Abstract
Two Escherichia coli isolates were recovered from the blood of two cancer patients and were demonstrated to produce high levels of the AmpC β-lactamase with isoelectric points of >9.0. The hypertranscription of ampC RNA was observed by Northern blot hybridization in both isolates. One isolate (isolate EC44) had a point mutation (G→A at position −28) and insertion of thymidine between positions −20 and −19 of the ampC promoter gene (GenBank accession no. AE000487). The single nucleotide insertion of T between positions −19 and −20 created an optimal distance (17 bp) in the Pribnow box for ampC hyperproduction. The other isolate (isolate EC38) had two point mutations (G→A at position −28 and C→T at position +58) and a 2-base (GT) insertion between positions −14 and −15. Although the insertion of GT between positions −14 and −15 may create a new promoter next to the original promoter, cloning of the ampC region with truncated nucleotides of the original −35 region of EC38 failed to verify the hypothesis that a new promoter would be created by such a nucleotide insertion. Instead, multiple start sites for ampC transcription at −1, +1, +2, and +3 were observed in an S1 nuclease protection assay. These results suggest that the RNA polymerase is flexible in the selection of a start site in ampC hypertranscription. In conclusion, nucleotide insertions between the −35 and −10 ampC promoter sequences was the mechanism for the hyperproduction of AmpC β-lactamase and resistance to oxyimino-cephalosporins. The failure of the two patients to respond to treatment with oxyimino-cephalosporins highlights the important role of such a resistance mechanism in the clinical setting.
The AmpC cephalosporinase gene in Escherichia coli and Shigella spp. is normally located on the chromosome and is weakly expressed because of a weak promoter and a weak transcriptional attenuator (11, 22). Wild-type strains produce a basal level of this enzyme which does not result in ampicillin and cephalosporin resistance. This enzyme can be hyperproduced due to genetic variation. Such hyperproduction of AmpC β-lactamase contributes to resistance to ampicillin and extended-spectrum cephalosporins (15). Although AmpC β-lactamases are responsible for the phenotypic expression of MICs, similar to extended-spectrum β-lactamases (ESBLs) (21), they are poorly inhibited by β-lactam-β-lactamase inhibitor combinations (21). Unlike other members of the family Enterobacteriaceae, ampC is not inducible since there is no ampR regulatory gene in E. coli (2). Genetically, promoter changes (5, 21-23), insertion of IS2 (13), an optimized distance (1, 12) in the Pribnow box (−35 and −10), and the presence of more than one copy of ampC (6, 7) have been described as crucial factors for ampC hyperproduction. Only one clinical isolate that hyperproduces AmpC due to nucleotide insertions (IS2) has been reported (13). Optimal distance (17 bp) in the Pribnow box has been reported to be the resistance mechanism only in an in vitro experiment (1), but hyperproduction due to the change of this distance has not been reported in the clinical setting. The study described here was performed to investigate two clinical isolates that hyperproduce AmpC β-lactamase due to two different nucleotide insertions in the Pribnow box.
MATERIALS AND METHODS
Bacterial strains.
The E. coli strains used in this study and the sources of the strains are listed in Table 1.
TABLE 1.
Strains used in this study
| Strain | Relevant properties | Source or reference |
|---|---|---|
| EC38 | Patients isolate with hyperproduction of AmpC | This study |
| EC44 | Patient isolate with hyperprodution of AmpC | This study |
| ATCC 25922 | Ampicillin-susceptible A control strain | ATCCa |
| JP995 | Rifampin-resistant E. coli recipient for conjugation | 26 |
| T-EC38 | Transconjugant of EC38 | This study |
| EC-IS1 | Transformant containing cloned vector (PCR-ScriptCamSK+) with full-length ampC sequence with −10 and −35 promoter sequences from ATCC 25922 | This study |
| EC-IS2 | Transformant containing cloned vector (PCR-ScriptCamSK+) with full-length ampC sequence with the promoter sequence from EC38 | This study |
| EC-IS3 | Transformant containing cloned vector (PCR-ScriptCamSK+) with full-length ampC sequence with truncation of the −35 promoter sequence from EC38 at position −26 | This study |
| XL10 | Epicurian Coli XL10-Gol Kan ultracompetent cells | Stratagene |
ATCC, American Type Culture Collection, Manassas, Va.
Susceptibility testing.
Antimicrobial susceptibility was determined by both broth microdilution and disk diffusion tests according to NCCLS guidelines (18, 19). The following antimicrobial agents were obtained as standard reference powders of known potency for susceptibility testing by the broth microdilution method: ampicillin (Sigma Chemical Co., St. Louis, Mo.), cefoxitin and cefazolin (Fujisawa, Osaka, Japan), cefixime (Merck Sharp & Dohme, Taipei, Taiwan), amoxicillin and clavulanic acid (SmithKline Beecham, Brockham Park, United Kingdom), cefotaxime (Hoechst Marion Roussel, Frankfurt, Germany), ceftazidime (Glaxo Group Research Limited, Greenford, United Kingdom), and amikacin and aztreonam (Bristol-Myers Squibb Laboratories, Princeton, N.Y.). All drugs were incorporated into Mueller-Hinton broth (BBL Microbiology Systems, Cockeysville, Md.) at serial twofold concentrations ranging from 0.025 to 64 μg/ml. Two control strains, E. coli ATCC 35218 and ATCC 25922, were included in each test run. The inoculated plates were incubated at 35°C for 16 to 18 h. The MIC of each antimicrobial agent was defined as the lowest concentration that inhibited visible growth of the organism (10).
Testing for ESBLs.
The double-disk synergy test, the Etest for ESBLs, and agar diffusion tests were used to screen the strains for ESBL production. In the double-disk synergy test, cefotaxime (30 μg), ceftazidime (30 μg), and aztreonam (30 μg) disks were placed on Mueller-Hinton agar (BBL Microbiology Systems) adjacent to an amoxicillin-clavulanic acid disk (20 μg of amoxicillin plus 10 μg of clavulanate). All disks were purchased from Becton Dickinson Microbiology Systems (Sparks, Md.). The procedures and interpretation of the results of the double-disk synergy test were as described previously (10).
The Etest (PDM Epsilometer; AB Biodisk, Solna, Sweden) used to screen for ESBL production, based on the recognition of a reduction of the ceftazidime or cefotaxime MIC in the presence of clavulanic acid, was performed according to the instructions of the manufacturer. The agar diffusion test was performed according to NCCLS guidelines. A ≥5-mm increase in the zone diameter for either ceftazidime-clavulanic acid (30/10 μg) or cefotaxime-clavulanic acid (30/10 μg) compared with the zone diameter when each antibiotic was tested alone was indicative of ESBL production (20).
Genomic fingerprinting by PFGE.
Total DNA was prepared, and pulsed-field gel electrophoresis (PFGE) was performed as described previously (25). The restriction enzyme XbaI (New England Biolabs, Beverly, Mass.) was used at the manufacturer's suggested temperature of 37°C. Restriction fragments were separated by PFGE in a 1% agarose gel (Bio-Rad, Hercules, Calif.) in 0.5× TBE buffer (45 mM Tris, 45 mM boric acid, 1.0 mM EDTA [pH 8.0]) by using the Bio-Rad CHEF-DRII apparatus (Bio-Rad Laboratories, Richmond, Calif.). The initial pulse time of 1 s was increased linearly to 35 s, and electrophoresis was performed for 20 h at 200 V and 4°C. The gels were then stained with ethidium bromide and photographed under UV light.
The band patterns were compared visually and were classified as indistinguishable (clonal), closely related (clonal variants, three band differences or less), possibly related (four to six band differences), and unrelated, according to previously described criteria (27).
Plasmid profile, transfer of resistance by conjugation, and IEF.
Plasmid profile analysis was performed by the alkaline extraction method (14). Conjugation was carried out by broth mating. A rifampin-resistant E. coli strain (strain JP995) was used as the recipient. The recipient and donor strains were separately inoculated into brain heart infusion broth and incubated at 37°C for 4 h. The recipient and donor strains were then mixed at a ratio of 1:1 (by volume) and incubated overnight at 37°C. The next morning, 0.01 ml of the mixture was spread onto a MacConkey agar plate containing rifampin (100 μg/ml) and ampicillin (50 μg/ml).
Isoelectric focusing (IEF) was performed as described previously (17). The bacteria were harvested from 20-h brain heart infusion broth cultures by centrifugation, and the pellet was resuspended in 1 ml of phosphate buffer (0.05 M; pH 7). Enzymes were released by two cycles of freezing (−70°C) and thawing (room temperature), followed by sonication for 5 min in a sonicator in ice-cold water. IEF was performed in an ampholine gel (pH 3.0 to 10.0; Pharmacia, Uppsala, Sweden). Preparations from standard strains known to harbor TEM-1, SHV-1, and SHV-5 were used as standards.
PCR for blaTEM and AmpC promoter region.
The following oligonucleotide primer sequences (Gibco BRL) were used for the PCR assays: 5′-ATAAAATTCTTGAAGACGAAA-3′ (primer A), 5′-GACAGTTACCAATGCTTAATCA-3′ (primer B), 5′-CTACGGTCTGGCTGCTA-3′ (primer C), and 5′-TGGAGCAAGAGGCGGTA-3′ (primer D). Primers A and B were specific for blaTEM (16). Primers C and D corresponded to nucleotide positions −61 to −45 and +92 to +108, respectively, of the structural ampC gene (21). The reactions were performed in 50-μl mixtures containing 2.5 U of Taq polymerase (Promega, Madison, Wis.); 1× buffer consisting of 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, and 50 mM KCl; 0.01 μg of gelatin; 200 μM (each) deoxynucleoside triphosphate; and 2 μM (each) oligonucleotide primer. Thirty-five cycles with the following temperature profile were performed for each reaction: 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min.
Sequencing was done by the method of Sanger et al. (24) with the corresponding primers specific for the blaTEM genes. An automated sequencer (ABI Prism 377; Perkin-Elmer, Norwalk, Conn.) was used.
AmpC expression assay.
Northern hybridization was performed as follows: 10 μg of total RNA was mixed with the same volume of RNA Sample Loading Buffer (Sigma), and the mixture was heated at 65°C for 15 min and chilled on ice before being run on a 1% agarose gel containing 2.2 M formaldehyde. The RNA sample was transferred onto a Nytran nitrocellulose membrane (Schleicher & Schuell, Inc., Keene, N.H.) according to the instructions in the manufacturer's laboratory manual. The membrane was automatically UV cross-linked with a UV cross-linker (Stratagene, La Jolla, Calif.) and prehybridized with 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.5% sodium dodecyl sulfate-5× Denhardt's solution (0.2% bovine serum albumin [Sigma], 0.2% Ficoll 400 [Sigma], 0.2% polyvinylpyrrolidone [Sigma]) containing 100 μg of denatured sheared salmon sperm DNA (Schleicher & Schuell, Inc.) at 42°C for 1 h before the addition of a denatured 32P-labeled probe. The probe was an ampC gene fragment generated by PCR with primers 5′-CTACGGTCTGGCTGCTA-3′ and 5′-CTAAGTGTAGATGACAGCAAGG-3′ designed from a published sequence (GenBank accession no. AE000487). This probe was labeled with [α-32P]ATP by using Ready-To-Go DNA labeling beads (Amersham-Pharmacia Biotech Inc., Little Chalfont, United Kingdom) and was purified according to the instructions of the manufacturer. After hybridization at 42°C overnight, the membrane was washed twice with 2× SSC-0.1% sodium dodecyl sulfate at room temperature for 5 min each time before being exposed to X-ray film.
Primer extension assay.
Primer 5′-AGCGTCGTTTTGAACATAGGGTC-3′, which corresponds to the E. coli ampC promoter sequence (bp 10418 to 10440 of the sequence with GenBank accession no. AE000487), was synthesized (Taiwan Genome Sciences, Inc., Taipei, Taiwan) and used to extend the ampC RNA transcripts. A total of 10 pmol of the primer was labeled with [γ- 32P]ATP by using the Ready-To-Go T4 polynucleotide kinase kit (Amersham-Pharmacia Biotech Inc.) for 1 h at room temperature and purified with a MicroSpin G-25 column (Amersham-Pharmacia Biotech Inc.) according to the instructions of the manufacturer. A total of 10 μg of RNA and 0.6 pmol of labeled primer were heated in 1× avian myeloblastosis virus reverse transcriptase buffer (Promega) at 72°C for 15 min, cooled to room temperature for 15 min, and then chilled on ice. The extension reaction was performed at 42°C for 1 h after the addition of sodium pyrophosphate (final concentration, 4 mM; Merck & Co., Inc., Rahway, N.J.), deoxynucleoside triphosphates (1 mM; Amersham-Pharmacia Biotech Inc.), and 30 U of avian myeloblastosis virus reverse transcriptase (Promega). The marker was generated by PCR with the same primer used for extension in the sequencing reaction (Sequenase [version 2.0] DNA sequencing kit; United States Biochemical Corp., Cleveland, Ohio) of a complete ampC gene (bp 10180 to 5316 of the sequence with GenBank accession no. AE000487) cloned into the pGEMT-EASY vector (Promega).
S1 nuclease protection assay.
The probe used for the S1 nuclease protection assay was a 76-mer oligonucleotide containing the promoter region of E. coli ampC (bp 10432 to 10512 of the sequence with GenBank accession no. AE000487) which was synthesized (MWG-Biotech Inc., Ebersberg, Germany), labeled with [γ-32P]ATP by using the Ready-To-Go T4 polynucleotide kinase kit (Amersham-Pharmacia Biotech Inc.) for 1 h at room temperature, and purified with a MicroSpin G-25 column (Amersham-Pharmacia Biotech Inc.) according to the instructions of the manufacturer. Nuclease protection assays were performed with the Multi-NPA RNA/DNA/Oligo Probe Protection Assay kit (Ambion) as suggested by the manufacturer. The marker was generated by PCR with a synthetic primer (Taiwan Genome Science, Inc., Taipei, Taiwan) whose sequence was identical to the last 22 bases at the 3′ end of the probe used in the sequencing reaction (Sequenase[version 2.0] DNA sequencing kit; United States Biochemical Corp.) of the complete ampC gene (bp 10180 to 9258 of the sequence with GenBank accession no. AE000487) and with the pGEMT-EASY vector (Promega).
Cloning of ampC resistance genes with different ISs.
The following DNA fragments were used: (i) insertion sequence (IS) IS1, a full-length ampC sequence with the original −10 and −35 promoter sequences from ATCC 25922; (ii) IS2, a full-length ampC sequence into which nucleotides G and T were inserted between positions −19 and −20 inside the promoter sequence from EC38; and (iii) IS3, a full-length ampC sequence into which nucleotides G and T were inserted between positions −19 and −20 but in which the −35 promoter sequence from EC38 was truncated at position −26. The fragments were cloned into a vector (PCR-ScriptCamSK+) according to the suggestions provided with the PCR-ScriptCam cloning kit (Stratagene). The insertion was located inside the site digested with the restriction enzyme SrfI. Cloned DNA fragments were reconfirmed by DNA sequencing. The primers used to amplify IS1, IS2, and IS3 were as follows: forward primer for IS1 and IS2, primer FP-1 (5′-AATGGGTTTTCTACGGTCTG-3′); forward primer for IS3, primer FP-3 (5′-TGATTGGTGTCGTTACAATCT-3′); reverse primers for IS1, IS2, and IS3, primer RP-2 (5′-TTACTGTAGAGCGTTGAGAAT-3′). IS1 was amplified from ATCC 25922. IS2 and IS3 were amplified from EC38.
RESULTS
Patient characteristics and bacterial strains.
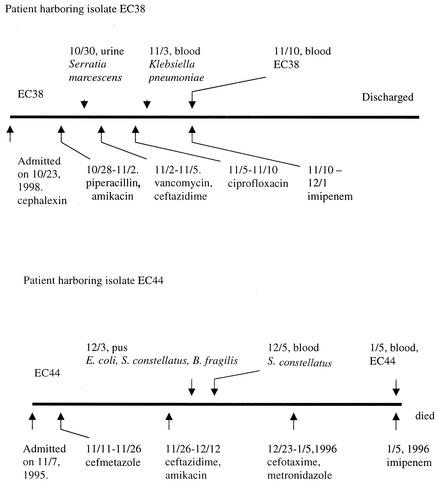
Two E. coli isolates (isolates EC38 and EC44) with antibiotic resistance profiles suggestive of the hyperproduction of the AmpC β-lactamase were isolated from blood specimens of patients in a medical center in Taiwan. EC38 was cultured from an acute myeloid leukemia patient on day 18 of hospitalization, when the patient was in a neutropenic state after chemotherapy. Four days prior to the isolation of EC38, the patient developed cefotaxime- and aztreonam- resistant but amoxicillin-clavulanate- and ciprofloxacin-susceptible Klebsiella pneumoniae bacteremia. Cefazolin, piperacillin, and amikacin had been given to the patient before these multiply resistant species of the family Enterobacteriaceae were isolated. Febrile neutropenia did not respond to empirical treatment with vancomycin and ceftazidime. Intravenous ciprofloxacin was then prescribed for the treatment of bacteremia. The patient finally recovered from the infection after subsequent use of imipenem (Fig. 1). EC44 was recovered from blood specimens of a 42-year-old male gastric cancer patient on day 59 of hospitalization. The patient had started chemotherapy and had undergone tumor resection 1 year prior to isolation of the organism. A drainage wound remained in his abdomen because of a recent intra-abdominal infection that was shown to be due to a mixed infection with Streptococcus constellatus, Bacteroides fragilis, and E. coli 1 month before the E. coli EC44 bacteremia developed. Empirical treatment with cefmetazole was given for the abdominal wound infection, and then ceftazidime plus amikacin and, finally, cefotaxime and metronidazole were prescribed according to the results of culture and susceptibility testing. An episode of septic shock occurred on day 13 of therapy with cefotaxime and metronidazole, despite the patient's improved initial condition with this combination therapy. The patient died of septic shock within 12 h of onset, and the duration of empirical imipenem use was less than 1 day (Fig. 1).
FIG. 1.
Flowcharts of antimicrobial therapies for the patients harboring isolates EC38 and EC44.
Susceptibility testing, plasmid profiling, and PFGE.
Isolates EC38 and EC44 were resistant to ampicillin, and the MICs of extended-spectrum cephalosporins (cefotaxime, ceftazidime, and ceftriaxone) were higher for these two strains than for ATCC 25922 (Table 2). The MICs of extended-spectrum cephalosporins met the criteria for further confirmatory tests for ESBL production. Different patterns of susceptibility to the other antibiotics tested were found for EC38 and EC44 (Table 2). One high-molecular-weight plasmid was identified in isolate EC38, but no plasmid was found in isolate EC44 (data not shown). These two isolates were different clones, according to the results of molecular typing by PFGE (data not shown).
TABLE 2.
In vitro susceptibilities of two clinical E. coli isolates and the E. coli ATCC 25922 control strain determined by the broth microdilution methoda
| Antibioticb | MIC (μg/ml)
|
||
|---|---|---|---|
| ATCC 25922 | EC38 | EC44 | |
| AMP | 2 | ≥64 | ≥64 |
| AMX + CLA | ≤0.25 | 16 | 16 |
| CFZ | ≤0.25 | 16 | 16 |
| FOX | ≤0.25 | ≥64 | 8 |
| CFX | ≤0.25 | ≥64 | 16 |
| ATM | ≤0.25 | 8 | 1 |
| CTR | ≤0.25 | ≤0.5 | ≤0.5 |
| CTR + CLA | ≤0.25 | ≤0.25 | ≤0.25 |
| CAZ | ≤0.25 | 4 | 4 |
| CAZ + CLA | ≤0.25 | 2 | 2 |
| CTX | ≤0.25 | 2 | 1 |
| CTX + CLA | ≤0.25 | 1 | 0.25 |
| IPM | ≤0.25 | <0.25 | <0.25 |
| AMK | ≤0.25 | 4 | 2 |
| CIP | 0.06 | 2 | 0.125 |
| TET | ≤1 | ≥64 | 2 |
Strain EC38 had two major bands with pIs of 5.4 and >9, and strain EC44 had one major band with a pI of >9.
AMP, ampicillin; ATM, aztreonam; CTR, ceftriaxone; CAZ, ceftazidime; CTX, cefotaxime; FOX, cefoxitin; AMX, amoxicillin; CFZ, cefazolin; CFX, cefixime; AMK, amikacin; IPM, imipenem; CIP, ciprofloxacin; TET, tetracycline; CLA, clavulanic acid at a fixed concentration 4 μg/ml when tested with ceftazidime, cefotaxime, and ceftriaxone and at a ratio of 2:1 when tested with amoxicillin.
IEF, confirmatory testing for ESBL production, and PCR for blaTEM.
IEF of extracts of donor isolates (isolates EC38 and EC44) obtained by sonication followed by nitrocefin screening revealed two major bands at pI 5.4 and pI >9 for EC38 and one major band at pI >9 for EC44. For the transconjugant of EC38 (T-EC38), only one major band at pI 5.4 was detected. PCR for blaTEM and sequencing results showed that the isolate with a pI 5.4 β-lactamase was positive for blaTEM and had a sequence identical to that of the TEM-1 β-lactamase (8). Both isolates were negative for ESBL production by the Etest, the agar diffusion test, and the double-disk synergy test. When clavulanic acid at a fixed concentration of 4 μg/ml was combined with ceftazidime, ceftriaxone, or cefotaxime individually for susceptibility testing, no more than a fourfold reduction in the MICs was observed (Table 2). The results obtained in these experiments confirmed that the isolates were not ESBL producers and had profiles indicating that they were AmpC hyperproducers.
RNA expression of ampC hyperproducer.
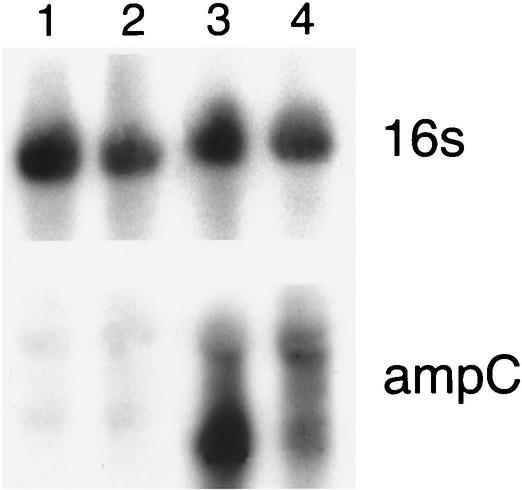
Total RNA extraction followed by hybridization with probes specific for ampC and 16S rRNA revealed a strong signal for hybridization with ampC for strains EC38 and EC44 but no hybridization signal for control strains ATCC 25922 and JP995. On the other hand, a strong signal for hybridization with the 16S rRNA-specific probe was observed for all strains, indicating that the quantities of total RNA in all strains were similar and were effectively transferred to the membrane (Fig. 2). In a comparison of EC38 and EC44 to ATCC 25922 by using the 16S RNA hybridization signal as an internal control, the estimated levels of β-lactamase transcription for EC38 and EC44 were 6 and 10 times higher, respectively, than that for ATCC 25922.
FIG. 2.
RNA expression assessed by hybridization with 16S rRNA- and ampC-specific probes. Lane 1, ATCC 25922; lane 2, JP995; lane 3, EC38; lane 4, EC44.
Sequence homology of ampC promoter.
Figure 3 shows the nucleotide sequence of the ampC promoter resulting from PCR with strains ATCC 25922, JP995, EC38, and EC44. The sequences of ATCC 25922 and JP995 were the same as the wild-type E. coli K-12 promoter sequence. For EC38, mutations at positions −28 and +58 and insertion of two bases (G and T) between positions −15 and −14 were identified. The insertion of GT suggested the possible creation of a new promoter: the GT insertion between positions −14 and −15 created a new nucleotide pattern similar to those of the −35 region (TTTACA) and the −10 region (TGTAAA), in which five nucleotides were identical to the optimal sequence of the −35 region (TTGACA) and four bases were identical to the optimal sequence of the −10 region (TATAAT) in prokaryotes (Pribnow box). In addition to the possible newly created −35 and −10 regions in EC38, a 17-bp distance between the −35 and −10 regions, which is the optimal distance for ampC expression, was observed (Fig. 3).
FIG. 3.
Sequence alignment of ampC promoter for E. coli K-12, ATCC 259922, EC38, and EC40. The documented and suspected new −35 and −10 promoter sequences are shown in boldface, and the attenuator is indicated by the arrows. There is a dot above the sequence every 20 nucleotides. The initiation codon is underlined.
Primer extension assay.
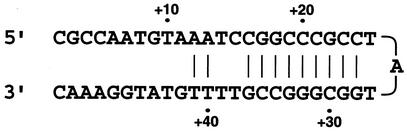
To demonstrate that the insertion of GT between positions −14 and −15 in isolate EC38 was the major factor responsible for the hyperproduction of AmpC, we used a primer extension assay to identify whether the double transcription sites were present due to an additional promoter and to estimate the quantities of the transcription products. The assay revealed that one major band (at about position +24) with a low molecular weight was present in wild-type strain ATCC 25922 and EC38. In addition, an extra weak band (position +1) was also identified in EC38 (Fig. 4). By using a DNA sequence alignment program (DNASTAR Inc., Madison, Wis.), a hairpin structure was identified downstream of the −35 and −10 regions (Fig. 5). This attenuator formed a hairpin structure during primer annealing, resulting in a major band with a very short sequence.
FIG. 4.
Primer extension assay for ATCC 25922 (lane 1) and EC38 (lane 2).
FIG. 5.
Hairpin structure identified and generated with a DNA sequence alignment program (DNASTAR Inc.).
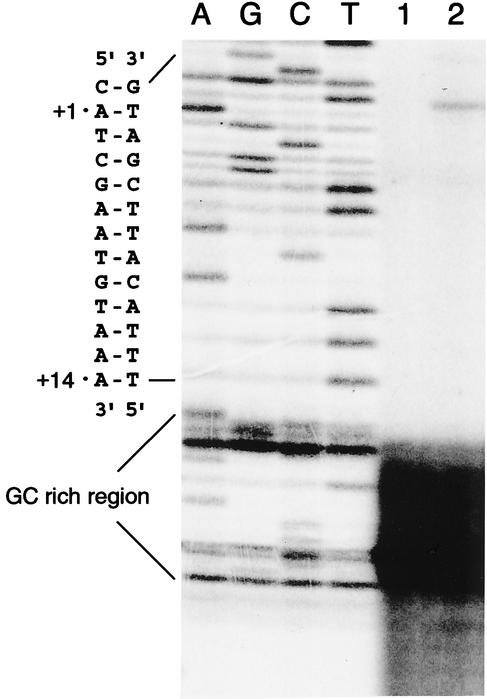
S1 nuclease protection assay.
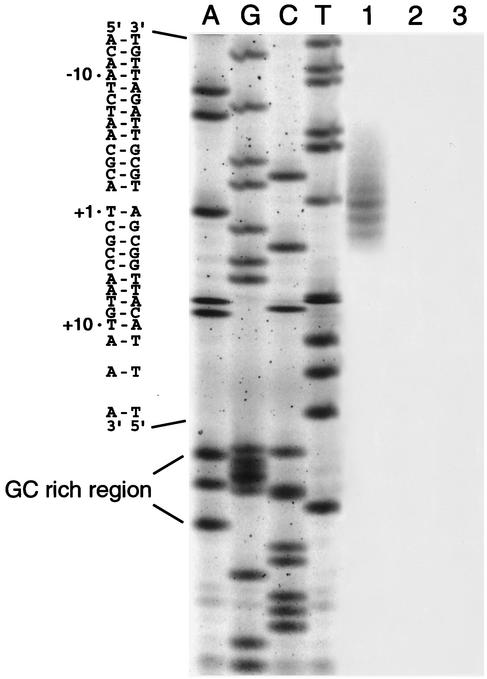
Since primer extension was blocked by the hairpin structure, the S1 nuclease protection assay was used to avoid the resulting difficulty in the detection of the transcription start site. In the S1 nuclease protection assay, no transcription signal was detected in ATCC 25922 or JP995. In contrast, a number of bands were detected in EC38. The major signals were located at positions −1, +1, +2, and +3. The results obtained by the nuclease protection assay revealed that the insertion of GT between positions −14 and −15 increased the level of expression of mRNA transcription (Fig. 6).
FIG. 6.
Nuclease protection assay. The lanes marked A, G, C, and T contain the ampC sequence. The position of the RNA start site was determined according to the molecular weight of the nuclease-protected fragment. Lane 1, EC38; lane 2, ATCC 25922; lane 3, JP995.
Cloning of ampC resistance genes with different ISs and estimation of β-lactamase activities.
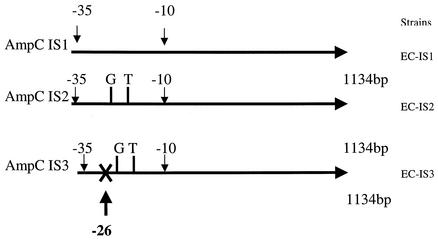
Three different DNA fragments that included IS1, IS2, and IS3 (Fig. 7) were cloned, and the clones were tested for their susceptibilities to various antibiotics. The strain resulting from cloning of the IS1 fragment of the full-length ampC sequence with the original −10 and −35 promoter sequences was resistant to ampicillin, amoxicillin-clavulanate, cefoxitin, cefixime, and cefazolin but was susceptible to aztreonam, ceftriaxone, ceftazidime, cefotaxime, and imipenem. When ceftriaxone, ceftazidime, and cefotaxime were combined with clavulanic acid at a fixed concentration of 4 μg/ml, no significant reductions in the MICs of ceftriaxone, ceftazidime, and cefotaxime were observed (Table 3). On the contrary, for the cloned strain with the IS2 fragment, the full-length ampC sequence into which GT was inserted between positions −19 and −20 inside the promoter sequence was resistant to ampicillin, amoxicillin-clavulanate, cefoxitin, cefixime, cefazolin, aztreonam, ceftazidime, and cefotaxime but was susceptible to ceftriaxone at 8 μg/ml and imipenem. Compared to the β-lactam MICs for the strain with the IS1 fragment, the MICs of many β-lactams for the cloned strain with the IS2 fragment were significantly increased. No significant reduction in the MIC was observed when ceftazidime, cefotaxime, or ceftriaxone was tested in combination with clavulanic acid at a fixed concentration of 4 μg/ml. The cloned strain with the IS3 fragment and the full-length ampC sequence in which GT was inserted between positions −19 and −20 but in which the −35 promoter sequence was truncated was susceptible to all antibiotics (Table 3).
FIG. 7.
Cloning of three different lengths of ampC insertion sequences into PCR-ScriptCamSK+ vector: EC-IS1, full-length ampC sequence with the original −10 and −35 promoter sequences; EC-IS2, full-length ampC sequence with insertion of nucleotides G and T between positions −19 and −20 inside the promoter sequence; and EC-IS3, full-length ampC sequence with insertion of nucleotides G and T between positions −19 and −20 but with truncation at position −26 of the −35 promoter sequence.
TABLE 3.
In vitro susceptibilities of the three E. coli clones EC-IS1, EC-IS2, and EC-IS3
| Antibiotica | MIC (μg/ml)
|
||
|---|---|---|---|
| EC-IS1 | EC-IS2 | EC-IS3 | |
| AMP | ≥32 | ≥32 | 8 |
| AMX + CLA | ≥32 | ≥32 | 0.25 |
| CFZ | ≥32 | ≥32 | 8 |
| FOX | ≥32 | ≥32 | 8 |
| CFX | 32 | ≥64 | 0.5 |
| ATM | 1 | ≥64 | <0.25 |
| CTR | 0.5 | 8 | 1 |
| CTR + CLA | 0.5 | 4 | 1 |
| CAZ | 2 | ≥32 | <0.25 |
| CAZ + CLA | 1 | ≥32 | <0.25 |
| CTX | 1 | ≥64 | <0.25 |
| CTX + CLA | 0.25 | 32 | <0.25 |
| IPM | 0.5 | 1 | 0.5 |
AMP, ampicillin; ATM, aztreonam; CAZ, ceftazidime; CTX, cefotaxime; CTR, ceftriaxone; FOX, cefoxitin; AMX, amoxicillin; CFZ, cefazolin; CFX, cefixime; AMK, amikacin; IPM, imipenem; CLA, clavulanic acid at a fixed concentration 4 μg/ml when tested with ceftriaxone, ceftazidime, and cefotaxime and at ratio of 2:1 when tested with amoxicillin.
DISCUSSION
In this study, the cephalosporin MICs for two clinical isolates, isolates EC44 and EC38, were not reduced in the presence of clavulanic acid, suggesting that the strains produced an AmpC β-lactamase. Subsequent study of ampC mRNA transcription revealed that ampC mRNA was highly expressed in isolates EC44 and EC38 compared to the levels of expression in isolates ATCC 25922 and JP995 (ampicillin-susceptible strains). These findings indicate that the elevated MICs of extended-spectrum cephalosporins were caused by the hyperproduction of the AmpC β-lactamase.
Previous studies showed that hyperproduction of β-lactamase can be achieved by several different mechanisms. The presence of mutations in the promoter region is the mechanism most frequently reported to be responsible for the hyperproduction of β-lactamases (5, 9, 12, 21, 22, 26). There have been a few reports of nucleotide insertion as a mechanism of new promoter creation (13, 23). Previous studies have documented high-level expression of ampC by identifying an optimal sequence of bases (17 bp) between the −35 and −10 regions (1, 9, 12). In this study, a mutation at position −28 and one base (T) insertion between positions −20 and −19 were observed in EC44, which is an optimal distance between the −35 and −10 regions for ampC expression.
For EC38, it was initially suspected that the insertion of two nucleotides between positions −14 and −15 would create a new promoter, since the optimal distance is not obtained after insertion of the nucleotides. A search for the double transcription start sites of ampC by the primer extension assay revealed two fragments with reverse-transcribed DNA signals. However, these bands might not have resulted from the double promoter start sites. The major signal at about +16 to +37 bp (the area in the hairpin structure with a high G+C content) was probably not the transcription start site but was more likely due to the stem-loop formation which stopped the primer extension (Fig. 5). As observed in EC38, an isolate with hypertranscription of ampC RNA, although the hairpin structure was self-forming during the annealing process, a limited number of RNA sequences with a non-stem-loop format still allowed primer extension and caused a weak signal at position +1 (the reverse transcription of DNA passed through the region of the hairpin) (Fig. 4). In this study, the double radioactive signals of primer extension occurred only with hypertranscription of ampC RNA. A recent study showed that the nucleotide at position +24, which was localized in the hairpin structure, and that the attenuator were involved in low-level transcription of the ampC gene (5). A mutation at this position may play an important role in ampC transcription. The mutation at position +24 may destabilize the hairpin formation and cause an increase in the level of ampC transcription (5). The results of our primer extension study suggest that the nucleotides at about positions +16 to +37 may be important for ampC transcription. However, additional experimental evidence is needed.
Further experiments to identify the transcription start sites by the S1 nuclease protection assay revealed the presence of multiple start sites at positions −1, +1, 2, and 3, indicating that the RNA polymerase is more flexible in the selection of start sites in high-level AmpC producers than in low-level AmpC producers, as has been discussed in a previous report (1). In addition, the results obtained for three different clones with promoter sequences showed that the insertion of GT between positions −19 and −20 definitely contributed to the hypertranscription of ampC and also showed that no other promoter was created downstream of the original −35 box after the GT insertion. The hyperproduction of ampC RNA was simply due to the increased distance of from 16 to 18 nucleotides between the −35 and −10 promoter sequences.
In contrast to many previous molecular studies with clinical isolates that hyperproduce chromosomal AmpC (3-5, 11, 23), the clinical responses of these AmpC hyperproducers have not been previously reported. The failure of treatment with oxyimino-cephalosporins in the two patients described here highlights the important clinical roles that such resistance mechanisms play in E. coli. In conclusion, this study reported on two AmpC-hyperproducing E. coli clinical isolates. The hyperproduction of AmpC β-lactamase in clinical isolates was demonstrated to be caused by the creation of an optimal distance for gene expression by the insertion of either one nucleotide or two nucleotides, which resulted in the increased hypertranscription of ampC RNA compared to the level of transcription in the wild-type strain.
Acknowledgments
This work was supported by a grant from the National Health Research Institutes of Taiwan.
REFERENCES
- 1.Aoyama, T., M. Takanami, E. Ohtsuka, Y. Taniyama, R. Marumoto, H. Sato, and M. Ikehara. 1983. Essential structure of E. coli promoter: effect of spacer length between the two consensus sequences on promoter function. Nucleic Acids Res. 11:5855-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett, P. M., and I. Chopra. 1993. Molecular basis of beta-lactamase induction in bacteria. Antimicrob. Agents Chemother. 37:153-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergstrom, S., and S. Normark. 1979. Beta-lactam resistance in clinical isolates of Escherichia coli caused by elevated production of the ampC-mediated chromosomal beta-lactamase. Antimicrob. Agents Chemother. 16:427-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caroff, N., E. Espaze, I. Berard, H. Richet, and A. Reynaud. 1999. Mutations in the ampC promoter of Escherichia coli isolates resistant to oxyiminocephalosporins without extended spectrum beta-lactamase production. FEMS Microbiol. Lett. 173:459-465. [DOI] [PubMed] [Google Scholar]
- 5.Caroff, N., E. Espaze, D. Gautreau, H. Richet, and A. Reynaud. 2000. Analysis of the effects of −42 and −32 ampC promoter mutations in clinical isolates of Escherichia coli hyperproducing ampC. J. Antimicrob. Chemother. 45:783-788. [DOI] [PubMed] [Google Scholar]
- 6.Edlund, T., T. Grundstrom, and S. Normark. 1979. Isolation and characterization of DNA repetitions carrying the chromosomal beta-lactamase gene of Escherichia coli K-12. Mol. Gen. Genet. 173:115-125. [DOI] [PubMed] [Google Scholar]
- 7.Edlund, T., and S. Normark. 1981. Recombination between short DNA homologies causes tandem duplication. Nature 292:269-271. [DOI] [PubMed] [Google Scholar]
- 8.Goussard, S., and P. Courvalin. 1991. Sequence of the genes blaT-1B and blaT-2. Gene 102:71-73. [DOI] [PubMed] [Google Scholar]
- 9.Hawley, D. K., and W. R. McClure. 1983. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 11:2237-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarlier, V., M. H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 11.Jaurin, B., T. Grundstrom, T. Edlund, and S. Normark. 1981. The E. coli beta-lactamase attenuator mediates growth rate-dependent regulation. Nature 290:221-225. [DOI] [PubMed] [Google Scholar]
- 12.Jaurin, B., T. Grundstrom, and S. Normark. 1982. Sequence elements determining ampC promoter strength in E. coli. EMBO J. 1:875-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaurin, B., and S. Normark. 1983. Insertion of IS2 creates a novel ampC promoter in Escherichia coli. Cell 32:809-816. [DOI] [PubMed] [Google Scholar]
- 14.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livermore, D. M. 1995. β-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mabilat, C., and S. Goussard. 1993. PCR detection and identification of genes for extended-spectrum β-lactamases, p. 553-563. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, D.C.
- 17.Matthew, M., and A. M. Harris. 1976. Identification of beta-lactamases by analytical isoelectric focusing: correlation with bacterial taxonomy. J. Gen. Microbiol. 94:55-67. [DOI] [PubMed] [Google Scholar]
- 18.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 19.National Committee for Clinical Laboratory Standards. 1999. Performance standards for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 20.National Committee for Clinical Laboratory Standards. 1999. Performance standards for antimicrobial susceptibility testing. Ninth informational supplement. M2-A6 and M7-A4, p. 19:36 (M100-S9). National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 21.Nelson, E. C., and B. G. Elisha. 1999. Molecular basis of AmpC hyperproduction in clinical isolates of Escherichia coli. Antimicrob. Agents Chemother. 43:957-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olsson, O., S. Bergstrom, F. P. Lindberg, and S. Normark. 1983. ampC beta-lactamase hyperproduction in Escherichia coli: natural ampicillin resistance generated by horizontal chromosomal DNA transfer from Shigella. Proc. Natl. Acad. Sci. USA 80:7556-7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olsson, O., S. Bergstrom, and S. Normark. 1982. Identification of a novel ampC beta-lactamase promoter in a clinical isolate of Escherichia coli. EMBO J. 1:1411-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schoonmaker, D., T. Heimberger, and G. Birkhead. 1992. Comparison of ribotyping and restriction enzyme analysis using pulsed-field gel electrophoresis for distinguishing Legionella pneumophila isolates obtained during a nosocomial outbreak. J. Clin. Microbiol. 30:1491-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siu, L. K., P. L. Ho, K. Y. Yuen, S. S. Wong, and P. Y. Chau. 1997. Transferable hyperproduction of TEM-1 beta-lactamase in Shigella flexneri due to a point mutation in the pribnow box. Antimicrob. Agents Chemother. 41:468-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]