 | 4 |
| 10 | 16 | 32 | 32 | 32 | 0.25 | 8 | 2 | 8 | 16 | 8 | 4 |
| 11 | 16 | 64 | 32 | 64 | 0.5 | 16 | 2 | 4 | 16 | 8 | 4 |
| 12 | 16 | 64 | 32 | 16 | 0.25 | 8 | 2 | 4 | 16 | 4 | 4 |
| Transformed colonies (with PCR fragment)c | |||||||||||
| 13 | 4 | 8 | 8 | 8 | ≤0.125 | 2 | 0.5 | 2 | 2 | 1 | 0.5 |
| 14 | 4 | 8 | 8 | 8 | ≤0.125 | 2 | 0.5 | 2 | 2 | 1 | 0.5 |
| 15 | 8 | 8 | 8 | 8 | ≤0.125 | 2 | 0.5 | 2 | 2 | 1 | 0.5 |
| 16 | 8 | 8 | 16 | 8 | ≤0.125 | 2 | 0.5 | 2 | 2 | 1 | 0.5 |
| 17 | 4 | 8 | 16 | 8 | ≤0.125 | 2 | 0.5 | 2 | 2 | 1 | 0.5 |
MIC determinations were repeated three times on different days by measuring all MICs at the same time. AMX, amoxicillin; AMC, ampicillin; ATM, aztreonam; CEF, cephalothin; IPM, imipenem; PEN, penicillin; CIP, ciprofloxacin; CHL, chloramphenicol; MTZ, metronidazole; RIF, rifampin; TET, tetracycline.
Naked genomic DNA was extracted from clinical resistant H. pylori IH-1.
PCR fragment included the C-terminal half of pbp-1A from clinical resistant H. pylori IH-1 (1,020 bp) (see the text).
Natural transformation of resistance determinants.
To examine whether the resistance to β-lactams as well as to other structurally and functionally unrelated antibiotics was transferable to susceptible H. pylori, natural transformation was performed using susceptible H. pylori ATCC 700392 as a recipient strain. The natural transformation frequency of the recipient H. pylori with naked genomic DNA extracted from the β-lactam-resistant clinical isolate IH-1 was 10−4 to 10−3. A negative control using no genomic DNA did not show growth. The transformed colonies (12 colonies) from the BHI agar plates containing amoxicillin (2 μg/ml) were used to determine MICs by the agar dilution method. All transformed colonies were resistant (MICs, ≥8 μg/ml) to all β-lactams tested except for imipenem. The MICs of the β-lactams for the transformed resistant colonies were either identical to or less than those for the original resistant isolate (Table 2). The MICs of ciprofloxacin, chloramphenicol, metronidazole, rifampin, and tetracycline for the transformed H. pylori colonies were also similar to those for the original isolate (Table 2). The results were essentially the same irrespective of whether the H. pylori cells were grown with or without amoxicillin before the MICs were measured. Natural transformation using H. pylori ATCC 43629 as a recipient strain was also performed, and we obtained results similar to those with H. pylori ATCC 700392 (data not shown). The stability of the resistance pattern during freezing or passage in nonselective agar plates was assessed using both the original resistant clinical isolates and the transformed resistant isolates. MICs were unchanged following storage at −80°C or following repetitive passage on nonselective agar plates. Overall results showed that (i) the clinical H. pylori isolates had high-level resistance to most β-lactams, (ii) resistance to the individual antibiotics was transferable to previously antibiotic-susceptible H. pylori, and (iii) β-lactam resistance appeared to be associated with the acquired multidrug resistance.
Genotypic relatedness of the high-level β-lactam-resistant H. pylori isolates.
Total genomic DNA was purified from the amoxicillin-resistant clinical isolates, the 12 transformed resistant H. pylori colonies, and the susceptible parental H. pylori ATCC 700392 strain. The genomic DNA was examined on 0.3 to 1% agarose gels, and no extrachromosomal DNA (e.g., R plasmid) was found in either the clinical resistant isolates or the transformed resistant H. pylori, suggesting that the high-level resistance to β-lactams and the acquired multidrug resistance were likely to be associated with chromosomal DNA. REP-PCR fingerprints were performed by using the purified genomic DNA from these isolates. The REP-PCR patterns of the four clinical isolates were very similar (Fig. 1, lanes 1 to 4), suggesting that the original isolates were closely related, possibly arising from a common ancestral strain. The REP-PCR patterns of the susceptible strain ATCC 700392 and the resistant clinical isolates were noticeably different. All the transformed resistant H. pylori colonies had similar fingerprinting patterns, which differed from both the susceptible parental H. pylori ATCC 700392 strain and the clinical resistant isolates (Fig. 1, lane 6), suggesting that the transformation resulted in alterations in the chromosomal DNA.
FIG. 1.
REP-PCR fingerprinting profiles of resistant clinical H. pylori isolates, susceptible H. pylori ATCC 700392, and its transformed resistant H. pylori ATCC 700392 strain. Lanes 1 to 4, fingerprinting profiles from genomic DNA of clinical resistant H. pylori isolates IH-1, IH-2, IH-3, and IH-4, respectively; lane 5, fingerprinting profile from genomic DNA of susceptible H. pylori ATCC 700392; lane 6, fingerprinting profile from genomic DNA of transformed resistant H. pylori ATCC 700392, which was transformed using naked genomic DNA extracted from the resistant clinical H. pylori IH-1 isolate; lane M, 100-bp DNA size marker from Roche. The REP-PCR amplifications were repeated three times to confirm reproducibility.
Role of β-lactamase in β-lactam resistance.
All colonies from the clinical resistant isolates and the transformed resistant H. pylori, including the susceptible parental H. pylori ATCC 700392 strain, showed negative reaction to assays for β-lactamase activity, suggesting that amoxicillin resistance was not due to the presence of β-lactamase activity. E. coli-harboring pUC19 (bla+) showed an immediately positive reaction for β-lactamase activity in the same β-lactamase assay. To ensure the absence of β-lactamase activity in the clinical resistant isolates and the transformed resistant H. pylori, fresh cells harvested from the two agar plates were ruptured by using a French press (600 lb; Aminco, Urbana, Ill.) as described previously (26) and the crude extracts of the cells were used to test for β-lactamase activity. No β-lactamase activity was detected after 18 h of incubation at room temperature.
Role of genes encoding putative PBPs and genes involved in cell wall synthesis in β-lactam resistance.
Several genes encoding putative PBPs and genes involved in cell wall synthesis have been identified in H. pylori (5, 20, 31, 39). We examined the 10 genes, described in Materials and Methods, for their possible involvement in β-lactam resistance. Genomic DNA from the three transformed resistant H. pylori colonies (colonies 1, 2, and 3 [Table 2]), including the susceptible parental H. pylori ATCC 700392 strain, was used to analyze DNA sequences for the 10 genes. Comparisons of each pair of genes between the transformed resistant colony and its susceptible parental H. pylori strain showed that only the gene encoding PBP 1A (pbp-1A) was significantly changed. DNA sequence analyses of pbp-1A from the three colonies showed that 36 nucleotides were substituted in the C-terminal portions (from nucleotide positions 960 to position 1980 [Fig. 2]), which is thought to comprise the penicillin binding domain (13). The substitutions were all identical among the three colonies. However, no nucleotide change was found in N-terminal portions of pbp-1A.
FIG. 2.
DNA sequence comparison of pbp-1A genes from H. pylori ATCC 700392, its transformed resistant H. pylori strain with genomic DNA extracted from clinical resistant H. pylori IH-1 (ATCC 700392/IH-1), and clinical resistant H. pylori IH-1. Nucleotides of the pbp-1A gene from transformed resistant H. pylori ATCC 700392/IH-1 were switched beginning at position 960 with those of clinical resistant H. pylori IH-1 (shaded DNA sequences) (see the text). The three asterisks indicate a stop codon of pbp-1A.
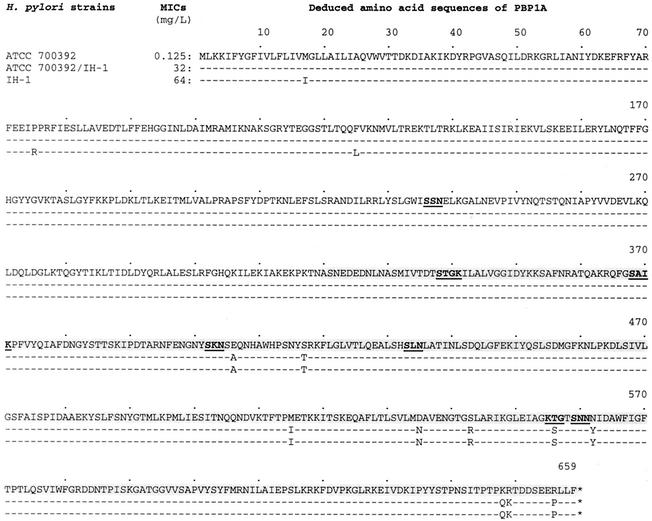
To compare DNA sequences of pbp-1A from clinical resistant H. pylori IH-1 with those of transformed resistant H. pylori, including susceptible parental H. pylori, a genomic library was constructed by using genomic DNA extracted from clinical resistant H. pylori IH-1. Two library clones for pbp-1A matched DNA sequences from nucleotide position 630688 to position 638051 (7,363 bp; Zap85) and from nucleotide position 626555 to position 633437 (6,882 bp; Zap229) of the complete genome sequence of H. pylori ATCC 700392 (43). The DNA sequence divergence between the pbp-1A genes from clinical resistant H. pylori IH-1 and susceptible H. pylori ATCC 700392 was 2.6% (51 of 1,980 nucleotides), producing 2% deduced amino acid divergence (13 of 659 amino acids). Thirty-six of the 51 nucleotide substitutions were in the C-terminal portion, and the others were in the N-terminal portion (Fig. 2). These nucleotide substitutions resulted in 13 amino acid changes, and 10 of the 13 amino acid changes were located in the C-terminal portion (Fig. 3). The 36 nucleotide substitutions resulting in 10 amino acid changes in the C-terminal portion were all transferred as a single event among all the transformed resistant H. pylori colonies tested. In contrast, the N-terminal portion of pbp-1A remained unchanged in the three transformed resistant H. pylori colonies (Fig. 2 and 3). The specific recombination event consistently occurred at the position of nucleotide 960 (change from G to A) (Fig. 2) and continued after the stop codon of pbp-1A at the position of nucleotide 1980 (Fig. 2). This specific feature was also observed in the three randomly selected transformed resistant H. pylori ATCC 43629 colonies (data not shown), suggesting that the C-terminal portion functions as a mosaic block in natural transformation.
FIG. 3.
Amino acid comparison of PBP 1A encoded by pbp-1A genes from H. pylori ATCC 700392, its transformed resistant H. pylori strain with genomic DNA extracted from clinical resistant H. pylori IH-1, and clinical resistant H. pylori IH-1. Amino acid sequences beginning from position 320 correspond to the nucleotide sequence switch of pbp-1A shown in Fig. 2 (shaded amino acid sequences). Amino acid sequence signature motifs of PBP 1A based on the report by Harris et al. (20) are shown in boldface type with underlining.
To examine the role of the mosaic block in β-lactam resistance, the mosaic block (from nucleotide position 960 to position 1980; 1,020 bp [Fig. 2]) for pbp-1A from clinical resistant H. pylori IH-1 and susceptible H. pylori ATCC 700392 was amplified by PCR and the PCR fragments were verified by DNA sequences. The two PCR-amplified fragments (1 μg each) were then used for natural transformation of susceptible H. pylori ATCC 700392 and ATCC 43629. The PCR fragment amplified from clinical resistant H. pylori IH-1 resulted in the two susceptible H. pylori strains being able to grow (frequencies of approximately 10−4) on BHI agar plates containing 2 μg of amoxicillin/ml. In contrast, the PCR fragment amplified from the susceptible H. pylori strains did not permit growth of the two susceptible H. pylori strains on agar plates containing 2 μg of amoxicillin/ml. Five randomly selected colonies grown on the amoxicillin-containing agar plates were used to measure MICs, and significantly increased MICs (2 to 16 μg/ml) of the β-lactams tested, except for imipenem, were found (Table 2). However, no change was found in the MICs of unrelated antibiotics (Table 2).
Role of genes known to be associated with antibiotic resistance in the acquired multidrug resistance.
Genes known to be involved in resistance to individual antibiotics, for example, gyrA for ciprofloxacin, rpoB for rifampin, rrn16S for tetracycline, and rdxA/frxA for metronidazole resistance, were examined by DNA sequence determination using three transformed resistant H. pylori colonies (colonies 1, 2, and 3 [Table 2]). The DNA sequences and the deduced amino acid sequences of GyrA, RpoB, RdxA, and FrxA from the three transformed resistant H. pylori colonies were unchanged compared with those for the susceptible parental H. pylori ATCC 700392 strain. The DNA sequences for the 16S rRNA gene were also unchanged compared with those for the susceptible parental H. pylori ATCC 700392 strain.
Role of membrane permeability.
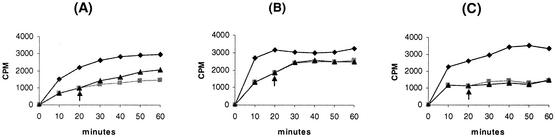
Antibiotic accumulation assays were performed using the clinical resistant H. pylori IH-1 isolate and one of the transformed resistant H. pylori colonies (colony 1 [Table 2]). The susceptible parental H. pylori ATCC 700392 strain was also included in the antibiotic accumulation assays. Accumulations of chloramphenicol, penicillin, and tetracycline were significantly decreased in transformed resistant H. pylori compared with what was seen with susceptible parental H. pylori (Fig. 4). The pattern of decreased accumulation of chloramphenicol, penicillin, and tetracycline in the clinical resistant IH-1 isolate was similar to that in the transformed resistant H. pylori (data not shown). Chloramphenicol accumulation increased slightly after the addition of CCCP (at 20 min) in both the clinical resistant isolates and the transformed resistant H. pylori. Accumulation of penicillin or tetracycline was unchanged by the addition of CCCP. Antibiotic accumulation assays using starved cells from clinical resistant H. pylori IH-1, transformed resistant H. pylori (colony 1 [Table 2]), and susceptible parental H. pylori ATCC 700392 were also performed to examine whether antibiotic accumulations changed in response to glucose. Starvation did not produce a change in outcome, including that for chloramphenicol accumulation (data not shown). Overall, the accumulations of chloramphenicol, penicillin, and tetracycline were all significantly decreased in both the clinical resistant isolate and the transformed resistant H. pylori.
FIG. 4.
Antibiotic accumulation assays for susceptible H. pylori ATCC 700392 and its transformed resistant H. pylori strain. Shown are accumulation patterns of chloramphenicol (A), penicillin (B), and tetracycline (C). Symbols: diamonds, susceptible H. pylori ATCC 700392; squares, one half of the transformed resistant H. pylori ATCC 700392, without treatment with 100 μM CCCP; triangles, the other half of the transformed resistant H. pylori ATCC 700392, with treatment with 100 μM CCCP at 20 min (arrows). Results are representative of five independent experiments.
Alterations of OMPs.
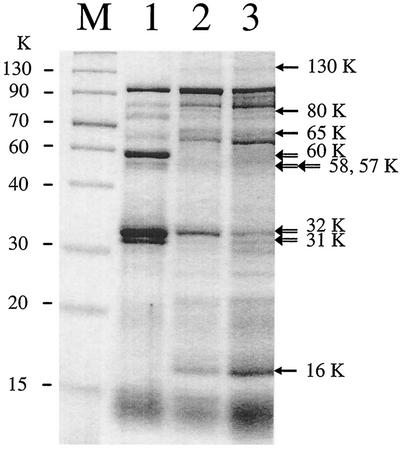
Decreased membrane permeability and active efflux of antibiotics are often associated with alterations of OMPs in gram-negative bacteria (34, 41). To test whether the high-level resistance to β-lactams and the acquired multidrug resistance were associated with alterations of OMPs, OMPs were purified from the clinical resistant isolates and the transformed resistant H. pylori (colonies 1, 2, and 3 [Table 2]). OMP profiles from the clinical resistant isolates were very similar to one another but different from the OMP profile of susceptible parental H. pylori ATCC 700392. OMP profiles from the three transformed resistant H. pylori colonies were also similar to one another but different from that of their susceptible parental H. pylori ATCC 700392 strain (Fig. 5). The differences between the susceptible parental H. pylori and the transformed resistant H. pylori included several OMP bands that either disappeared or were present at very low levels in the latter (e.g., bands at a molecular weight of approximately 60,000 [60K], 58K, 57K, 32K, and 31K) (Fig. 5, lanes 1 and 2). These OMP bands were also absent, or present at very low levels, in the clinical resistant isolates (Fig. 5, lane 3). In contrast, a number of OMP bands were only found (or were present at higher levels) in the clinical isolates and the transformed resistant H. pylori (e.g., bands at approximately 130, 80, 65, and 16K) (Fig. 5, lanes 2 and 3). These OMP bands were either absent or present at very low levels in susceptible parental H. pylori ATCC 700392.
FIG. 5.
OMP profiles for susceptible H. pylori ATCC 700392 (lane 1), transformed resistant H. pylori (lane 2), and resistant clinical H. pylori IH-1 (lane 3). Lane M contains a size marker from Gibco BRL. Open arrows indicate OMPs (approximately 60, 58, 57, 32, and 31K) that disappeared (or were at very low levels) in transformed resistant H. pylori ATCC 700392 and resistant clinical H. pylori IH-1 but were clearly seen in susceptible H. pylori ATCC 700392, and closed arrows indicate OMPs (approximately 130, 80, 65, and 16K) that were present in both transformed resistant H. pylori ATCC 700392 and resistant clinical H. pylori IH-1 but were not present (or were at very low levels) in susceptible H. pylori ATCC 700392.
DISCUSSION
Antibiotic resistance in bacteria can be categorized as intrinsic or acquired resistance. Intrinsic resistance is a genetic property of most bacterial strains and typically evolved independently of the clinical use of antimicrobial agents. Acquired resistance implies that a susceptible organism has developed resistance to antimicrobial agents to which it was previously susceptible. In this report, we performed detailed phenotypic and genotypic examination of previously reported amoxicillin-resistant clinical H. pylori isolates (Dore et al., letter). The clinical isolates showed low to high levels of resistance to β-lactams. The clinical isolates also showed low- or moderate-level resistance to several structurally and functionally unrelated antibiotics, suggesting that the β-lactam resistance may be part of the phenomenon of acquired multidrug resistance. The pattern of multidrug resistance was stable during both passage and freezing. Acquired multidrug resistance was also transferable from the resistant clinical isolates to susceptible H. pylori by using naked genomic DNA. None of the transformed resistant H. pylori colonies was resistant only to β-lactams. The MICs of the β-lactams (except imipenem) for the transformed resistant colonies varied between 8 and 64 μg/ml, suggesting that several factors may be involved in resistance to β-lactams.
The genotypic relatedness of four clinical isolates was assessed by REP-PCR fingerprinting analysis (1). The REP-PCR fingerprints were very similar among the clinical isolates, suggesting that they may have arisen from a common ancestral strain and that the β-lactam resistance may have been vertically disseminated among clinical H. pylori isolates. The fingerprinting patterns of the transformed resistant H. pylori colonies were also significantly different from those of the susceptible parental H. pylori, which is evidence against simple point mutations as the cause of antibiotic resistance, as was shown previously (27, 39). Total genomic DNA analysis suggested that β-lactam resistance and acquired multidrug resistance were not mediated by extrachromosomal DNA, such as an R plasmid. However, these experiments cannot fully exclude the possibility that transposable elements involved in resistance may reside in the chromosomal DNA. Indeed, in other bacteria, a huge DNA fragment carrying multiple genes responsible for individual antibiotic resistance (called an integron) has been reported (42). We performed DNA sequencing analysis of the known genes responsible for individual antibiotic resistance by using transformed resistant H. pylori. We found significant alterations only in pbp-1A, suggesting that the resistance mechanism is unlikely to be associated with an integron carrying individual antibiotic resistance determinants. β-Lactamase was not present, which is consistent with previous observations that β-lactam resistance in H. pylori was not mediated by β-lactamase (5, 6). In general, non-β-lactamase-producing bacteria acquire resistance to β-lactams by (i) alterations in the preexisting PBPs or acquisition of a novel PBP insensitive to β-lactams, (ii) decreased membrane permeability that allows accumulation of only low levels of β-lactams, (iii) energy-coupled active efflux of β-lactams, or (iv) combinations of these resistance strategies.
The PBPs are a set of enzymes involved in the synthesis of the peptidoglycan layer of the bacterial cell wall and play a role as transpeptidases, transglycosylases, endopeptidases, or carboxypeptidases (13). The covalent binding of β-lactams to various PBPs results in the inability of the bacterium to build a complete cell wall and ultimately leads to cell lysis and death. Alterations in PBPs resulting in reduced binding ability can lead to resistance to β-lactams. Examples include alterations of PBPs 3a and 3b of Haemophilus influenzae (38), PBPs 2b and 2x of Streptococcus pneumoniae (17), and PBP 1A of Proteus mirabilis (33). Several putative PBPs have been described in H. pylori, including three high-molecular-weight PBPs and six putative low-molecular-weight PBPs (20, 21). Alterations in PBP 1A have been reported in relation to increased MICs of β-lactams (5, 11, 35, 39). In this study, alterations of PBP 1A were also related to β-lactam resistance. However, several differences were found in comparison to the results of previous reports. For example, higher levels of β-lactam resistance were associated with more alterations of PBP 1A. Susceptible H. pylori transformed with the PCR fragment containing the 10 amino acid changes increased the MICs of amoxicillin up to 8 μg/ml. In contrast, previous reports showed that susceptible H. pylori transformed with altered PBP 1A containing one to four amino acid changes increased the MICs of amoxicillin up to 2 μg/ml (5, 11, 35, 39). The clinical resistant H. pylori isolates, however, showed much higher levels of β-lactam resistance than did the transformed resistant H. pylori colonies. These results suggest that an additional resistance mechanism(s) is involved in the production of the higher levels of β-lactam resistance seen in the clinical resistant isolates (see below). In this study, we also found a mosaic block of pbp-1A that contained 10 amino acid changes and six homologous motifs identified in H. pylori PBP 1A (20). In particular, changes of Glu406 to Ala and Asn562 to Tyr were directly adjacent to the SKN and KTG or SNN homologous motifs, respectively. Acquisition of β-lactam resistance by susceptible H. pylori was consistently associated with transfer of the mosaic block rather than transfer of individual altered amino acids, as shown previously. These observations suggest that the higher levels of β-lactam resistance are related to the 10 amino acid changes seen in the mosaic block.
Presence of a mosaic block (or structure) in PBPs has been also reported among naturally transformable gram-positive bacteria (e.g., PBPs 1a, 2x, and 2b in S. pneumoniae) (19). The mosaic blocks differ as much as 25% at the nucleotide level, resulting in approximately 10% amino acid substitutions (19). The mosaic block of H. pylori PBP 1A showed divergences at 4% of the nucleotides, which resulted in changes of 3% of the amino acids. Based on the amino acid sequence signature for the transpeptidase domain of E. coli (13), the mosaic block included the transpeptidase domain of H. pylori PBP 1A. In H. pylori, the motifs SXXK and KS(T)G are separated by 219 amino acids, which is similar to the transpeptidase of E. coli PBP 2. The origin of the mosaic block is not clear. However, it is possible that a point mutation has accumulated in the transpeptidase domain and that the altered-transpeptidase domain was transferred to susceptible H. pylori under β-lactam selective pressure. Another possibility is that the mosaic block arose by introduction from closely related β-lactam-resistant bacterial species (7).
We also examined whether decreased membrane permeability and/or energy-coupled active efflux mechanisms were associated with β-lactam resistance. Although we did not find clear evidence for energy-coupled active efflux in the resistant strains, our data suggest a role for decreased membrane permeability as antibiotic accumulations were significantly decreased in the resistant strains. These observations demonstrate that the combination of both decreased membrane permeability and alterations of PBP 1A may be involved in production of the high-level resistance to β-lactams as well as in production of the low- to moderate-level resistance to several structurally and functionally unrelated antibiotics in H. pylori. Finally, we found alterations of OMP patterns in which the pattern of the transformed resistant H. pylori became similar to that of the resistant clinical isolates. These results suggest that the specific OMP alterations may play a role in multidrug resistance. Indeed, several investigators have reported that alterations of OMP pattern in gram-negative bacteria are related to acquisition of multidrug resistance (34, 41). The complete genomic sequence of H. pylori shows the presence of large families of OMPs, suggesting that a number of repertoires for OMP patterns may be present in H. pylori. Overall, the results are consistent with the notion that high-level resistance to β-lactams in the multidrug-resistant clinical H. pylori isolates is mediated by both alterations in PBP 1A and decreased membrane permeability.
Acknowledgments
The material presented in this study is based upon work supported in part by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs, and by Public Health Service grant DK56338, which funds the Texas Gulf Coast Digestive Diseases Center.
REFERENCES
- 1.Burucoa, C., V. Lhomme, and J. L. Fauchere. 1999. Performance criteria of DNA fingerprinting methods for typing of Helicobacter pylori isolates: experimental results and meta-analysis. J. Clin. Microbiol. 37:4071-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charvalos, E., Y. Tselentis, M. M. Hamzehpour, T. Kohler, and J. C. Pechere. 1995. Evidence for an efflux pump in multidrug-resistant Campylobacter jejuni. Antimicrob. Agents Chemother. 39:2019-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen, S. P., L. M. McMurry, D. C. Hooper, J. S. Wolfson, and S. B. Levy. 1989. Cross-resistance to fluoroquinolones in multiple-antibiotic-resistant (Mar) Escherichia coli selected by tetracycline or chloramphenicol: decreased drug accumulation associated with membrane changes in addition to OmpF reduction. Antimicrob. Agents Chemother. 33:1318-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Debets-Ossenkopp, Y. J., A. J. Herscheid, R. G. Pot, E. J. Kuipers, J. G. Kusters, and C. M. Vandenbroucke-Grauls. 1999. Prevalence of Helicobacter pylori resistance to metronidazole, clarithromycin, amoxicillin, tetracycline and trovafloxacin in The Netherlands. J. Antimicrob. Chemother. 43:511-515. [DOI] [PubMed] [Google Scholar]
- 5.DeLoney, C. R., and N. L. Schiller. 2000. Characterization of an in vitro-selected amoxicillin-resistant strain of Helicobacter pylori. Antimicrob. Agents Chemother. 44:3368-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dore, M. P., D. Y. Graham, and A. R. Sepulveda. 1999. Different penicillin-binding protein profiles in amoxicillin-resistant Helicobacter pylori. Helicobacter 4:154-161. [DOI] [PubMed] [Google Scholar]
- 7.Dowson, C. G., A. Hutchison, J. A. Brannigan, R. C. George, D. Hansman, J. Linares, A. Tomasz, J. M. Smith, and B. G. Spratt. 1989. Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 86:8842-8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards, D. I. 1993. Nitroimidazole drugs—action and resistance mechanisms. I. Mechanisms of action. J. Antimicrob. Chemother. 31:9-20. [DOI] [PubMed] [Google Scholar]
- 9.Filip, C., G. Fletcher, J. L. Wulff, and C. F. Earhart. 1973. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J. Bacteriol. 115:717-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.George, A. M., R. M. Hall, and H. W. Stokes. 1995. Multidrug resistance in Klebsiella pneumoniae: a novel gene, ramA, confers a multidrug resistance phenotype in Escherichia coli. Microbiology 141:1909-1920. [DOI] [PubMed] [Google Scholar]
- 11.Gerrits, M. M., D. Schuijffel, A. A. van Zwet, E. J. Kuipers, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2002. Alterations in penicillin-binding protein 1A confer resistance to β-lactam antibiotics in Helicobacter pylori. Antimicrob. Agents Chemother. 46:2229-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerrits, M. M., M. R. De Zoete, N. L. Arents, E. J. Kuipers, and J. G. Kusters. 2002. 16S rRNA mutation-mediated tetracycline resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 46:2996-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goffin, C., and J. M. Ghuysen. 1998. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev. 62:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodwin, A., D. Kersulyte, G. Sisson, S. J. Veldhuyzen van Zanten, D. E. Berg, and P. S. Hoffman. 1998. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol. Microbiol. 28:383-393. [DOI] [PubMed] [Google Scholar]
- 15.Graham, D. Y. 1998. Antibiotic resistance in Helicobacter pylori: implications for therapy. Gastroenterology 115:1272-1277. [DOI] [PubMed] [Google Scholar]
- 16.Graham, D. Y. 2000. Therapy of Helicobacter pylori: current status and issues. Gastroenterology 118:S2-S8. [DOI] [PubMed] [Google Scholar]
- 17.Grebe, T., and R. Hakenbeck. 1996. Penicillin-binding proteins 2b and 2x of Streptococcus pneumoniae are primary resistance determinants for different classes of beta-lactam antibiotics. Antimicrob. Agents Chemother. 40:829-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haas, R., T. F. Meyer, and J. P. van Putten. 1993. Aflagellated mutants of Helicobacter pylori generated by genetic transformation of naturally competent strains using transposon shuttle mutagenesis. Mol. Microbiol. 8:753-760. [DOI] [PubMed] [Google Scholar]
- 19.Hakenbeck, R. 1999. Beta-lactam-resistant Streptococcus pneumoniae: epidemiology and evolutionary mechanism. Chemotherapy 45:83-94. [DOI] [PubMed] [Google Scholar]
- 20.Harris, A. G., S. L. Hazell, and A. G. Netting. 2000. Use of digoxigenin-labelled ampicillin in the identification of penicillin-binding proteins in Helicobacter pylori. J. Antimicrob. Chemother. 45:591-598. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda, F., Y. Yokota, Y. Mine, and M. Tatsuta. 1990. Activity of cefixime against Helicobacter pylori and affinities for the penicillin-binding proteins. Antimicrob. Agents Chemother. 34:2426-2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacoby, G. A., and G. L. Archer. 1991. New mechanisms of bacterial resistance to antimicrobial agents. N. Engl. J. Med. 324:601-612. [DOI] [PubMed] [Google Scholar]
- 23.Kato, M., Y. Yamaoka, J. J. Kim, R. Reddy, M. Asaka, K. Kashima, M. S. Osato, F. A. El-Zaatari, D. Y. Graham, and D. H. Kwon. 2000. Regional differences in metronidazole resistance and increasing clarithromycin resistance among Helicobacter pylori isolates from Japan. Antimicrob. Agents Chemother. 44:2214-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, J. J., R. Reddy, M. Lee, J. G. Kim, F. A. El-Zaatari, M. S. Osato, D. Y. Graham, and D. H. Kwon. 2001. Analysis of metronidazole, clarithromycin and tetracycline resistance of Helicobacter pylori isolates from Korea. J. Antimicrob. Chemother. 47:459-461. [DOI] [PubMed] [Google Scholar]
- 25.Kwon, D. H., F. A. K. El-Zaatari, J. S. Woo, C. L. Perng, D. Y. Graham, and M. F. Go. 1998. REP-PCR fragments as biomarkers for differentiating gastroduodenal disease-specific Helicobacter pylori strains. Dig. Dis. Sci. 43:980-987. [DOI] [PubMed] [Google Scholar]
- 26.Kwon, D. H., F. A. K. El-Zaatari, M. Kato, M. S. Osato, R. Reddy, Y. Yamaoka, and D. Y. Graham. 2000. Analysis of rdxA and involvement of additional genes encoding NAD(P)H flavin oxidoreductase (FrxA) and ferredoxin-like protein (FdxB) in metronidazole resistance of Helicobacter pylori. Antimicrob. Agents Chemother. 44:2133-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwon, D. H., K. Hulten, M. Kato, J. J. Kim, M. Lee, F. A. K. El-Zaatari, M. S. Osato, and D. Y. Graham. 2001. DNA sequence analysis of rdxA and frxA from 12 pairs of metronidazole-sensitive and -resistant clinical Helicobacter pylori isolates. Antimicrob. Agents Chemother. 45:2609-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livermore, D. M. 1995. β-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Logan, R. P., P. A. Gummett, H. D. Schaufelberger, R. R. Greaves, G. M. Mendelson, M. M. Walker, P. H. Thomas, J. H. Baron, and J. J. Misiewicz. 1994. Eradication of Helicobacter pylori with clarithromycin and omeprazole. Gut 35:323-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Megraud, F. 1998. Antibiotic resistance in Helicobacter pylori infection. Br. Med. Bull. 54:207-216. [DOI] [PubMed] [Google Scholar]
- 31.Mittl, P. R., L. Luthy, P. Hunziker, and M. G. Grutter. 2000. The cysteine-rich protein A from Helicobacter pylori is a beta-lactamase. J. Biol. Chem. 275:17693-17699. [DOI] [PubMed] [Google Scholar]
- 32.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 33.Neuwirth, C., E. Siebor, J. M. Duez, A. Pechinot, and A. Kazmierczak. 1995. Imipenem resistance in clinical isolates of Proteus mirabilis associated with alterations in penicillin-binding proteins. J. Antimicrob. Chemother. 36:335-342. [DOI] [PubMed] [Google Scholar]
- 34.Nikaido, H. 1994. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 264:382-387. [DOI] [PubMed] [Google Scholar]
- 35.Okamoto, T., H. Yoshiyama, T. Nakazawa, I. D. Park, M. W. Chang, H. Yanai, K. Okita, and M. Shirai. 2002. Change in PBP1 is involved in amoxicillin resistance of clinical isolates of Helicobacter pylori. J. Antimicrob. Chemother. 50:849-856. [DOI] [PubMed] [Google Scholar]
- 36.Osato, M. S., R. Reddy, S. G. Reddy, R. L. Penland, and D. Y. Graham. 2001. Comparison of the Etest and the NCCLS-approved agar dilution method to detect metronidazole and clarithromycin resistant Helicobacter pylori. Int. J. Antimicrob. Agents 17:39-44. [DOI] [PubMed] [Google Scholar]
- 37.Osato, M. S., R. Reddy, S. G. Reddy, R. L. Penland, H. M. Malaty, and D. Y. Graham. 2001. Pattern of primary resistance of Helicobacter pylori to metronidazole or clarithromycin in the United States. Arch. Intern. Med. 161:1217-1220. [DOI] [PubMed] [Google Scholar]
- 38.Parr, T. R., Jr., and L. E. Bryan. 1984. Mechanism of resistance of an ampicillin-resistant, beta-lactamase-negative clinical isolate of Haemophilus influenzae type b to β-lactam antibiotics. Antimicrob. Agents Chemother. 25:747-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paul, R., S. Postius, K. Melchers, and K. P. Schafer. 2001. Mutations of the Helicobacter pylori genes rdxA and pbp1 cause resistance against metronidazole and amoxicillin. Antimicrob. Agents Chemother. 45:962-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perreten, V., F. V. Schwarz, M. Teuber, and S. B. Levy. 2001. Mdt(A), a new efflux protein conferring multiple antibiotic resistance in Lactococcus lactis and Escherichia coli. Antimicrob. Agents Chemother. 45:1109-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rowe-Magnus, D. A., A. M. Guerout, and D. Mazel. 2002. Bacterial resistance evolution by recruitment of super-integron gene cassettes. Mol. Microbiol. 43:1657-1669. [DOI] [PubMed] [Google Scholar]
- 43.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenny, L. M. Fitzegerald, N. M. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hays, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 44.Trieber, C. A., and D. E. Taylor. 2002. Mutations in the 16S rRNA genes of Helicobacter pylori mediate resistance to tetracycline. J. Bacteriol. 184:2131-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vester, B., and S. Douthwaite. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob. Agents Chemother. 45:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, G., T. J. M. Wilson, Q. Jiang, and D. E. Taylor. 2001. Spontaneous mutations that confer antibiotic resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 45:727-733. [DOI] [PMC free article] [PubMed] [Google Scholar]