Abstract
The current classification of sporadic parathyroid neoplasia, specifically the distinction of adenoma from multiple gland neoplasia (double adenoma and nonfamilial primary hyperplasia) is problematic and results in a relatively high rate of clinical error. Oligonucleotide microarrays (Affymetrix U133A) were used to evaluate parathyroid samples from 61 patients; 35 adenomas, 10 nonfamilial multiple gland neoplasia, 3 familial primary hyperplasia, 8 renal-induced hyperplasia, and 5 from patients without parathyroid disease (normals). A multiclass comparison using supervised clustering identified distinct gene signatures for each class of parathyroid samples. We developed a predictor model that correctly identified 34 of 35 cases of adenoma, 9 of 10 cases of nonfamilial multiple gland neoplasia, and identified a minimum set of 11 genes for the distinction of adenoma versus multiple gland neoplasia. All methods of unsupervised clustering showed two related but different types of parathyroid adenomas that we have arbitrarily designated as type 1 and type 2 adenomas. Multiple gland parathyroid neoplasia, which represents either synchronous or asynchronous autonomous growth in two, three, or all four parathyroid glands, is a distinct molecular entity and does not represent the molecular pathogenesis of adenoma occurring in multiple glands.
The challenge of cancer pathology has always been to further define meaningful subsets of tumors to maximize treatment strategies. Virtually all classes of tumors, both benign and malignant, have seen dramatic changes in their pathological classification throughout the past 10 to 20 years. One exception to this is parathyroid neoplasia, which still defies to a large extent, the most basic pathological classification of adenoma (single gland neoplasia) versus neoplasia occurring in multiple glands either synchronously or asynchronously (double adenoma and nonfamilial primary hyperplasia). The development of new high-throughput measurements of gene expression, such as gene chips, have provided a powerful tool for functional genomic analysis in the context of tumor classification. This technology has been applied to other classes of tumors and the results have been rewarding in the context of both class discovery and class prediction in regards to pathological classification.1–14
The distinction of parathyroid adenoma (single gland neoplasia) from hyperplasia (multiple gland neoplasia) is extremely problematic and generally unreliable with the current standards of practice in pathology and many pathologists refuse to make this distinction because of the known high rate of clinical error. This distinction is important because the former diagnosis is cured by removal of a single gland while the latter requires the excision of multiple glands for equal results. This error in pathological classification is often discovered only when recurrent hyperparathyroidism occurs in a case originally designated as parathyroid adenoma, for which the initial treatment was less than a three and one-half gland parathyroidectomy. To add another level of complexity to this issue, some patients with multiple gland neoplasia present with an asynchronous pattern of growth in the four parathyroid glands or as double adenomas.
Using gene expression profiling, we have evaluated a large group of parathyroid samples for both class prediction and class discovery in the context of a molecular classification of parathyroid neoplasia. The primary goal of this study was to distinguish single from multiple gland neoplasia and to predict cases of recurrent hyperparathyroidism. To refine this process we sought to define the minimum number of genes that effectively allows for the separation of single from multiple gland neoplasia. In addition, we evaluated cases of sporadic parathyroid neoplasia for class discovery of previously unrecognized subsets of parathyroid neoplasia as defined by the current standards of pathological classification.
Materials and Methods
A more in-depth description of all methods and results are available as Morr_et_al_Supplementary_Info.doc at http://ajp.amjpathol.org. The online details are intended to meet the guidelines put forth by the Microarray Gene Expression Data Society (www.mged.org/miame) for the necessary requirements for publication as published in the Minimum Information about a Microarray Experiment (MIAME) (www.mged.org/Workgroups/MIAME).
Patients
Tissue samples were obtained from 61 patients: 35 sporadic parathyroid adenomas with no evidence of recurrent disease, 5 recurrent hyperparathyroidism initially diagnosed as adenoma and subsequently classified as double adenoma (4 cases) or primary hyperplasia (1 case), 5 nonfamilial primary hyperplasia, 3 familial primary hyperplasia, 8 renal-induced hyperplasia, and 5 from patients without parathyroid disease (normals). The latter were collected as remnant specimens attached to total thyroidectomies for benign disease and from patients with no evidence of hyperparathyroidism. Clinical follow-up ranged from 12 to 24 months with an average of 17.5 months. This study was part of an institutional review board-approved protocol at the Ohio State University College of Medicine.
Pathological Evaluation
The 35 sporadic parathyroid adenomas all showed a single enlarged (≥0.7 cm) hypercellular parathyroid gland with or without a rim of normal parathyroid tissue and a biopsy of at least one other parathyroid gland with findings consistent with normal parathyroid tissue. The five nonfamilial hyperplasias, three familial hyperplasias, and eight renal-induced hyperplasias all showed hypercellular parathyroid tissue involving three or more glands, with the exception of one case of nonfamilial hyperplasia that showed only a modest increase in cellularity in all four glands. For the five cases of recurrent hyperparathyroidism the initial surgery showed a single enlarged hypercellular parathyroid gland without a rim of normal parathyroid tissue and a biopsy of at least one other parathyroid gland with findings consistent with normal parathyroid tissue. A second surgery at the time of recurrent hyperparathyroidism in four of these cases revealed a second enlarged hypercellular gland, that would at least meet the criteria of double adenomas by the traditional standards of classification. For the fifth case, the remaining glands show mild enlargement and hypercellularity beyond the expected limits of normal meeting the minimal criteria for the diagnosis of nonfamilial primary hyperplasia. For all five cases of recurrent hyperparathyroidism the second surgical intervention resulted in a resolution of hyperparathyroidism.
RNA Extraction, Amplification, and Hybridization
Total RNA was extracted by standard methods (Trizol; Life Technologies, Grand Island, NY) and then purified using the Qiagen RNeasy method according to the manufacturer’s directions (Qiagen, Valencia, CA). cRNA was synthesized and labeled according to the Affymetrix protocol (Affymetrix Inc., Santa Clara, CA) for GeneChip experiments. For microarray analysis, each sample was hybridized to U133A GeneChips (Affymetrix Inc.) following the manufacturer’s protocols. For the five cases of recurrent hyperparathyroidism tissue was used from the initial parathyroid surgery.
Data Analysis
Normalization of the absolute expression values for a single sample compared to all samples was done and quality control measurements were performed using the Affymetrix platform (GeneChip Software). The normalized microarray expression data were then evaluated using GeneCluster (version 2.1.6 β, http://www-genome.wi.mit.edu/cancer/), BRB-ArrayTools Version 3.1 (http://linus.nci.nih.gov/BRB-ArrayTools.html), and Spotfire Decision Site for Functional Genomics (Spotfire, Inc., Somerville, MA). Data filtering for supervised analysis of differentially expressed genes was done using GeneCluster, and the following parameters were used for the final analysis applied in sequential order: a maximum normalized signal intensity value of 20,000 units and a minimum value of 20 units, a Max/Min fold change of 3 or 20, and a 1000 units Max-Min absolute variation across the data set. Each data set was then normalized by standardizing each row (gene) to mean = 0 and variance = 1. Unless specified as otherwise, all results are reported using the above parameters with a Max/Min fold change of 20. Data filtering for class prediction was done using BRB-ArrayTools using a univariate t-test at an α of 0.0001.
Supervised Analysis
All parathyroid samples were initially divided into one of six groups: adenoma (35 samples), familial hyperplasia (3 samples), nonfamilial primary hyperplasia (5 samples), recurrent hyperparathyroidism (5 samples), renal-induced hyperplasia (8 samples), and normal parathyroids (5 samples). The five cases of recurrent hyperparathyroidism were compared to the five cases of nonfamilial primary hyperplasia by supervised clustering, and because of a lack of significant difference and the unequivocal clinicopathological evidence of multiple gland involvement, were subsequently grouped together and referred to as nonfamilial multiple gland neoplasia for a comparison to the other groups. Genes were ranked according to their differential expression in a supervised multiclass analysis by using a signal-to-noise feature15 that looks at the means in each class scaled by the sum of the standard deviations. Permutations of each sample (500 each) according to class distinction were then performed to compare these correlations to what would be expected by chance alone.
Prediction of Class Distinction
To assess whether the gene expression signatures for adenoma (single gland neoplasia) and nonfamilial hyperplasia (multiple gland neoplasia) were robust enough by supervised clustering to predict the class label of an unknown sample, we developed a multivariate predictor implementing a compound covariate prediction method as developed by Radmacher and colleagues.16 Because of the absence of an independent data set for validation, we used a leave-one-out cross validation method. The approach involved selecting differentially expressed genes using a univariate t-test on the original data set, and then constructing a multivariate predictor using a linear combination of the selected gene log ratios weighted by the t-statistic obtained from the univariate analysis. Various leave-one-out cross validation algorithms were constructed at different levels of data filtering to test for the minimal number of genes that gave the least number of errors. For each algorithm a cross-validated estimate of prediction error was developed using a univariate t-test at an α of 0.0001 to select for the most highly significant differentially expressed genes in regards to class prediction, but not necessarily biological importance. Each algorithm involved gene selection from the test data set of all of the samples excluding one, followed by construction of the multivariate predictor using the test data set, and then prediction of the tumor class of the sample left out of the training. To avoid overestimating the prediction accuracy we did 2000 random permutations of the class labels to simulate a purely coincidental relationship for each algorithm that allowed for the evaluation of the probability of obtaining a similar or smaller cross-validated error rate.17 The proportion of misclassifications by random permutation testing to the observed rate can then be calculated to estimate any bias in the original classification of the class labels. If the proportion of misclassifications is larger than the observed rate this will result in a large P value, and an indication that the observed occurred with a high probability by chance. Conversely, if the proportion of misclassifications is smaller than the observed rate this will result in a small P value, and an indication that the observed occurred with a low probability by chance. The errors and proportion of misclassifications to the observed rate for each predictor model were then summarized, and compared to produce the predictor model with the lowest leave-one-out cross-validated error rate and total number of genes used for prediction.
Unsupervised Clustering
Unsupervised clustering refers to clustering of the data set without a priori knowledge of the individual clinicopathological patient data and implies that the analyst does not impose any structure to the classification, but rather allows the data to provide the classification. There are many methods of unsupervised clustering, both hierarchical and nonhierarchical, and we chose to compare three different methods including classical agglomerative hierarchical clustering, self-organizing maps (SOMs), and principle component analysis.18–20 Because the clinical distinction of importance is that of single versus nonfamilial multiple gland neoplasia, we restricted our analysis of unsupervised clustering to these groups.
Real-Time Semiquantitative Reverse Transcriptase-Polymerase Chain Reaction (PCR)
Real-time semiquantitative PCR was done on all cases using the SYBR Green I dye chemistry using an ABI Prism 7700 Sequence Detection System (PE Applied Biosystems, Weiterstadt, Germany) per the manufacturer’s protocol for 2 of the discriminator genes identified in the 11 gene predictor model, neurotrimin (NTM) and ectodermal neural cortex (ENC-1). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal amplification control and complete details of the PCR conditions, primers, and analysis are available in the online supplemental files (Morr_et_al_Supplementary_Info at http://ajp.amjpathol.org). The real-time PCR results were evaluated by the delta-delta CT (ΔΔCT) method that allows for the determination of transcription difference for an individual case (Pfaffl MW, Gene Quantification web page. http://www.wzw.tum.de/gene-quantification/.). To determine the transcription difference between all samples in a group wise comparison the Relative Expression Software Tool (REST) was used, to produce a factor of up-regulation (UF) or down-regulation (DF) (Pfaffl MW, Gene Quantification web page. http://www.wzw.tum.de/gene-quantification/.). Where appropriate Fisher’s (two-tailed) unpaired exact test was used, and P values less than 0.05 were considered to indicate statistical significance.
Results
Clinical
There were five cases of recurrent hyperparathyroidism, all of which were previously diagnosed as parathyroid adenoma. These five cases recurred from a time period of 3 to 12 months after an initial one and one-half gland parathyroidectomy. For two of these cases, there was a less than complete correction of the hypercalcemia and elevated parathyroid hormone levels in the immediate postoperative period after the initial surgery, whereas for the other three cases, there was a complete return to normal levels for both of these measurements. Clinically, these five cases were recognized on recurrence as cases for which the initial diagnosis of parathyroid adenoma was incorrect.
Supervised Analysis
Because the five cases of recurrent hyperparathyroidism by the current standards of pathological classification would be classified as multiple gland neoplasia we first sought to determine whether there were any differences between this group and those cases identified as nonfamilial hyperplasia at the initial surgery. As seen in Figure 1a, no distinct gene signature was identified in the comparison of these two groups of samples. Only 11 differentially expressed genes with a P value <0.05 were identified in this comparison. The five cases of recurrent hyperparathyroidism and the five cases of nonfamilial hyperplasia were then grouped and compared to an equal number of adenomas randomly chosen. In this manner there is a distinct gene signature for the adenomas and combined groups of multiple gland neoplasia as shown in Figure 1b. There were 100 differentially expressed genes with a P value <0.05 by this comparison. Likewise (Figure 1, c and d), both groups of multiple gland neoplasia showed a distinct gene signature in comparison to the entire group of adenomas. These results show that any type of multiple gland parathyroid neoplasia, regardless of pathologically recognized involvement of two, three, or all four glands is uniquely different from parathyroid adenoma (single gland neoplasia), and consistent with the clinical interpretation and outcome. Because of these results we then grouped the five cases of recurrent hyperparathyroidism with the five cases of nonfamilial hyperplasia for comparison to all groups by supervised clustering, and refer to these cases collectively as nonfamilial multiple gland neoplasia.
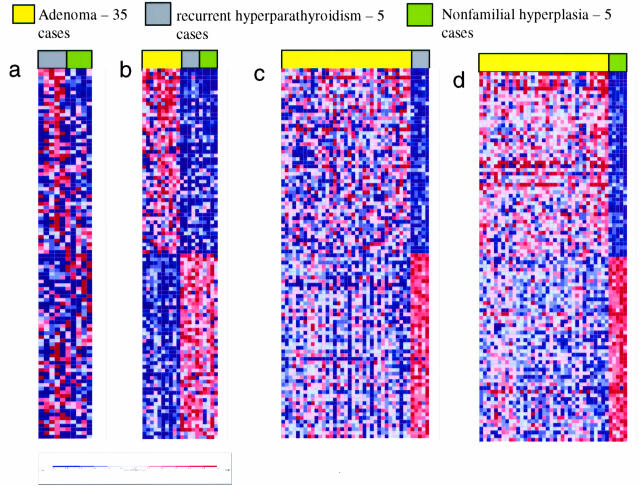
Figure 1.
Supervised analysis. Results of supervised clustering with gene ranking by differential expression using a signal-to-noise feature in a comparison of cases of recurrent hyperparathyroidism versus cases initially diagnosed as nonfamilial primary parathyroid hyperplasia. Individual gene expression values are shown as rows and individual patients as columns. The color of each square in the color matrix represents the mean centered value normalized to zero across all samples with red indicating relative overexpression and blue relative underexpression. The ranking of each gene from top to bottom within a given gene signature reflects the level of confidence for overexpression of this gene in comparison to random permutation testing. The color scale at the bottom indicates relative expression in standard deviations from the mean. Supervised clustering shows the top 50 overexpressed genes for each group of samples. a: The five cases of recurrent hyperparathyroidism, all of which were originally assigned erroneously to the adenoma group, failed to show a distinct gene signature in comparison to the cases initially diagnosed as primary parathyroid hyperplasia. b: The five cases of recurrent hyperparathyroidism are grouped with the five cases of nonfamilial hyperplasia and in comparison to an equal number of adenomas show a distinct gene signature for each group. c: The five cases of recurrent hyperparathyroidism show a distinct gene signature in comparison to the entire group of adenomas. d: In a similar manner the five cases initially diagnosed as primary parathyroid hyperplasia show a distinct gene signature in comparison to all cases of adenoma.
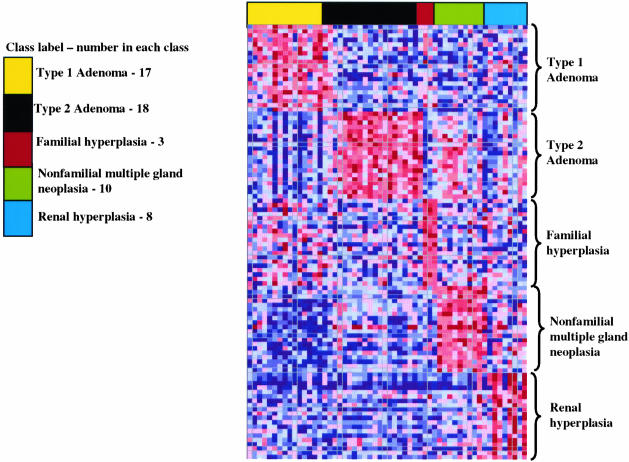
By multiclass supervised clustering with all classes of parathyroid samples (adenoma, nonfamilial multiple gland neoplasia, familial hyperplasia, renal-induced hyperplasia, and normals), there were obvious gene signatures for each class. These results are displayed in Figure 2 for the top 50 overexpressed genes in each class of samples. Gene ranking by random permutation testing showed a large number of statistically significant overexpressed genes for each class (P < 0.05) with the exception of familial hyperplasia, for which the signal-to-noise scores were never greater than the 1% permutation values because of the small number of cases (n = 3). Table 1 summarizes the results for the number of significantly overexpressed genes (P < 0.05) in each class of samples. As shown in Table 1 there were far fewer statistically significant differentially expressed genes for the comparison of the cases of recurrent hyperparathyroidism versus the cases of nonfamilial hyperplasia (11 versus average 296, P < 0.001), in contrast to the remaining comparisons. The complete details of this list can be viewed as Permutation Files at http://ajp.amjpathol.org.
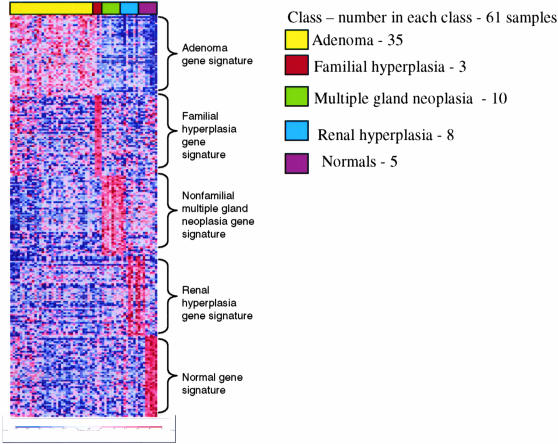
Figure 2.
Supervised analysis. Results of supervised clustering with gene ranking by differential expression using a signal-to-noise feature in a multiclass comparison of all parathyroid classes with the cases of recurrent hyperparathyroidism grouped with the cases of nonfamilial hyperplasia and represented as nonfamilial multiple gland neoplasia. Each class of samples is assigned a set of random colors for easier visualization of the data. The data are presented as the same format as described in Figure 1. Results represent the top 50 overexpressed genes (gene signature) for each class. Note that cases of recurrent hyperparathyroidism grouped with the cases of nonfamilial hyperplasia show a distinct gene signature similar to their individual representation in Figure 1.
Table 1.
Results of Number of Significantly Overexpressed Transcripts (P < 0.05) for Each Class of Parathyroid Samples in a Multi-Class Supervised Comparison
| Comparison | RecHPT versus NFH | Adenoma versus all others | FH versus all others | Nonfamilial MGN versus all others | Renal-induced hyperplasia versus all others | Normals versus all others |
|---|---|---|---|---|---|---|
| No. of transcripts >5% permutation level for each class designation | 11 | 313 | 250* | 362 | 56 | >500 |
RecHPT = five cases of recurrent hyperparathyroidism; NFH = five cases initially diagnosed as primary parathyroid hyperplasia (nonfamilial hyperplasia); nonfamilial MGN = cases of recurrent hyperparathyroidism and nonfamilial hyperplasia grouped together; FH = three cases of MEN1 = related hyperparathyroidism.
In consideration of the fact that normal parathyroids contain a considerable amount of adipose tissue that is generally not present in more than minimal amounts in the other classes of parathyroid samples, the above analysis was done excluding this group of samples. Supervised clustering by the same analysis with the remaining classes of parathyroid samples excluding the normals (adenoma, familial hyperplasia, nonfamilial multiple gland neoplasia, and renal-induced hyperplasia) showed a similar distinct gene signature for each group of samples (see Supplemental_Figure1.ppt at http://ajp.amjpathol.org)and likewise, a large number of significantly overexpressed genes (P < 0.05) for each class.
Class Prediction
The results for multiple leave-one-out cross-validation predictor models for the distinction of single versus multiple gland neoplasia are summarized in Table 2. Figure 3 shows that as few as two genes make relatively few errors in this group of 45 cases (total errors = 5), but the most accurate class predictor model using the minimal number of genes to distinguish single from multiple gland neoplasia used a total of 11 transcripts representing 10 genes (Table 3). With this set of genes (Figure 4), the distinction was correctly made in 9 of 10 cases of nonfamilial multiplegland neoplasia (90%), and in 34 of 35 cases of single glandneoplasia (97%) (P < 0.0001). The one case of single gland neoplasia incorrectly classified using this model was incorrectly predicted with every model constructed. Importantly, as summarized by the different P values of the compound covariate predictor listed in Table 2, permutation analysis for each model constructed was highly significant. Thus, the null hypothesis that there is no difference in single and multiple gland neoplasia can be rejected, and there is no evidence of overfitting of the data. The complete results for each model can be viewed as Class Prediction Files at http://ajp.amjpathol.org. There was no difference in age (P = 0.81), race (P = 0.52), or sex (P = 0.69) between the cases of single versus multiple gland neoplasia.
Table 2.
Summary of Results for Various Predictor Models Using a Compound Covariate Prediction Method with a Leave-One-Out Cross Validation
| No. genes passing filter | No. genes in classifier | Compound covariate predictor, P value | Correct all samples | No. correct single gland neoplasia | No. correct multiple gland neoplasia |
|---|---|---|---|---|---|
| 3629 | 36 | 0.001 | 96% | 34/35 | 9/10 |
| 1912 | 28 | 0.001 | 96% | 34/35 | 9/10 |
| 973 | 22 | <0.0005 | 96% | 34/35 | 9/10 |
| 192 | 11 | <0.0005 | 96% | 34/35 | 9/10 |
| 78 | 7 | <0.0005 | 93% | 33/35 | 9/10 |
| 12 | 2 | <0.0005 | 89% | 31/35 | 9/10 |
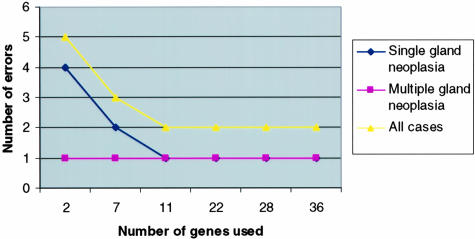
Figure 3.
Class distinction prediction models. The error rate of various predictor models by a leave-one-out cross validation for the class distinction of single versus multiple gland neoplasia is depicted for each group classification as well as the sum of errors for both groups. The predictor models illustrated used from 2 to 36 genes for classification. Shown are the number of errors for the 45 cases of sporadic parathyroid neoplasia (35 single and 10 multiple gland neoplasia). Note that whereas as few as two genes used in the model resulted in as few as five total errors, the best predictor model using 11 genes made only two errors.
Table 3.
Eleven Transcripts Represented in Predictor Model with Highest Degree of Accuracy and Least Number of Genes Used in Model
| Probe set | Description | Gene symbol |
|---|---|---|
| 206018_at | Forkhead box G1B | FOXG1B |
| 218087_s_at | Sorbin and SH3 domain containing 1 | SORBS1 |
| 205478_at | Protein phosphatase 1, regulatory (inhibitor) subunit 1A | PPP1R1A |
| 205795_at | Neurexin 3 | NRXN3 |
| 220794_at | Hypothetical protein FLJ21195 similar to protein related to DAC and cerberus | FLJ21195 |
| 201341_at | Ectodermal-neural cortex (with BTB-like domain) | ENC1 |
| 222020_s_at | Neurotrimin | HNT |
| 206193_s_at | Corneodesmosin | CDSN |
| 206192_at | Corneodesmosin | CDSN |
| 206258_at | Sialyltransferase 8E (alpha-2, 8-polysialyltransferase) | SIAT8E |
| 213808_at | Homo sapiens clone 23688 mRNA sequence |
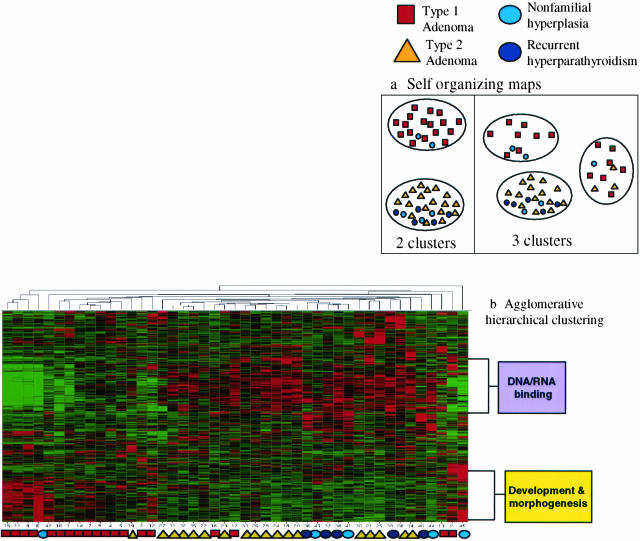
Figure 4.
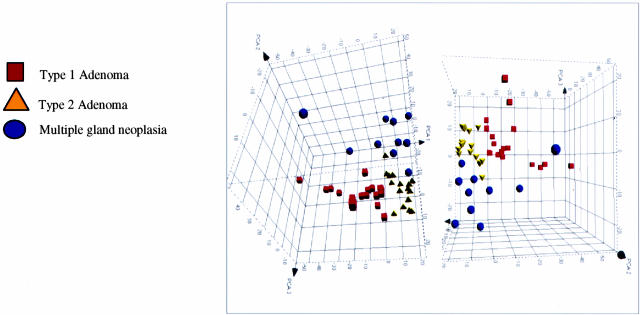
Unsupervised clustering by hierarchical clustering and SOMs. Results of unsupervised analysis by hierarchical clustering and SOMs of the cases of sporadic parathyroid neoplasia. All methods of clustering were done at a 20-fold maximum/minimum variation of gene expression that included slightly more than 2000 genes in the comparison. Results of clustering by SOMs (a) identified two groups of parathyroid adenoma (for complete details see supplemental data Cluster_SOM.xls at http://pathology.osu.edu/parathyroid) that we have designated type 1 and type 2 adenoma. There was no distinct separation of the cases of multiple gland neoplasia from the cases of adenoma when the latter were considered one group. b: Agglomerative hierarchical clustering showed results similar to those by SOMs in regards to the distribution of samples. The dendrogram of samples again shows a division of the adenomas into two groups, with a distinction of type 1 and type 2 adenomas. The cases of nonfamilial multiple gland neoplasia tended to cluster together and with the type 2 adenoma samples. Two gene clusters were identified that showed differential expression in type 1 adenoma versus type 2 adenomas and multiple gland neoplasia.
Unsupervised Clustering
We then clustered all cases of sporadic parathyroid neoplasia (adenomas and nonfamilial multiple gland neoplasia) because this is the clinical distinction of most importance, by SOMs. Regardless of the number of SOMs used for clustering there was a separation of parathyroid adenoma into at least two groups (Figure 4a) (for complete details see Cluster_SOM.xls at http://ajp.amjpathol.org). There was no clear separation of multiple gland neoplasia from the entire group of adenomas. For all clusters the variance within the clusters was less than the minimum between cluster variance (0.125 versus 0.3269 and 0.178 versus 0.291 for two and three clusters, respectively). At the present time, we have arbitrarily designated these two groups of parathyroid adenoma as simply type 1 and type 2 adenoma. There was no difference in age (P = 0.37), race (P = 0.77), or sex (P = 0.85) between the cases of type 1 and type 2 adenoma.
A similar analysis by agglomerative hierarchical clustering (Figure 4b), done with 1736 transcripts, showed results similar to those by SOMs in regards to the distribution of samples. The dendrogram of samples again shows a division of the adenomas into two groups, with a distinction of type 1 and type 2 adenomas. The cases of nonfamilial multiple gland neoplasia tended to cluster together and with the type 2 adenoma samples. From this analysis there were two obvious gene clusters. One cluster consisting of 320 genes is dominated by genes involved in development and morphogenesis and is overexpressed in type 1 adenomas and underexpressed in type 2 adenomas and nonfamilial multiple gland neoplasia. The other cluster consisting of 332 genes is dominated by genes involved in DNA/RNA binding and is underexpressed in the type 1 adenomas and overexpressed in type 2 adenomas and nonfamilial multiple gland neoplasia. Additional details of these gene clusters are available as supplemental data Hier_clus_devmorph.xls and Hier_clus_DRNAbinding.xls at http://ajp.amjpathol.org.
The results of PCA are displayed in Figure 5 and shows three clusters by this analysis that are consistent with distribution of samples by SOMs and agglomerative hierarchical clustering, but with a more clear distinction of the cases of nonfamilial multiple gland neoplasia from the adenoma groups. There is a separation of type 1 and type 2 adenoma and note that much like the results of agglomerative hierarchical clustering, the type 2 adenoma cases are more closely related to the nonfamilial multiple gland neoplasia cases than are the type 1 adenoma cases. The distinction of the cases of nonfamilial multiple gland neoplasia from the adenoma groups is consistent with the results of supervised clustering and confirm that single gland neoplasia (adenoma) has a different molecular profile from nonfamilial multiple gland neoplasia.
Figure 5.
Unsupervised clustering by principle component analysis. Results of unsupervised analysis by principle component analysis (PCA) for all cases of sporadic parathyroid neoplasia. There is a clear distinction of the cases of multiple gland neoplasia from the cases of parathyroid adenoma, with the latter group again showing what appear to be two different groups of samples.
Supervised Clustering of Type 1 and Type 2 Parathyroid Adenomas
Using this information of two types of parathyroid adenomas, we then reclassified the adenoma group of parathyroid samples into type 1 and type 2 and repeated our previous supervised clustering by the nearest neighbor analysis (Figure 6). By this analysis, parathyroid adenoma is comprised of two distinct classes of tumors with distinct gene signatures. In this multiclass comparison of all parathyroid class labels and with two classes of adenomas (type 1 adenoma, type 2 adenoma, nonfamilial multiple gland neoplasia, familial hyperplasia, renal-induced hyperplasia), there were more than 500 genes for both types of adenomas that were strongly and distinctly overexpressed (P < 0.01). Complete details of these results can be reviewed as PERM_AdT1_allclass_500probelist and PERM_AdT2_allclass_500probelist at http://ajp.amjpathol.org.
Figure 6.
Supervised analysis with two adenoma classes. Results of supervised clustering with gene ranking by differential expression using a signal-to-noise feature in a multiclass comparison of all parathyroid class labels excluding normals and with two classes of adenomas (type 1 adenoma, type 2 adenoma, nonfamilial multiple gland neoplasia, familial hyperplasia, renal-induced hyperplasia). Results are shown for the top 20 overexpressed genes (gene signature) for each group. There is an obvious and distinctly separate gene signature for the type 1 and 2 adenomas, as well as for the other classes of parathyroid samples.
Clinically or pathologically, there were no distinguishing features of either group of parathyroid adenomas, but this is not a surprising finding because parathyroid neoplasia in general is an extremely homogenous group of tumors. In the context of molecular classification of parathyroid neoplasia, the identification of two types of parathyroid adenomas was a completely unexpected finding and displays how class discovery will most likely be an important part of any future molecular classification of parathyroid neoplasia.
Real-Time Semiquantitative Reverse Transcriptase-PCR
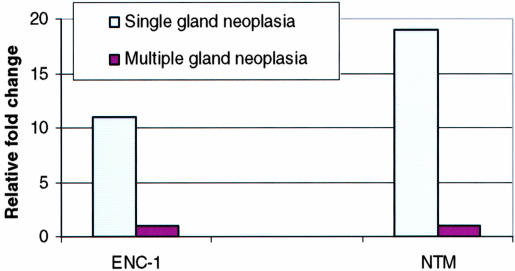
As a group the parathyroid adenomas showed a significant overexpression of ENC-1 and NTM, two of the genes identified in the 11 gene predictor model, in comparison to nonfamilial multiple gland neoplasia (11- and 19-fold, respectively; P < 0.001; Figure 7), or renal-induced hyperplasia and normals (11- and 6-fold, respectively; P < 0.001). Using the ΔΔCT method to compare individual case results there was a significant overexpression in parathyroid adenomas for ENC-1 (P < 0.001) and NTM (P < 0.001) compared to nonfamilial multiple gland neoplasia, or renal-induced hyperplasia and normals. There was no difference in expression of ENC-1 (P = 0.177) or NTM (P = 0.477) for the comparison of recurrent hyperparathyroidism to nonfamilial hyperplasia, confirming that the original classification of these cases as adenomas was incorrect and consistent with the expected clinical outcome for this error.
Figure 7.
Real-time semiquantitative PCR. Fold change for ENC-1 and NTM by real-time PCR. Corroborative evidence by real-time PCR shows that 2 of the genes in the 11 gene predictor model for the distinction of single versus multiple gland neoplasia are significantly overexpressed in the former.
Renal-Induced Parathyroid Hyperplasia
Renal-induced parathyroid hyperplasia showed a recognizable gene signature by supervised clustering (Figure 1a or Figure 6), but in both instances there seemed to be more heterogeneity within the signature compared to the other groups. Likewise, there were far fewer statistically significant overexpressed genes (56 versus average 356, P < 0.001), for this category of parathyroid samples in a multiclass comparison irrespective of whether or not the normal parathyroids were included in the comparison (full details are available as Gene Ranking and Permutation Testing by Supervised Clustering in Morr_et_al_Supplementary_Info.doc at http://ajp.amjpathol.org). It is quite possible, in fact even likely, that the samples classified as renal-induced hyperplasia in this study are a mixture of physiological hyperplastic glands and neoplastic glands.
Biologically Relevant Differentially Expressed Genes
There were many overexpressed genes in each category of parathyroid neoplasia, but the list presented here is that for sporadic parathyroid neoplasia (adenoma and nonfamilial multiple gland neoplasia) because these were the largest groups examined. As shown in Table 4 parathyroid adenoma over- or underexpressed a number of genes involved in DNA repair or cell cycle progression. Most notable among this group was the down-regulation of BRCA1 (OMIM: 113705), BRCA2 (OMIM: 600185), and p95 (NBS1; OMIM: 602667). Sporadic multiple gland parathyroid neoplasia showed a remarkable overexpression of a number of genes related to central nervous central development or disease. The majority of these genes have previously only been described as being expressed in the central nervous system, and remarkably the majority of this list has been previously implicated in some type of an inherited neurodegenerative disorder.
Table 4.
List of Potentially Biological Relevant Genes in Sporadic Parathyroid Neoplasia
| Change | Symbol | Gene name | Summary | |
|---|---|---|---|---|
| Adenoma | ↑ | CHK2 | Checkpoint kinase 2 | DNA repair |
| ↑ | GADD45G | Growth arrest and DNA-damage-inducible, gamma | DNA repair | |
| ↑ | GADD45A | Growth arrest and DNA-damage-inducible, alpha | DNA repair | |
| ↑ | GADD45B | Growth arrest and DNA-damage-inducible, beta | DNA repair | |
| ↑ | CHEK1 | CHK1 checkpoint homolog (S. pombe) | DNA repair; signal transduction resulting in cell cycle arrest | |
| ↑ | NBS1 | Nijmegen breakage syndrome 1 (nibrin) | Cell cycle checkpoint; damaged DNA binding; double-strand break repair | |
| ↓ | CCNA2 | Cyclin A2 | Promotes both cell cycle G1/S and G2/M transitions | |
| ↓ | CDC25C | Cell division cycle 25C | Triggers entry into mitosis | |
| ↓ | CDC14A | CDC14 cell division cycle 14 homolog A (S. cerevisiae) | Involved in the exit of cell mitosis and initiation of DNA replication | |
| ↓ | CDKN1C | Cyclin-dependent kinase inhibitor 1C (p57, Kip2) | Negative regulation of cell proliferation | |
| ↓ | BRCA1 | Breast cancer 1, early onset | DNA repair | |
| ↓ | BRCA2 | Breast cancer 2, early onset | DNA repair | |
| Multiple gland neoplasia | ↑ | NEFH | Neurofilament, heavy polypeptide 200 kd | Abnormalities in neural migration in brain development in Down Syndrome |
| ↑ | NRP1 | Neuropilin 1 | Regulate VEGF-induced angiogenesis | |
| ↑ | PRX | Periaxin | Dejerine-Sottas neuropathy | |
| ↑ | PAFAH1B3 | Platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29 kd | Mental retardation | |
| ↑ | GLB1 | Galactosidase, beta 1 | GM1-gangliosidosis | |
| ↑ | IDUA | Iduronidase, alpha-L | Mucopolysaccharidosis Type I | |
| ↑ | HEXB | Hexosaminidase B (beta polypeptide) | Sandhoff disease (GM2-gangliosidosis type II) | |
| ↑ | GAA | Glucosidase, alpha; acid | Pompe’s disease | |
| ↑ | MANBA | Mannosidase, beta A, lysosomal | Beta-mannosidosis | |
| ↑ | NAGLU | N-acetylglucosaminidase, alpha | Sanfilippo syndrome, type B | |
| ↑ | GUSB | Glucuronidase, beta | Mucopolysaccharidosis type VII | |
| ↑ | MAN2B1 | Mannosidase, alpha, class 2B, member 1 | Mannosidosis, alpha Types I and II |
Discussion
Undoubtedly the most important aspect of this study is the molecular classification of parathyroid adenoma (single gland neoplasia) and hyperplasia (multiple gland neoplasia) by their distinctly different gene expression signatures. The issue of whether parathyroid hyperplasia is actually the same process as parathyroid adenoma but occurring in multiple glands, either synchronously or asynchronously, has never been resolved.21–28
By using supervised analysis this study gives strong support to the concept that parathyroid adenoma and hyperplasia are distinct entities. By unsupervised clustering using principle component analysis soft-organizing maps, and hierarchical clustering, similar results were shown. The results by agglomerative hierarchical clustering were not as straightforward, but not totally unexpected. Agglomerative hierarchical clustering is very sensitive at identifying variation in individual comparisons, but less so at the highest levels of partitioning because the broadest branches are totally dependent on the lower links. As a result such methods of clustering are often not in agreement with biologically complex specimens such as tissue samples in comparison to studies in yeast or cell lines.29,30 Although we have shown that neoplasia in two or more glands is distinctly different from neoplasia in one gland, our number of cases of hyperplasia are most likely too small to definitively prove that multiple gland neoplasia is a single entity. Nonetheless, the clinical distinction of importance is the distinction of parathyroid neoplasia involving a single gland versus involvement of multiple glands.
It is possible that our near perfect classification of sporadic parathyroid neoplasia by the leave-one-out method of cross-validation is overly optimistic because of the lack of an independent validation set and the criteria set for an acceptable length of follow-up in regards to recurrent hyperparathyroidism. The lack of an independent validation set is not unique to this study and is generally related to economic feasibility and scarcity of tissue resources in many studies using high-throughput measurements of gene expression. As such, this is a preliminary model that will require validation by an independent set of samples. As our predictor model used a supervised analysis it is suspect to the correct classification of the samples. The compound covariate prediction method we used with a leave-one-out cross validation is a rigorous test of robustness and it was reassuring to see that for every predictor model tested there was no evidence of bias in the original classification of the samples. Previous studies have shown that without random permutation of the class labels many predictor models tend to overestimate the prediction accuracy and fail in an independent validation set.18 All of the predictor models we developed showed the number of misclassifications by random permutation testing were much smaller than the observed rate, indicating that our assignment of the class labels occurred with a low probability by chance alone.
At the present time the only true gold standard of parathyroid adenoma is clinical follow-up to exclude multiple gland neoplasia, but there are no well-defined criteria for an acceptable length of follow-up. Currently, the standard of care to rule out multiple gland neoplasia is clinical follow-up at 6 months.31,32 Our study had a minimal follow-up of 12 months (average, 17.5 months) which certainly exceeds the standard of care. Although this does not totally exclude the presence of multiple gland neoplasia among our group of 35 adenomas,33 the fact that close to 90% of these cases would be adenoma by chance alone, coupled with a clinically acceptable period of follow-up, makes this possibility extremely unlikely.
Although not feasible for economic reasons that gene expression profiling will be used to make the distinction of single gland neoplasia versus multiple gland neoplasia, it is highly probable in the near future a biomarker(s) identified by such studies will be used to accurately make this distinction in a preoperative setting. Most importantly this biomarker(s) will need to use serum protein(s) so this distinction can be made before surgery. The use of such a biomarker would have immediate impact in clinical scenarios in which preoperative localization studies are less than definitive for a single neoplastic gland. Such a biomarker could also correctly identify patients with a need for limited to no follow-up postoperatively. Because of the relatively high incidence of parathyroid neoplasia, particularly in the elderly female population in which it is estimated to be between 1 to 2%,34,35 a highly accurate molecular classification of parathyroid neoplasia would improve overall patient care as well as prove to be highly cost-effective for the health care system. Although postoperative blood calcium and parathyroid hormone levels would have warranted close follow-up of two of the patients with recurrent hyperparathyroidism in this study, the other three would have been missed. In addition, a single elevated parathyroid hormone level after parathyroidectomy is an unreliable indicator of recurrent hyperparathyroidism in the period of time discussed here.36
The list of genes presented in Table 4 as putative biologically relevant genes involved in sporadic parathyroid neoplasia may shed new light on the underlying pathogenesis of these tumors. It is not surprising that parathyroid adenomas showed under- or overexpression of a number of genes involved in DNA repair and cell cycle progression. These tumors, despite their usual lack of malignancy, show a preponderance of chromosomal changes by comparative genomic hybridization or loss of heterozygosity analysis.37–40 Many of the changes seen in these genes, including CHK2, GADD45, CHEK1, CCNA2, and CDC25 are consistent with regulatory processes opposed to cell cycle progression in the presence of DNA damage and as such fail to explain the underlying pathogenesis of neoplasia. What is intriguing is the down-regulation of BRCA1, BRCA2, and p95, which together form part of a large multisubunit protein complex of tumor suppressors, DNA damage sensors, and signal transducers41 that has been aptly named BASC, for “BRCA1-associated genome surveillance complex.” Dysregulation of this complex at least potentially may explain the high rate of chromosomal damage previously noted in adenomas.
In contrast, multiple gland parathyroid neoplasia showed a surprising finding of overexpression of a number of genes related to central nervous central development or disease. Most of these overexpressed genes encode large proteins that undergo extensive posttranslational processing and are involved in lysosomal activity or expressed on the cell surface. The members of this group of overexpressed genes are indeed impressive in regards to their importance for neuropathology, but few, if any, have a link to neoplasia. Although it is difficult to explain the pathogenesis of this group of overexpressed genes in regards to parathyroid neoplasia, their expression is not totally unexplainable as they are neuroendocrine tumors. In this context, overexpression of genes generally thought of as having an expression restricted to the central nervous system may reflect a more primitive state of differentiation in multiple gland neoplasia. The relative lack of expression of these genes in parathyroid adenoma adds support to our findings that multiple gland neoplasia is a distinct entity and not merely the occurrence of multiple adenomas in a single patient.
Although our results were quite distinct for all classes of parathyroid neoplasia, the results for renal-induced parathyroid hyperplasia in regards to gene ranking and permutation testing were not as statistically significant in a comparative sense. The reasons for this most likely relate to the fact that renal-induced parathyroid hyperplasia is a very heterogeneous disease both clinically and pathologically, with a number of cases representing true hyperplasia and others neoplasia.
To summarize, gene expression profiling of sporadic parathyroid neoplasia has the potential to change the pathological classification of parathyroid neoplasia. It is even possible that serum markers for the distinction of adenoma versus hyperplasia, and the similar detection of autonomous functioning parathyroid glands in renal-induced hyperplasia, will be available before parathyroidectomy in the near future. In addition, recent increased awareness of the potential long-term, accelerated irreversible detrimental cardiovascular effects of uncorrected hyperparathyroidism42–44 will most likely bring identification and correct classification of parathyroid neoplasia to the forefront of medicine in the near future.
Acknowledgments
We thank Dr. Richard Simon and Amy Peng for developing the BRB ArrayTools used during the performance of analyses.
Footnotes
Address reprint requests to Carl Morrison, M.D., Department of Anatomic Pathology, The Ohio State University, 310 West 10th Ave., M-417 Starling Loving Hall, Columbus, OH 43210. E-mail: morrison-4@medctr.osu.edu.
Supported by the corresponding author’s (C.M.) developmental funds of the Department of Pathology, Ohio State University Medical Center.
Supplemental information can be found at http://ajp.amjpathol.org.
References
- Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang CH, Angelo M, Ladd C, Reich M, Latulippe E, Mesirov JP, Poggio T, Gerald W, Loda M, Lander ES, Golub TR. Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci USA. 2001;98:15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, Behm FG, Raimondi SC, Relling MV, Patel A, Cheng C, Campana D, Wilkins D, Zhou X, Li J, Liu H, Pui CH, Evans WE, Naeve C, Wong L, Downing JR. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;2:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, Tamayo P, Renshaw AA, D’Amico AV, Richie JP, Lander ES, Loda M, Kantoff PW, Golub TR, Sellers WR. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaiya AA, Franzen B, Hagman A, Dysvik B, Roblick UJ, Becker S, Moberger B, Auer G, Linder S. Molecular classification of borderline ovarian tumors using hierarchical cluster analysis of protein expression profiles. Int J Cancer. 2002;98:895–899. doi: 10.1002/ijc.10288. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, Weber G, Mark EJ, Lander ES, Wong W, Johnson BE, Golub TR, Sugarbaker DJ, Meyerson M. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen TO, West RB, Linn SC, Alter O, Knowling MA, O’Connell JX, Zhu S, Fero M, Sherlock G, Pollack JR, Brown PO, Botstein D, van de Rijn M. Molecular characterisation of soft tissue tumours: a gene expression study. Lancet. 2002;359:1301–1307. doi: 10.1016/S0140-6736(02)08270-3. [DOI] [PubMed] [Google Scholar]
- Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RC, Gaasenbeek M, Angelo M, Reich M, Pinkus GS, Ray TS, Koval MA, Last KW, Norton A, Lister TA, Mesirov J, Neuberg DS, Lander ES, Aster JC, Golub TR. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Giordano TJ, Thomas DG, Kuick R, Lizyness M, Misek DE, Smith AL, Sanders D, Aljundi RT, Gauger PG, Thompson NW, Taylor JM, Hanash SM. Distinct transcriptional profiles of adrenocortical tumors uncovered by DNA microarray analysis. Am J Pathol. 2003;162:521–531. doi: 10.1016/S0002-9440(10)63846-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DR, Kardia SL, Shedden KA, Kuick R, Michailidis G, Taylor JM, Misek DE, Wu R, Zhai Y, Darrah DM, Reed H, Ellenson LH, Giordano TJ, Fearon ER, Hanash SM, Cho KR. Gene expression in ovarian cancer reflects both morphology and biological behavior, distinguishing clear cell from other poor-prognosis ovarian carcinomas. Cancer Res. 2002;62:4722–4729. [PubMed] [Google Scholar]
- Barden CB, Shister KW, Zhu B, Guiter G, Greenblatt DY, Zeiger MA, Fahey TJ., III Classification of follicular thyroid tumors by molecular signature: results of gene profiling. Clin Cancer Res. 2003;9:1792–1800. [PubMed] [Google Scholar]
- Mischel PS, Shai R, Shi T, Horvath S, Lu KV, Choe G, Seligson D, Kremen TJ, Palotie A, Liau LM, Cloughesy TF, Nelson SF. Identification of molecular subtypes of glioblastoma by gene expression profiling. Oncogene. 2003;22:2361–2373. doi: 10.1038/sj.onc.1206344. [DOI] [PubMed] [Google Scholar]
- El-Naggar AK, Kim HW, Clayman GL, Coombes MM, Le B, Lai S, Zhan F, Luna MA, Hong WK, Lee JJ. Differential expression profiling of head and neck squamous carcinoma: significance in their phenotypic and biological classification. Oncogene. 2002;21:8206–8219. doi: 10.1038/sj.onc.1206021. [DOI] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Radmacher MD, McShane LM, Simon R. A paradigm for class prediction using gene expression profiles. J Comput Biol. 2002;9:505–511. doi: 10.1089/106652702760138592. [DOI] [PubMed] [Google Scholar]
- Simon R, Radmacher MD, Dobbin K, McShane LM. Pitfalls in the use of DNA microarray data for diagnostic and prognostic classification. J Natl Cancer Inst. 2003;95:14–18. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- Knudsen, S. Knudsen S, editor. New York: John Wiley & Sons, Inc.,; A Biologist’s Guide to Analysis of DNA Microarray Data. 2002:pp 41–59. [Google Scholar]
- Parmigiani G, Garrett ES, Irizarry RA, Zeger SL. Parmigiani G, Garrett ES, Irizarry RA, Zeger SL, editors. New York: Springer,; The Analysis of Gene Expression DataMethods and Software. 2003:pp 1–36. [Google Scholar]
- Hastie T, Tibshirani R, Friedman J. Hastie T, Tibshirani R, Friedman J, editors. New York: Springer,; The Elements of Statistical LearningData Mining, Inference, and Prediction. 2001:pp 1–39, 437–500. [Google Scholar]
- Albright F, Bloomberg E, Castleman B, Churchill ED. Hyperparathyroidism due to diffuse hyperplasia of all parathyroid glands rather than adenoma of one: clinical studies on three such cases. Arch Intern Med. 1934;54:315–329. [Google Scholar]
- Castleman B, Mallory TB. The pathology of the parathyroid gland in hyperparathyroidism. A study of 25 cases. Am J Pathol. 1935;11:1–71. [PMC free article] [PubMed] [Google Scholar]
- Larian B, Alavi S, Roesler J, Namazie A, Blackwell K, Calcaterra TC, Wang MB. The role of hyperplasia in multiple parathyroid adenomas. Head Neck. 2001;23:134–139. doi: 10.1002/1097-0347(200102)23:2<134::aid-hed1008>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Baloch ZW, LiVolsi VA. Double adenoma of the parathyroid gland: does the entity exist? Arch Pathol Lab Med. 2001;125:178–179. doi: 10.5858/2001-125-0178-DAOTPG. [DOI] [PubMed] [Google Scholar]
- Attie JN, Bock G, Auguste LJ. Multiple parathyroid adenomas: report of thirty-three cases. Surgery. 1990;108:1014–1019. [PubMed] [Google Scholar]
- Verdonk CA, Edis AJ. Parathyroid “double adenomas”: fact of fiction? Surgery. 1981;90:523–526. [PubMed] [Google Scholar]
- Tezelman S, Shen W, Siperstein AE, Duh QY, Clark OH. Persistent or recurrent hyperparathyroidism in patients with double adenomas. Surgery. 1995;118:1115–1122. doi: 10.1016/s0039-6060(05)80122-9. [DOI] [PubMed] [Google Scholar]
- Tezelman S, Shen W, Shaver JK, Siperstein AE, Duh QY, Klein H, Clark OH. Double parathyroid adenomas. Clinical and biochemical characteristics before and after parathyroidectomy. Ann Surg. 1993;218:300–307. doi: 10.1097/00000658-199309000-00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somorjai RL, Dolenko B, Baumgartner R. Class prediction and discovery using gene microarray and proteomics mass spectroscopy data: curses, caveats, cautions. Bioinformatics. 2003;19:1484–1491. doi: 10.1093/bioinformatics/btg182. [DOI] [PubMed] [Google Scholar]
- Bloom G, Yang IV, Boulware D, Kwong KY, Coppola D, Eschrich S, Quackenbush J, Yeatman TJ. Multi-platform, multi-site, microarray-based human tumor classification. Am J Pathol. 2004;164:9–16. doi: 10.1016/S0002-9440(10)63090-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa JA, Powe NR, Levine MA, Udelsman R, Zeiger MA. Profile of a clinical practice: thresholds for surgery and surgical outcomes for patients with primary hyperparathyroidism: a national survey of endocrine surgeons. J Clin Endocrinol Metab. 1998;83:2658–2665. doi: 10.1210/jcem.83.8.5006. [DOI] [PubMed] [Google Scholar]
- Sackett WR, Barraclough B, Reeve TS, Delbridge LW. Worldwide trends in the surgical treatment of primary hyperparathyroidism in the era of minimally invasive parathyroidectomy. Arch Surg. 2002;137:1055–1059. doi: 10.1001/archsurg.137.9.1055. [DOI] [PubMed] [Google Scholar]
- Rudberg C, Akerstrom G, Palmer M, Ljunghall S, Adami HO, Johansson H, Grimelius L, Thoren L, Bergstrom R. Late results of operation for primary hyperparathyroidism in 441 patients. Surgery. 1986;99:643–651. [PubMed] [Google Scholar]
- Heath DA. Primary hyperparathyroidism. Clinical presentation and factors influencing clinical management. Endocrinol Metab Clin North Am. 1989;18:631–646. [PubMed] [Google Scholar]
- Palmer M, Jakobsson S, Akerstrom G, Ljunghall S. Prevalence of hypercalcaemia in a health survey: a 14-year follow-up study of serum calcium values. Eur J Clin Invest. 1988;18:39–46. doi: 10.1111/j.1365-2362.1988.tb01163.x. [DOI] [PubMed] [Google Scholar]
- Denizot A, Pucini M, Chagnaud C, Botti G, Henry JF. Normocalcemia with elevated parathyroid hormone levels after surgical treatment of primary hyperparathyroidism. Am J Surg. 2001;182:15–19. doi: 10.1016/s0002-9610(01)00664-x. [DOI] [PubMed] [Google Scholar]
- Agarwal SK, Schrock E, Kester MB, Burns AL, Heffess CS, Ried T, Marx SJ. Comparative genomic hybridization analysis of human parathyroid tumors. Cancer Genet Cytogenet. 1998;106:30–36. doi: 10.1016/s0165-4608(98)00049-1. [DOI] [PubMed] [Google Scholar]
- Correa P, Juhlin C, Rastad J, Akerstrom G, Westin G, Carling T. Allelic loss in clinically and screening-detected primary hyperparathyroidism. Clin Endocrinol (Oxf) 2002;56:113–117. doi: 10.1046/j.0300-0664.2001.01436.x. [DOI] [PubMed] [Google Scholar]
- Garcia JL, Tardio JC, Gutierrez NC, Gonzalez MB, Polo JR, Hernandez JM, Menarguez J. Chromosomal imbalances identified by comparative genomic hybridization in sporadic parathyroid adenomas. Eur J Endocrinol. 2002;146:209–213. doi: 10.1530/eje.0.1460209. [DOI] [PubMed] [Google Scholar]
- Imanishi Y, Tahara H, Palanisamy N, Spitalny S, Salusky IB, Goodman W, Brandi ML, Drueke TB, Sarfati E, Urena P, Chaganti RS, Arnold A. Clonal chromosomal defects in the molecular pathogenesis of refractory hyperparathyroidism of uremia. J Am Soc Nephrol. 2002;13:1490–1498. doi: 10.1097/01.asn.0000018148.50109.c0. [DOI] [PubMed] [Google Scholar]
- Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 2000;14:927–939. [PMC free article] [PubMed] [Google Scholar]
- Vestergaard P, Mollerup CL, Frokjaer VG, Christiansen P, Blichert-Toft M, Mosekilde L. Cardiovascular events before and after surgery for primary hyperparathyroidism. World J Surg. 2003;27:216–222. doi: 10.1007/s00268-002-6541-z. [DOI] [PubMed] [Google Scholar]
- Qunibi WY, Nolan CA, Ayus JC. Cardiovascular calcification in patients with end-stage renal disease: a century-old phenomenon. Kidney Int Suppl. 2002;82:S73–S80. doi: 10.1046/j.1523-1755.62.s82.15.x. [DOI] [PubMed] [Google Scholar]
- Goodman WG, Goldin J, Kuizon BD, Yoon C, Gales B, Sider D, Wang Y, Chung J, Emerick A, Greaser L, Elashoff RM, Salusky IB. Coronary-artery calcification in young adults with end-stage renal disease who are undergoing dialysis. N Engl J Med. 2000;342:1478–1483. doi: 10.1056/NEJM200005183422003. [DOI] [PubMed] [Google Scholar]