Abstract
Ovarian cancer remains the fifth leading cause of cancer death for women in the United States. In this study, the gene expression of 20 ovarian carcinomas, 17 ovarian carcinomas metastatic to the omentum, and 50 normal ovaries was determined by Gene Logic Inc. using Affymetrix GeneChip HU_95 arrays containing ∼12,000 known genes. Differences in gene expression were quantified as fold changes in gene expression in ovarian carcinomas compared to normal ovaries and ovarian carcinoma metastases. Genes up-regulated in ovarian carcinoma tissue samples compared to more than 300 other normal and diseased tissue samples were identified. Seven genes were selected for further screening by immunohistochemistry to determine the presence and localization of the proteins. These seven genes were: the β8 integrin subunit, bone morphogenetic protein-7, claudin-4, collagen type IX α2, cellular retinoic acid binding protein-1, forkhead box J1, and S100 calcium-binding protein A1. Statistical analyses showed that the β8 integrin subunit, claudin-4, and S100A1 provided the best distinction between ovarian carcinoma and normal ovary tissues, and may serve as the best candidate tumor markers among the seven genes studied. These results suggest that further exploration into other up-regulated genes may identify novel diagnostic, therapeutic, and/or prognostic biomarkers in ovarian carcinoma.
Ovarian cancer is the leading cause of gynecological malignancy in North American women. Each year in the United States, ∼24,000 new cases of ovarian cancer are diagnosed and 14,000 deaths are attributed to this disease.1 Contributing to the poor prognosis is the lack of symptoms in the early stages of the disease.2,3 More than 75% of diagnoses are made in stage III and IV, after distant metastasis has occurred. The 5-year survival rate for women diagnosed with late-stage disease is 25%, compared to more than 90% for women diagnosed with stage I of the disease.1
In recent years, large-scale gene expression analyses have been performed to identify differentially expressed genes in ovarian carcinoma.4–22 A common goal of these studies was to identify potential tumor markers for the diagnosis of early-stage ovarian cancer, as well as to use these markers as targets for improved therapy and treatment of the disease during all stages. These earlier studies compared the gene expression profiles of tissues or cell lines derived from ovarian cancer samples, normal ovaries, other normal samples, and other types of tumors.4–22 A major problem in identifying genes up-regulated in ovarian carcinoma is that normal ovary epithelial cells are very difficult to obtain in large enough numbers to perform gene microarray experiments. Although some groups have analyzed gene expression of the cells that are on the surface of normal ovaries, it is still controversial whether these cells truly serve as the normal counterpart for ovarian epithelial tumors.23 The cumulative results of these gene expression studies reveal more than 150 potentially up-regulated genes that are associated with ovarian cancer. However, only a small portion of the genes reported as up-regulated in ovarian carcinoma were further validated by a second technique such as immunohistochemical analysis or reverse transcriptase-polymerase chain reaction. A number of the genes that show promise as biomarkers based on their secondary validation include: ApoJ, claudin-3, claudin-4, COL3A1, HE4, CD24, LU, progesterone-binding protein, mucin 1, ryudocan, E16, osteoblast-specific factor-2, prostatin, and secretory protein P1.B.4–12,24 Finally, proteomics and two-dimensional electrophoresis protein analysis are also being used in an attempt to identify protein patterns that are unique to ovarian cancer.25,26
In this study we sought to improve on earlier studies by comparing the gene expression of ovarian carcinoma tissue samples to more than 300 other tissue samples. By examining a large number of other types of tissues, it was possible to identify genes relatively specific to ovarian carcinoma, without relying entirely on the gene expression profile of normal ovary epithelial cells. Seven known genes that were overexpressed in ovarian carcinoma tissues were selected for further analysis: bone morphogenetic protein-7 (BMP-7), the β8 integrin subunit, claudin-4, cellular retinoic acid-binding protein-1 (CRABP-1), collagen type IX α2 (COL IX α2), forkhead box J1 (FOX J1), and S100A1. To verify the corresponding protein expression of these seven genes, immunohistochemical staining was performed on normal ovaries, ovarian carcinoma tissues, and ovarian carcinoma tumors metastatic to the omentum. Statistical analyses were conducted to determine how well the expression of each gene/protein distinguishes ovarian carcinoma from normal ovarian tissues.
Materials and Methods
Tissue Samples
Tissues were obtained from the University of Minnesota Cancer Center’s Tissue Procurement Facility on approval by the University of Minnesota Institutional Review Board. Tissue Procurement Facility employees obtained signed consent from each patient, allowing procurement of excess waste tissue and access to medical records. Bulk tumor and normal tissues were identified, dissected, and snap-frozen in liquid nitrogen within 15 to 30 minutes of resection from the patient. Tissue sections were made from each sample, stained with hematoxylin and eosin (H&E), and examined by a pathologist by light microscopy to confirm the pathological state of each sample. Later, a second pathologist confirmed the diagnosis of each sample, documented the percent tumor (typically 100%), and documented any necrosis (typically none).
Tissue samples from 50 normal ovaries (women ranging in age from 32 to 79 years with a mean age of 51.0 years), 20 serous papillary ovarian carcinoma tumors (age range of 29 to 79 years with a mean age of 57.6 years), 17 metastases of serous papillary ovarian carcinoma to the omentum (age range of 29 to 79 years with a mean age of 59.7 years), and 24 other sets of tissue samples were provided to Gene Logic Inc. (Gaithersburg, MD) for microarray analysis as part of a collaboration with the University of Minnesota. The majority of ovarian tumor samples were classified as stage 3 tumors, whereas the tumor grade varied among the samples. None of the patients had been treated with chemotherapy before surgical resection of the tissue. The 24 other tissue sets that encompassed 321 different tissue samples were: 12 normal adipose tissue, 4 normal breast (from which adipose tissue was removed), 7 normal cervix, 24 normal colon, 11 normal kidney, 12 normal liver, 24 normal lung, 43 normal myometrium, 7 normal omentum, 12 normal skeletal muscle, 9 normal skin, 8 normal small intestine, 55 normal thymus, 11 normal tonsil, 11 tonsils with lymphoid hyperplasia, 3 endometrial hyperplasia, 3 squamous cell carcinoma of the cervix, 7 colon adenocarcinoma, 7 endometrial adenocarcinoma, 8 kidney cell carcinoma, 7 lung adenocarcinoma, 9 squamous carcinoma of the lung, 8 gall bladder with chronic inflammation, and 19 leiomyoma. On receipt of the tissue samples at Gene Logic Inc., a third pathologist examined the H&E-stained slides to verify the diagnosis.
A portion of the ovarian tissues were embedded in O.C.T. by the Tissue Procurement Facility and provided to us for the purpose of immunohistochemical analysis; specifically, 10 normal ovaries, 10 serous papillary ovarian carcinoma tissues, and 10 serous papillary ovarian tumors metastatic to the omentum. Fifteen additional tissues (five each of normal ovaries, serous papillary ovarian carcinoma tumors, and serous papillary ovarian carcinoma tumors metastatic to the omentum) were also embedded in O.C.T. by the Tissue Procurement Facility and provided to us for the purpose of immunohistochemical analysis. These 15 additional tissues were not among the tissues analyzed by Gene Logic Inc.
Gene Expression Analysis
All tissue samples underwent stringent quality control measures to verify the integrity of the RNA before use in gene array experiments. Namely, RNA was isolated, the quantity was determined spectrophotometrically, and the quality was assessed on agarose gels. Tissue samples were not used if the RNA yield was low or RNA degradation was evident. Gene expression was determined by Gene Logic Inc. using Affymetrix HU_95 arrays containing ∼12,000 known genes and 48,000 expressed sequence tags as we have previously described.27,28 Briefly, RNA was obtained from 20 serous ovarian carcinoma tissues, 17 ovarian carcinoma tumors metastatic to the omentum, 50 normal ovaries, and 321 other tissue samples. Gene expression analysis was performed with the Gene Logic GeneExpress Software System using the Gene Logic normalization algorithm. Sample sets were created in which each sample set contained gene expression data from all of the tissues of a particular organ or tissue type. Gene signature analyses were then performed, and genes were defined as being present in a sample set if more than 75% of the samples expressed the gene above background levels.
Fold change analyses were performed in which the ratio of the geometric means of the expression intensities for each gene fragment was computed, and the ratio was reported in terms of the fold change (up or down). Confidence intervals and P values on the fold change were also calculated using a two-sided Welch modified two-sample t-test. Differences were considered significant if the P value was ≤0.05. Gene fragments that were most discriminatory between sample sets were also identified by Contrast Analysis using the Gene Logic GeneExpress software system. A subset of gene fragments was then further analyzed by performing e-Northerns using the Gene Logic GeneExpress software system. The e-Northerns provide a visual display of the gene expression values for each of the 408 tissue samples belonging to a sample set.
Hierarchical cluster analyses were performed using Eisen cluster software.29 Data were normalized and the genes were clustered using the complete linkage clustering algorithm. Graphical displays of the gene expression data were obtained by using Tree View Software (available at http://rana.lbl.gov/EisenSoftware.htm).
Antibodies
Primary antibodies were used at the following concentrations: 1 μg/ml of purified mouse IgG (mIgG) (Sigma, St. Louis, MO) was used as a negative control; 1 μg/ml of monoclonal antibody (mAb) P5D2 against the β1 integrin subunit (provided by Dr. Leo Furcht, University of Minnesota, Minneapolis, MN) was used as a positive control; 5 μg/ml of purified mouse mAb against the β8 integrin subunit (provided by Dr. Stephen Nishimura, University of California, San Francisco, CA); 5 μg/ml of purified rabbit polyclonal antibody (Ab) against BMP-7 (Biotrend, Cologne, Germany); 1 μg/ml of purified mouse mAb against claudin-4 (Zymed Laboratories, San Francisco, CA); a dilution of 1:250 of mouse mAb against CRABP-1 (Affinity BioReagents, Golden, CO); a dilution of 1:1000 of rabbit polyclonal Ab against COL IX α2 (Calbiochem, San Diego, CA); a ready-to-use solution of unknown concentration of purified mouse mAb against hepatocyte nuclear factor-3/FOX J1 (Lab Vision, Fremont, CA); and a dilution of 1:50 of purified rabbit polyclonal Ab against S100A1 (DAKO, Carpinteria, CA). Secondary antibodies used in the immunohistochemical staining procedure were purified, biotinylated anti-mouse or anti-rabbit IgG (Vector Laboratories, Burlingame, CA).
Immunohistochemical Staining
Immunohistochemistry was performed as we have previously described30,31 with minor modifications. Glass slides were incubated in a 0.01% poly-l-lysine solution (Sigma) for 5 minutes at room temperature to enhance stabilization of tissues onto the slides. O.C.T.-embedded tissues were cut on a cryostat into 5-μm sections, affixed onto poly-l-lysine-coated glass slides, and submerged in acetone for 10 minutes at room temperature to fix the tissues onto the slides. Slides were then rinsed in an excess of phosphate-buffered saline (PBS), pH 7.4, and blocked for 1 hour in PBS containing 3% ovalbumin and 1% normal goat serum (Pierce, Rockford, IL). Slides were rinsed again in excess PBS and incubated with 250 μl of the primary Ab in PBS containing 3% ovalbumin and 1% normal goat serum for 1 hour at room temperature.
The slides were again rinsed in excess PBS, followed by the addition of 250 μl of 0.03% H2O2 in PBS for 10 minutes at room temperature to quench endogenous peroxidase. After another rinse in excess PBS, the slides were incubated for 1 hour at room temperature in 250 μl of a 1:500 dilution of the anti-mouse or anti-rabbit biotinylated secondary Ab to visualize the primary antibodies. After rinsing in excess PBS, the slides were incubated with 250 μl of Vectastain ABC solution (Vector Laboratories) for 1 hour at room temperature. After another rinse in excess PBS, the slides were incubated in 250 μl of 3,3′-diaminobenzidine solution (Vector Laboratories) for 5 to 8 minutes at room temperature. After rinsing with tap water, the slides were incubated in hematoxylin counterstain solution (Vector Laboratories) for ∼2 to 3 minutes. On drying at room temperature for 20 minutes, glass coverslips were applied to the slides using Cytoseal aqueous mounting media (Richard-Allan Scientific, Kalamazoo, MI).
Quantitation of Tissue Staining Intensity
On completion of immunohistochemical staining of the tissue samples, a pathologist examined the tissue slides in a blinded manner and documented the intensity and localization of staining. The classifications of intensity were based on a five-point scale: +++, maximum positive staining; ++, moderate positive staining; +, weak but positive staining; ±, faint or questionable staining; and −, a complete lack of staining. All staining was compared to the positive control, the β1 integrin subunit, which received a score of +++.
Statistical Analysis
To determine which gene markers were best for distinguishing ovarian carcinoma tissue from normal ovarian tissue, the specificity, sensitivity, and Youden’s misclassification index were calculated for each gene marker via pairwise tissue comparisons. Associations between gene frequency and staining classifications were analyzed using the Wilcoxon-Mann-Whitney test. Linear and logistic regression analyses were used to evaluate associations between patient demographic characteristics (age, alcohol use, smoking history, and tumor grade) and gene frequencies or staining classification.
Results
Gene Expression Analysis
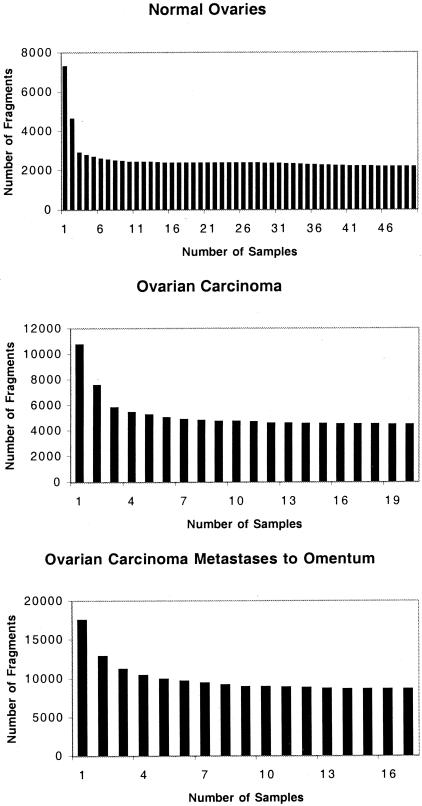
RNA was prepared and gene expression was performed on all samples using Affymetrix HU_95 arrays. Gene signature analyses were performed to identify genes that were expressed (present) in more than 75% of the samples in each sample set. Using a threshold of 75% 11,679 gene fragments were present in the set of 50 normal ovary samples, 12,651 gene fragments were present in the sample set of 20 serous papillary ovarian carcinomas, and 15,294 gene fragments were present in the sample set of 17 serous papillary ovarian carcinomas metastatic to the omentum. The dependence of the number of gene fragments present in all of the samples of a sample set is shown as a function of the number of samples analyzed in Figure 1. The number of gene fragments defined as present in all of the samples in each sample set did not vary greatly, provided that eight or more samples of the set were included in the analysis (Figure 1).
Figure 1.
Dependence of the number of gene fragments present in all samples on the number of samples analyzed. The number of fragments present in all samples of a sample set is shown as a function of the number of samples. Normal ovaries (top), serous papillary ovarian carcinoma (middle), and serous papillary ovarian carcinoma metastatic to the omentum (bottom).
Fold Differences
The relative intensity of gene expression in the ovarian carcinomas compared to the normal ovary samples was determined (Table 1). One hundred thirty-seven gene fragments were expressed at ≥10-fold different levels in the ovarian carcinoma sample set compared with the set of normal ovaries (Table 1). An additional 427 gene fragments were expressed at ≥5-fold to 10-fold different levels between the two sample sets, and a total of 4322 gene fragments were expressed at ≥2-fold different levels between the two sample sets (Table 1).
Table 1.
Fold Change Analysis of Gene Expression in Ovarian Carcinoma Tissues Compared to Normal Tissue Counterpart
| Fold change range‡ | Fold change of ovarian carcinoma versus normal ovary*
|
Fold change of ovarian carcinoma versus ovarian carcinoma metastases to the omentum†
|
||||
|---|---|---|---|---|---|---|
| Up in ovarian carcinoma | Down in ovarian carcinoma | Total gene fragments changed | Up in ovarian carcinoma | Down in ovarian carcinoma | Total gene fragments changed | |
| Over 100 | 1 | 0 | 1 | 0 | 0 | 0 |
| 10 to 100 | 90 | 46 | 136 | 0 | 3 | 3 |
| 5 to 10 | 210 | 217 | 427 | 1 | 10 | 11 |
| 4 to 5 | 155 | 170 | 325 | 3 | 14 | 17 |
| 3 to 4 | 401 | 347 | 748 | 19 | 45 | 64 |
| 2 to 3 | 1499 | 1186 | 2685 | 136 | 393 | 529 |
| 1 to 2 | 23,592 | 21,348 | 44,940 | 17,378 | 27,365 | 44,743 |
| Unchanged | — | — | 923 | — | — | 1336 |
A fold change analysis of gene expression in the set of 20 serous papillary ovarian carcinoma tissues compared with that in the set of 50 normal ovary tissues.
A fold change analysis of gene expression in the set of 20 serous papillary ovarian carcinoma tissues compared with that in the set of 17 serous papillary ovarian carcinoma that had metastasized to the omentum.
The number of gene fragments in each indicated range of fold change.
The relative intensity of gene expression in the ovarian carcinoma samples compared to the omental metastatic samples was also determined (Table 1). Only three gene fragments were expressed at ≥10-fold different levels in the ovarian carcinoma set compared with the set of omental metastases (Table 1). Also, a total of 624 gene fragments were expressed at ≥2-fold different levels between the two sample sets (Table 1). These results suggest that the ovarian carcinoma samples are much more similar to each other than to the normal ovary samples.
Contrast Analysis and E-Northerns
The 4322 gene fragments that were expressed greater than twofold more in ovarian carcinomas compared to normal ovaries were analyzed by Contrast Analysis using the Gene Logic GeneExpress software system to identify those gene fragments that were the most discriminatory between ovarian carcinoma and normal ovaries. The 400 gene fragments that were more highly expressed in the ovarian carcinoma samples and most discriminatory between the two sample sets were then further analyzed by performing e-Northerns using the Gene Logic GeneExpress software system. The e-Northern analysis provides a graphic representation of the level of gene expression values for each sample in the sets of normal ovaries, ovarian carcinomas, metastases of ovarian carcinoma to the omentum, and 321 other tissue samples from 24 different sites.
Forty known genes were preferentially up-regulated in ovarian carcinomas compared to all of the other tissue types examined (Table 2). The results of the e-Northern analyses are listed in the last three columns of Table 2 as the percentage of ovarian carcinoma tissues (n = 20), metastatic ovarian carcinoma tissues (n = 17), and normal ovaries (n = 50) that express each of the 40 genes (ie, scored as present using the Gene Logic GeneExpress software system). The majority of the ovarian carcinoma tissues and the metastatic ovarian carcinoma tissues expressed the 40 genes listed in Table 2, whereas only a few normal ovaries expressed these genes.
Table 2.
Genes Specifically Up-Regulated in Ovarian Carcinoma Tissue Samples Compared to More than 350 Other Tissue Samples as Determined by Gene Microarrays
| Known gene symbol | Known gene name | Function |
|---|---|---|
| ALDH3B2 | Aldehyde dehydrogenase 3 family, member B2 | Enzyme, metabolism |
| BHLHB3 | Basic helix-loop-helix domain containing, class B, 3 | Cell signaling, differentiation |
| BMP7 | Bone morphogenetic protein 7 (osteogenic protein 1) | Cytokine |
| CAPS | Calcyphosine | Ion transport, Ca2+ |
| CCNA1 | Cyclin A1 | Cell cycle |
| CCNE1 | Cyclin E1 | Cell cycle |
| CDH6 | Cadherin 6, type 2, K-cadherin (fetal kidney) | Cell-cell adhesion receptor |
| CDKN2A | Cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | Cell cycle |
| COL9A2 | Collagen, type IX, alpha 2 | Cell adhesion, ECM |
| CP | Ceruloplasmin (ferroxidase) | Ligand-binding protein, copper |
| DAG1 | Dystroglycan 1 (dystrophin-associated glycoprotein 1) | Cell-ECM adhesion receptor |
| EGFL6 | EGF-like-domain, multiple 6 | Cytokine |
| EYA2 | Eyes absent homolog 2 (Drosophila) | Transcriptional co-activator |
| FOLR1 | Folate receptor 1 (adult) | Cell signaling, receptor, transporter |
| FOXJ1 | Forkhead box J1 | Transcriptional activator |
| HOXD1 | Homeo box D1 | Transcription factor |
| HTR3A | 5-hydroxytryptamine (serotonin) receptor 3A | Ion transport, cations |
| ITGB8 | Integrin, beta 8 subunit | Cell-ECM adhesion receptor |
| KLK5 | Kallikrein 5 | Protease, serine |
| KLK6 | Kallikrein 6 (neurosin, zyme) | Protease, serine |
| KLK7 | Kallikrein 7 (chymotryptic, stratum corneum) | Protease, serine |
| KLK8 | Kallikrein 8 (neuropsin/ovasin) | Protease, serine |
| MSLN | Mesothelin | Cell adhesion receptor |
| MUC1 | Mucin 1, transmembrane | Tumor antigen, epithelial cell antigen |
| NMU | Neuromedin U | Cell signaling, muscle contraction |
| PAX8 | Paired box gene 8 | Transcription factor |
| PNOC | Prepronociceptin | Cell signaling, neurotransmitter |
| PRAME | Preferentially expressed antigen in melanoma | Tumor antigen |
| PRKCI | Protein kinase C, iota | Cell signaling |
| PRSS21 | Protease, serine, 21 (testisin) | Protease, serine |
| S100A1 | S100 calcium binding protein A1 | Ligand-binding protein, divalent cations |
| SALL4 | Sal-like 4 (Drosophila) | Transcription factor |
| SCARA3 | Scavenger receptor class A, member 3 | Scavenger receptor |
| SCGB2A1 | Secretoglobin, family 2A, member 1 | Ligand-binding protein |
| SGPL1 | Sphingosine-1-phosphate lyase 1 | Enzyme, metabolism |
| SLC4A11 | Solute carrier family 4, sodium bicarbonate transporter-like, member 11 | Ion transport |
| SOX11 | SRY (sex determining region Y)-box 11 | Transcription factor |
| SPON1 | Spondin 1, (f-spondin) extracellular matrix protein | Cell adhesion, ECM |
| TMPRSS3 | Transmembrane protease, serine 3 | Protease, serine |
| UBE2H | Ubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast) | Enzyme, ubiquitination |
| (Table continues) |
The known gene fragments expressed as ≥2-fold higher levels in the set of ovarian carcinoma than in the set of normal ovaries and other tissue sets were analyzed by Contrast Analysis. Genes were ranked based on the degree of increase in expression, and e-Northern analysis was performed on the top 400 genes. This table is an alphabetical listing of the 40 genes most specific to ovarian carcinoma, as well as the cellular function of their gene products.
Table 2.
(continued)
| Previously referenced as up-regulated in ovarian carcinoma
|
Previously referenced as expressed in other types of cancers | Tissue samples expressing gene (%)
|
|||
|---|---|---|---|---|---|
| Determined by gene arrays | Determined by another technique (e.g. IH, PCR) | Ovarian carcinoma (n = 20) | Ovarian carcinoma metastases (n = 17) | Normal ovaries (n = 50) | |
| − | − | − | 65 | 82 | 4 |
| − | − | * | 85 | 100 | 16 |
| − | − | * | 60 | 82 | 4 |
| − | − | − | 95 | 88 | 38 |
| − | − | * | 85 | 94 | 14 |
| − | * | * | 60 | 82 | 2 |
| * | * | * | 80 | 88 | 2 |
| − | * | * | 75 | 88 | 0 |
| − | − | * | 30 | 41 | 0 |
| * | * | * | 70 | 76 | 0 |
| − | − | * | 10 | 29 | 0 |
| − | − | * | 80 | 100 | 8 |
| * | − | − | 90 | 88 | 14 |
| * | * | − | 100 | 100 | 14 |
| − | − | * | 85 | 52 | 4 |
| − | − | * | 35 | 64 | 2 |
| − | − | − | 70 | 94 | 14 |
| * | − | − | 60 | 82 | 0 |
| − | * | * | 40 | 47 | 0 |
| * | * | * | 95 | 88 | 12 |
| − | * | − | 95 | 100 | 12 |
| − | * | * | 95 | 100 | 0 |
| * | * | − | 100 | 100 | 8 |
| * | * | * | 96 | 94 | 18 |
| − | − | − | 75 | 100 | 2 |
| * | − | * | 90 | 100 | 2 |
| * | − | − | 45 | 76 | 2 |
| * | − | * | 80 | 88 | 22 |
| − | − | * | 85 | 94 | 48 |
| − | * | * | 75 | 70 | 0 |
| * | − | * | 95 | 100 | 2 |
| − | − | − | 55 | 100 | 4 |
| − | − | − | 80 | 58 | 20 |
| * | − | * | 95 | 100 | 16 |
| − | − | − | 100 | 100 | 86 |
| − | − | − | 35 | 29 | 0 |
| − | − | * | 30 | 100 | 0 |
| * | − | − | 100 | 100 | 96 |
| − | * | − | 95 | 76 | 0 |
| − | − | − | 75 | 82 | 12 |
The gene products of these 40 genes spanned a large spectrum of functional activity, including: nine enzymes, six cell adhesion molecules/receptors, six transcription factors, five cell-signaling proteins, three ligand-binding proteins, three cell cycle/cell proliferation proteins, three ion transport proteins, two cytokines, two tumor antigens, and one scavenger receptor (Table 2). As a testament to the validity of our approach and selection criteria, 14 of the 40 genes listed in Table 2 that we found to be specifically up-regulated in ovarian carcinoma had previously been shown by others using gene array technology to be up-regulated in ovarian carcinoma.4–10,13,22,32 To date, only 6 of these 14 genes have been validated by a second technique. An additional 7 of the 40 genes listed in Table 2 have been previously shown by other techniques to be up-regulated in ovarian carcinoma. Furthermore, 10 of the 40 genes have not been previously implicated in ovarian carcinoma, but have been implicated in other types of cancer. Thus, 9 of the 40 genes listed in Table 2 have not previously been identified as being up-regulated in ovarian carcinoma or any other type of cancer.
Another set of 26 known genes was up-regulated in the ovarian carcinoma samples compared to normal ovaries (Table 3). However, by e-Northern analysis, we found that these 26 genes were also expressed by one or more other types of tissue. These 26 genes included a variety of proteins, including: five cell adhesion molecules/receptors, four transcription factors, three cell cycle/cell proliferation proteins, three accessory proteins, three ion transport proteins, two enzymes, two cell-signaling proteins, one tumor antigen, one ligand-binding protein, one histone, and one unknown. Again, as a testament to the validity of our approach and selection criteria, 4 of the 26 genes listed in Table 3 that we found to be up-regulated in ovarian carcinoma had previously been shown by others using gene array technology to be up-regulated in ovarian carcinoma.6,12,13,19,22 To date, only two of these four genes have been validated by a second technique. An additional 3 of the 26 genes listed in Table 3 have been previously shown by other techniques to be up-regulated in ovarian carcinoma. Furthermore, 9 of the 26 genes have not been previously implicated in ovarian carcinoma, but have been implicated in other types of cancer. Thus, 10 of the 26 genes have not previously been identified as being up-regulated in ovarian carcinoma or any other type of cancer. Although these 26 genes may play important roles in the development of ovarian carcinoma, they are not as specific to ovarian carcinoma as the genes listed in Table 2.
Table 3.
Genes Up-Regulated in Ovarian Carcinoma Tissue Samples with Limited Expression in Other Tissues as Determined by Gene Microarrays
| Known gene symbol | Known gene name | Function | Previously referenced as upregulated in ovarian carcinoma
|
Previously referenced as expressed in other types of cancers | |
|---|---|---|---|---|---|
| Determined by gene arrays | Determined by another technique (e.g. IH, PCR) | ||||
| ATP6V1B1 | ATPase, H+ transporting, lysosomal 56/58 kDa, V1 subunit B, isoform 1 | Ion transport, H+ | − | − | − |
| C20orf1 | Chromosome 20 open reading frame 1 | Cell cycle | − | − | * |
| CD47 | CD47 antigen (Rh-related antigen, integrin-associated signal transducer) | Cell adhesion receptor | − | * | − |
| CENPF | Centromere protein F, 350/400 ka (mitosin) | Cell cycle, mitosis | − | − | − |
| CLDN4 | Claudin 4 | Cell-cell adhesion receptor | * | * | * |
| COL8A2 | Collagen, type VIII, alpha 2 | Cell adhesion, ECM | − | − | * |
| CRABP1 | Cellular retinoic acid binding protein 1 | Accessory protein | * | − | * |
| CRABP2 | Cellular retinoic acid binding protein 2 | Accessory protein | − | − | * |
| DD96 | Epithelial protein up-regulated in carcinoma, membrane associated protein 17 | Tumor antigen | − | − | * |
| ETV4 | Ets variant gene 4 (E1A enhancer binding protein, E1AF) | Transcriptional activator | − | * | * |
| FOXM1 | Forkhead box M1 | Transcriptional activator | − | − | * |
| GPR49 | G protein-coupled receptor 49 | Cell signaling, receptor | * | − | − |
| GRB7 | Growth factor receptor-bound protein 7 | Accessory protein, adaptor protein | − | − | * |
| H2BFB | H2B histone family, member B | Histone | − | − | − |
| IFRG28 | 28 kD interferon responsive protein | Unknown | − | − | − |
| ITGB4 | Integrin, beta 4 | Cell-cell/ECM adhesion receptor | − | * | * |
| KLK10 | Kallikrein 10 | Protease, serine | * | * | * |
| KNSL6 | Kinesin-like 6 (mitotic centromere-associated kinesin) | Motor protein, mitosis | − | − | − |
| LNIR | Ig superfamily receptor LNIR | Cell adhesion receptor | − | − | − |
| SCNN1A | Sodium channel, nonvoltage-gated 1 alpha | Ion transport, Na+ | − | − | − |
| SIAHBP1 | Fuse-binding protein-interacting repressor | Ligand-binding protein, Ro RNP | − | − | * |
| SLC34A2 | Solute carrier family 34 (sodium phosphate), member 2 | Ion transport, phosphate | − | − | − |
| TFAP2A | Transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) | Transcriptional activator | − | − | * |
| TFAP2C | Transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) | Transcriptional activator | − | − | * |
| USP18 | Ubiquitin specific protease 18 | Protease | − | − | − |
| XPR1 | Xenotropic and polytropic retrovirus receptor | Cell signaling, receptor | − | − | − |
The known gene fragments expressed as ≥2-fold higher levels in the set of serous papillary ovarian cancer than in the set of normal ovaries were analyzed by Contrast Analysis. Genes were ranked based on the degree of increase in expression, and e-Northern analysis was performed on the top 400 genes. The 26 genes listed alphabetically in this table were highly expressed in ovarian carcinoma samples, but were also expressed by one or more other tissues. Thus, these genes may not be as specific to ovarian cancer as those in Table 2.
Clustering
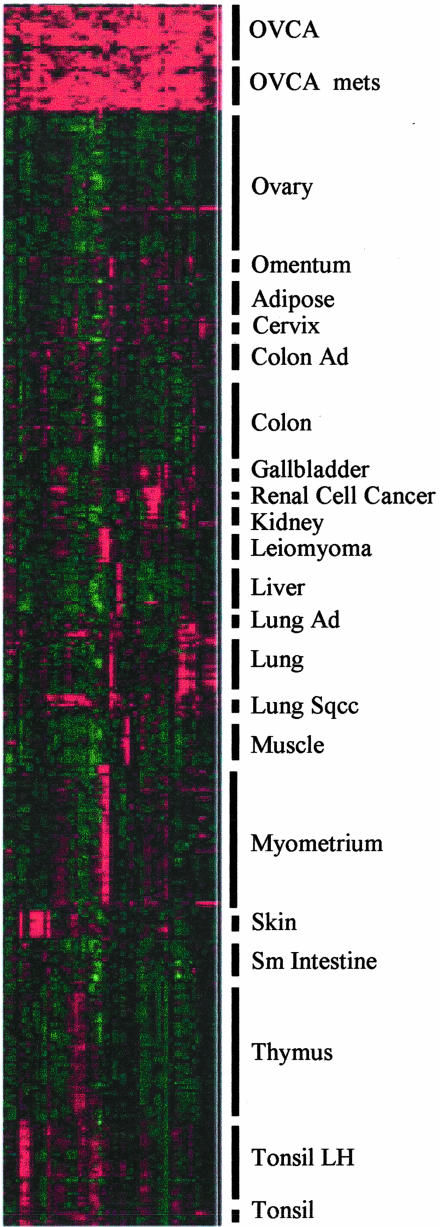
The Eisen clustering software, Cluster, was used as another means of displaying the gene expression data for the set of 40 known genes preferentially expressed by the ovarian carcinoma sample set (Table 2). By this technique, the ovarian carcinoma samples had intensely positive gene expression values (shown in red in Figure 2). The gene expression values for the normal ovary samples as well as all of the other tissue sample sets (shown in green or black in Figure 2) were much less intense and very distinct from the ovarian carcinoma samples. The differences in intensity of the squares in Figure 2 are indicative of the biological heterogeneity that exists among ovarian carcinomas.
Figure 2.
Differentially expressed genes in ovarian carcinoma, normal ovaries, and 20 other tissues. Eisen Cluster software was used to graphically display the intensity of gene expression values for each of the 40 genes listed in Table 2 for the 391 different tissue samples. The color of each square represents the ratio of the gene expression in the indicated sample relative to the average signal of expression of all genes examined. Red indicates gene expression above the median; black, equal to the median; and green, below the median. The intensity of the color reflects the magnitude of divergence from the median. Columns represent individual cDNAs for the 40 genes listed in Table 2, and rows represent the indicated tissue samples, as described in the Material and Methods section.
Criteria for Selecting a Subset of Genes for Protein Analysis
To determine whether the differentially expressed gene fragments that were unique to ovarian carcinoma corresponded to protein expression, a subset of the genes listed in Tables 2 and 3 were selected for analysis of their protein counterparts via immunohistochemistry. The following criteria were used to select the genes. First, the genes must be up-regulated at least twofold or greater in ovarian carcinoma tissues compared to normal ovary tissues. Second, the genes should either be completely absent or expressed at significantly lower levels in normal ovarian tissues. Third, increased expression of the genes should be solely characteristic of ovarian carcinoma, and minimal expression should be detected in any other tissues in the body. Fourth, the genes should be present in the vast majority of ovarian carcinoma samples. Finally, genes were selected if antibodies against their corresponding proteins were available. Based on these selection criteria, seven genes were chosen for further analysis: the β8 integrin subunit, BMP-7, CRABP-1, claudin-4, COL IX α2, FOX J1, and S100 calcium-binding protein A1 (S100A1). Only one of these seven genes, claudin-4, has been previously characterized for both gene and protein expression in ovarian cancer.6,13 Although the other 59 genes listed in Tables 2 and 3 would have been just as interesting to study and met our first four criteria, we did not select them at this time because antibodies were not commercially available for many of them.
Tissue Distribution of the Differentially Expressed Genes by E-Northerns
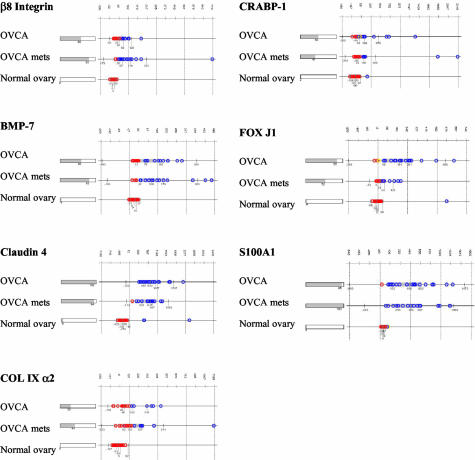
We compared the expression of the seven selected genes in ovarian tissues and other tissues. E-Northerns were generated using the Gene Logic GeneExpress software system to display the gene expression values for each sample in the sets of normal ovaries, ovarian carcinomas, ovarian carcinomas metastatic to the omentum, and 321 other tissue samples from 24 different sites. The percentage of samples expressing detectable levels of each gene fragment is shown as a bar graph on the left side and the intensity of gene expression in each sample of the set is indicated on the right side of Figure 3.
Figure 3.
E-Northern analysis of differentially expressed gene fragments in human tissues. The expression of each indicated gene fragment was examined in 20 serous papillary ovarian carcinomas, 17 serous papillary ovarian carcinomas metastatic to the omentum, and 50 normal ovaries. The bar graph on the left depicts the percentage of samples that express detectable levels of the indicated gene fragment. The intensity of gene expression in each sample is plotted as average expression value on a linear scale on the right; median ± 2 SD of expression values are shown.
A representative portion of an e-Northern of a β8 integrin subunit gene fragment is shown in Figure 3. Sixty percent of the ovarian carcinoma tissues and 82% of the omental metastatic tissues expressed detectable levels of this gene fragment, whereas none of the normal ovaries expressed it. The β8 integrin gene fragment was not significantly expressed in more than 90% of the 321 other tissues examined. Notably, three of the eight kidney cell carcinoma tissues, three of the nine squamous cell lung carcinomas, and three of the seven endometrial adenocarcinoma tissues expressed low levels of the β8 integrin subunit; in most cases, the intensity of expression was less than that in the ovarian carcinoma tissue samples.
A representative portion of an e-Northern of a BMP-7 gene fragment is shown in Figure 3. Sixty percent of the ovarian carcinoma tissues and 82% of the omental metastatic tissues expressed measurable levels of this gene fragment, whereas only 4% of the normal ovary tissues expressed this BMP-7 gene fragment. Several other tissues also expressed this gene fragment, but in general, fewer than 20% of the samples in each tissue type expressed detectable levels. Exceptions to this finding were that eight of the nine skin tissue samples, five of the seven normal cervix samples, and three of the squamous cell carcinomas of the cervix samples expressed the BMP-7 gene fragment, although at much lower intensities than the ovarian carcinoma tissues. The intensity of gene expression in the majority of the other tissues was considerably lower compared to the ovarian carcinoma tissues. Exceptions to this finding included high BMP-7 intensities in 2 of the 24 normal lungs and 3 of the 9 squamous cell lung carcinoma tissues.
An e-Northern of the claudin-4 gene fragment revealed that 100% of the ovarian carcinoma tissues and 94% of the omental metastatic tissues expressed detectable levels of this gene fragment, whereas only 6% of the normal ovary tissues expressed measurable levels (Figure 3). The majority of other tissue types also expressed this gene fragment, and the intensity of expression varied across all tissue types.
An e-Northern of the COL IX α2 gene fragment shows that 30% of ovarian carcinoma tissues and 41% of omental metastatic tissues expressed measurable levels of the gene fragment, whereas none of the normal ovaries expressed detectable levels of the COL IX α2 gene fragment (Figure 3). This COL IX α2 gene fragment was minimally expressed in a few of the other tissues, including 1 of the 7 colon adenocarcinomas, 3 of the 24 normal lungs, 1 of the 12 normal skeletal muscles, and 1 of the 43 normal myometrium. Notably, more than 95% of the other tissues did not express detectable levels of the COL IX α2 gene fragment.
A representative e-Northern of a CRABP-1 gene fragment indicates that 50% of the ovarian carcinoma tissues and 41% of the omental metastatic tissues expressed measurable levels of this gene fragment, whereas only 1 of the 50 normal ovary tissues expressed the CRABP-1 gene fragment (Figure 3). Five of the nine normal skin tissues, three of the four normal breast tissues, and three of the seven endometrial adenocarcinoma tissues expressed the CRABP-1 gene fragment. Otherwise, only six other tissue samples minimally expressed this gene fragment, including one chronically inflamed gall bladder tissue, one normal kidney, one normal tonsil, one endometrial hyperplasia, and two leiomyoma tissues. Therefore, compared to the ovarian carcinoma tissues, more than 90% of the other tissues did not express this CRABP-1 gene fragment.
An e-Northern of the FOX J1 gene fragment shows that 85% of ovarian tissues and 52% of omental metastatic tissues expressed detectable levels of this gene fragment, whereas only 4% of the normal ovary tissues expressed measurable levels of the FOX J1 gene fragment (Figure 3). Some of the other tissue types also expressed FOX J1; notably normal lung, lung adenocarcinoma, endometrial hyperplasia, and normal cervix. However, fewer of the samples within these tissue sets expressed the gene, and the gene was expressed at a lower intensity compared to the ovarian carcinoma tissues. Interestingly, six of the seven endometrial adenocarcinoma tissues expressed this gene at levels comparable to the omental metastases. Overall, 90% of the other tissue samples did not express detectable levels of the FOX J1 gene fragment.
An e-Northern of the S100A1 gene fragment revealed that 95% of the ovarian carcinoma tissues and 100% of the omental metastatic tissues expressed this gene fragment, whereas only 1 of the 50 normal ovary tissues expressed the S100A1 gene fragment (Figure 3). The S100A1 gene fragment was also expressed in 71% of adipose tissues, 80% of kidney carcinoma tissues, 68% of normal kidney tissues, 93% of skeletal muscle tissues, all four of the normal breast tissues, 42% of the endometrial adenocarcinoma tissues, and 44% of skin tissues. All of these tissues, except the skeletal muscle tissues, expressed the gene fragment at much lower intensities compared to the ovarian carcinoma tissues, whereas more than 80% of the other tissue samples did not express the S100A1 gene fragment at all.
Immunohistochemical Staining of the Differentially Expressed Gene Products
The protein expression of the seven differentially expressed genes was analyzed by immunohistochemistry in 45 ovarian tissues. Gene expression data were available from Gene Logic Inc. for 30 of the 45 ovarian tissues screened, including: 10 normal ovaries, 10 serous papillary ovarian carcinoma tissues, and 10 serous papillary ovarian carcinoma tumors metastatic to the omentum. The remaining 15 tissues that were analyzed by immunohistochemistry were: 5 normal ovaries, 5 serous papillary ovarian carcinoma tumors, and 5 serous papillary ovarian carcinoma tumors metastatic to the omentum.
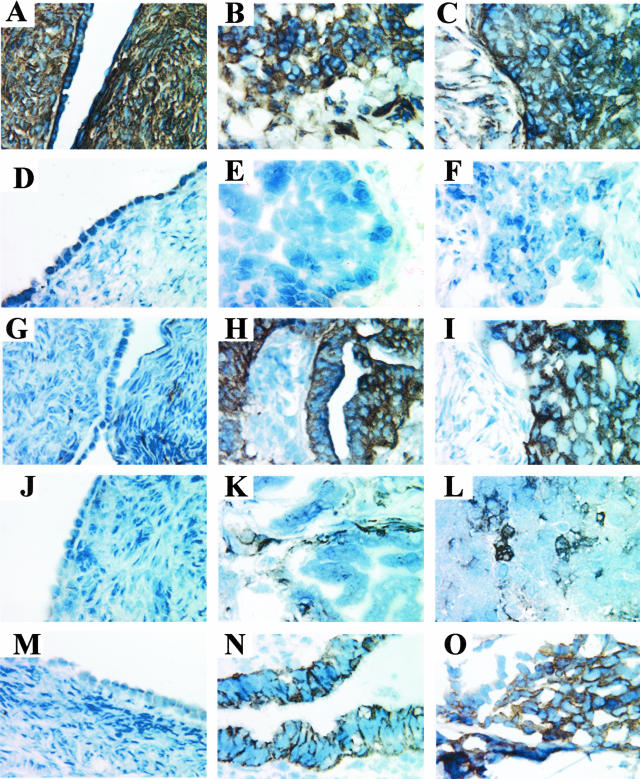
Monoclonal and polyclonal antibodies against the seven proteins were used, as well as normal mouse IgG (negative control) and a mAb against the β1 integrin subunit (positive control). The β1 integrin subunit was used as a positive control because it is a cell adhesion molecule known to be ubiquitously expressed on the surface of most cells except hematopoietic cells. The β1 integrin subunit was expressed in normal ovary (Figure 4A), ovarian carcinoma (Figure 4B), and ovarian carcinoma metastatic to the omentum (Figure 4C). All three tissue types exhibited a strong, membranous staining pattern for β1 integrin. As expected, normal mouse IgG did not stain normal ovary (Figure 4D), ovarian carcinoma (Figure 4E), or ovarian carcinoma metastatic to the omentum (Figure 4F).
Figure 4.
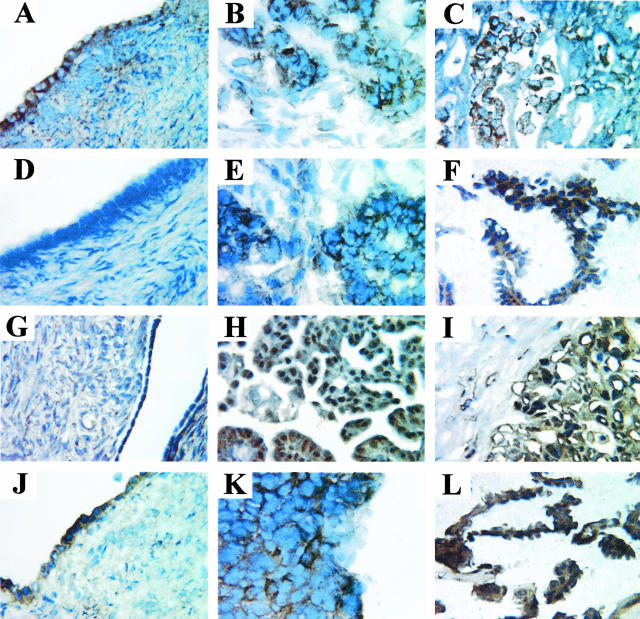
Immunohistochemical staining of differentially expressed gene products. Normal ovaries (A, D, G, J, M), serous papillary ovarian carcinoma (B, E, H, K, N), and serous papillary ovarian carcinoma metastatic to the omentum (C, F, I, L, O) tissues were stained with a mAb against the β1 integrin subunit (A–C), normal mouse IgG (D–F), and antibodies against: the β8 integrin subunit (G–I), BMP-7 (J–L), and claudin-4 (M–O). Original magnifications, ×60.
The β8 integrin subunit has been previously described as a cell surface molecule.33 In this study, the β8 integrin subunit exhibited a strong membranous staining pattern in the ovarian tumors (Figure 4, H and I), but was not detected in normal ovaries (Figure 4G). The vast majority of ovarian tumors examined exhibited a membranous staining pattern for the β8 integrin subunit, whereas the majority of normal ovaries did not express the β8 integrin protein.
The second protein we studied was BMP-7, a cytokine that was expected to be localized in the cytoplasm and in the extracellular matrix (ECM) on secretion.34 Interestingly, BMP-7 staining was variable across all ovarian tumors studied. The protein was detected in the tumor cells in some ovarian carcinoma tissues and in the surrounding stroma in other ovarian carcinoma tissues. Figure 4 shows BMP-7 expression in the stroma of one ovarian tumor (Figure 4K) and BMP-7 expression in patches of tumor cells in one omental metastatic tumor (Figure 4L). In the majority of cases, normal ovaries did not express BMP-7 (Figure 4J).
The third protein of interest, claudin-4, is a tight junction protein located on the cell surface.35 Claudin-4 was detected on the cell surface in all ovarian tumor tissues examined. Figure 4 shows a strong membranous staining pattern for claudin-4 in both the ovarian tumor (Figure 4N) and the metastatic omental tumor (Figure 4O). Claudin-4 was not detected in normal ovaries (Figure 4M).
The fourth protein whose localization we examined, COL IX α2, is an adhesion molecule found in the ECM.36 The COL IX α2 protein was observed as an intercellular epitope in most ovarian tumors (Figure 5, B and C), but was also detected in the cytoplasm in some cases. Also, COL IX α2 was frequently expressed in the stromal tissue surrounding the ovarian tumor nests, as well as in the stroma of normal ovaries (Figure 5A). This high background-staining pattern may be attributable to nonspecific staining of the Ab against COL IX α2 used in the immunohistochemical analyses.
Figure 5.
Immunohistochemical staining of differentially expressed gene products. Normal ovaries (A, D, G, J), serous papillary ovarian carcinoma (B, E, H, K), and serous papillary ovarian carcinoma metastatic to the omentum (C, F, I, L) tissues were stained with antibodies against: COL IX α2 (A–C), CRABP-1 (D–F), FOX J1 (G–I), and S100A1 (J–L). Original magnifications, ×60.
The fifth protein we studied was CRABP-1, a transport protein found in the cytoplasm.37 CRABP-1 expression was variable across all tissues examined, and it was localized to the cell membrane in some tissues and in the cytoplasm in other tissues. Representative examples show a membranous staining pattern for CRABP-1 in the ovarian tumor (Figure 5E), a cytoplasmic staining pattern for CRABP-1 in the omental metastatic tumor (Figure 5F), and no detection of CRABP-1 in the normal ovary (Figure 5D).
The sixth protein we selected was FOX J1, a transcription factor with expected localization to the nucleus and possibly the cytoplasm.38 Interestingly, only a few of the ovarian tumor samples demonstrated a nuclear FOX J1 staining pattern (Figure 5, H and I). Instead, most tumor samples examined exhibited cytoplasmic and membranous staining patterns for FOX J1. In addition, more than half of the normal ovary samples exhibited some degree of FOX J1 expression, as shown in Figure 5G in which a normal ovary exhibits slight nuclear staining of FOX J1 in the surface epithelium. This high background staining observed for FOX J1 may be in part attributable to nonspecific staining of the anti-FOX J1 antibody used in the immunohistochemical analyses.
Our final protein of interest was S100A1, a protein involved in the cell cycle and localized to the cytoplasm.39 Most of the ovarian tumor tissues examined exhibited either a cytoplasmic or membranous S100A1 staining pattern. Figure 5K shows an ovarian tumor with both cytoplasmic and membranous staining for S100A1, whereas Figure 5L shows an omental metastatic tumor with cytoplasmic staining for S100A1. The majority of normal ovaries examined did not express the S100A1 protein. An example of a normal ovary with S100A1 expression in the cytoplasm of the surface epithelial cells is shown in Figure 5J.
In summary, the majority of ovarian tumor tissues exhibited positive staining for the β8 integrin subunit, claudin-4, COL IX α2, FOX J1, and S100A1, and negative staining for BMP-7 and CRABP-1. The majority of the normal ovary tissues exhibited negative staining for the β8 integrin subunit, BMP-7, claudin-4, CRABP-1, and S100A1, and positive staining for COL IX α2 and FOX J1.
Statistical Analysis
Linear and logistic regression analyses were performed to assess whether associations exist between gene and protein expression of the seven genes and various patient characteristics such as age, alcohol use, smoking status, and tumor grade. The results of these analyses indicated no evidence of such associations.
To determine which of the seven gene markers were the strongest candidates for distinguishing normal ovarian tissue from ovarian carcinoma tissue, the specificity, sensitivity, and Youden’s misclassification index were calculated via pairwise tissue comparisons. For the purpose of these statistical measures, tissue staining intensities were classified as either positive (+, ++, +++) or negative (−, ±). Youden’s misclassification index (J), which is based on the specificity and sensitivity, indicates the overall probability that the protein classifications correctly distinguish each tissue being compared. Genes with a J value of 0.5 or greater were considered to be predictive of disease state.
The results of the comparison between normal ovary and ovarian carcinoma tissues are shown in Table 4. The β8 integrin subunit, claudin-4, and S100A1 all had a J value greater than 0.5 and are thus considered to be the best markers for distinguishing ovarian tumor tissue from normal ovary tissue. We then compared the immunohistochemistry results obtained for normal ovary tissues and ovarian tumors metastatic to the omentum. Again, the β8 integrin subunit, claudin-4, and S100A1 all had a J value greater than 0.5 in this comparison and were thus considered to be the best markers for distinguishing metastatic ovarian tumor tissue from normal ovary tissue (Table 4). Finally, a comparison was made between ovarian tumor tissues and metastatic ovarian tumor tissues; none of the seven genes were able to distinguish between them (Table 4). This result is not surprising because both types of ovarian carcinoma tissues exhibited similar gene expression profiles, as evidenced by the fold-change analysis (Table 1).
Table 4.
Specificity, Sensitivity, and Youden’s Misclassification Index (J) for Seven Genes Selected from Gene Microarray Analysis as Potential Candidates for Specific Biomarkers of Ovarian Carcinoma Tissues
| Gene name | Comparison I: normal ovary tissues versus ovarian carcinoma tissues
|
Comparison II: normal ovary tissues versus metastatic ovarian carcinoma tissues
|
Comparison III: ovarian carcinoma tissues versus metastatic ovarian carcinoma tissues
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Specificity* | Sensitivity† | J‡ | Specificity* | Sensitivity† | J‡ | Specificity§ | Sensitivity§ | J | |
| β8 integrin | 0.867 | 0.8 | 0.667 | 0.867 | 0.8 | 0.667 | 0.2 | 0.8 | 0 |
| BMP-7 | 0.667 | 0.6 | 0.267 | 0.667 | 0.6 | 0.267 | 0.4 | 0.867 | 0.267 |
| Claudin-4 | 0.933 | 0.993 | 0.867 | 0.933 | 1 | 0.933 | 0.067 | 1 | 0.067 |
| COL IX α2 | 1 | 0.2 | 0.2 | 1 | 0.2 | 0.2 | 0.067 | 1 | 0.067 |
| CRABP-1 | 0.533 | 0.533 | 0.067 | 1 | 0.133 | 0.133 | 0.933 | 0.133 | 0.067 |
| FOX J1 | 0.333 | 0.933 | 0.267 | 0.333 | 1 | 0.333 | 0.067 | 1 | 0.067 |
| S100A1 | 0.6 | 1 | 0.6 | 0.6 | 0.933 | 0.533 | 0.667 | 0.333 | 0 |
Seven genes were analyzed for their ability to distinguish the following: comparison I, 15 normal ovary tissues from 15 serous papillary ovarian carcinoma tissues; comparison II, 15 normal ovary tissues from 15 serous papillary ovarian carcinoma tissues metastatic to the omentum; and comparison III, 15 serous papillary ovarian tumor tissues from 15 serous papillary ovarian tumor tissues metastatic to the omentum. All three statistical measures were based on the protein staining intensities as determined by immunohistochemical analysis. For the purpose of these statistical measures, tissue staining intensities were classified as positive if they had scored +, ++, or +++, and they were classified as negative if they had scored − or +/−.
Specificity indicates the probability that a negative protein classification correctly identifies the tissue as normal ovary tissue (ie, not as carcinoma).
Sensitivity indicates the probability that a positive protein classification correctly identifies the tissue as ovarian carcinoma tissue (ie, tumor being present).
Youden’s misclassification index (J) indicates the overall probability of correct classifications. Genes with a J value of 0.5 or greater (in bold) were considered to be predictive of cancer.
ROC curves (plots of sensitivity versus specificity) were constructed to determine the optimal staining intensities for each gene at which the two tissue types could be distinguished from each other. Specificity and sensitivity indicate the probability that the optimal staining intensities could correctly distinguish ovarian carcinoma from ovarian carcinoma metastases to the omentum.
To determine the associations between gene expression of the seven genes and protein staining classifications, Wilcoxon-Mann-Whitney statistical tests were performed. Gene expression values were defined as being present or absent by use of the Gene Logic GeneExpress software system using the Gene Logic normalization algorithm. For the purpose of this statistical test, protein staining intensities for the 30 tissues we stained by immunohistochemistry were again classified as positive (+, ++, +++) or negative (−, ±). Statistically significant P values indicate the gene fragments whose expression values are higher in tissues that stained positive than in tissues that stained negative, indicating that these gene fragments are truly associated with the protein staining classifications. The β8 integrin subunit and claudin-4 gene fragments demonstrated a statistically significant (P < 0.001) association with the protein staining classifications. Statistically significant associations were not found between the presence of the five other genes by microarray techniques and the presence of the protein by immunohistochemistry (data not shown). However, protein expression by immunohistochemistry is semiquantitative, and thus an association with gene expression cannot be ruled out. In addition, different mAbs might provide a more accurate assay.
Discussion
In this study, 66 genes were identified by microarray technology to be differentially expressed by ovarian carcinoma tissue samples compared with normal ovarian tissue samples. Nineteen of the 66 genes were reported here for the first time to be up-regulated in cancerous tissues. Twenty-eight of the 66 genes had been previously reported to be up-regulated in ovarian carcinoma by gene array technology or other techniques, whereas an additional 19 of the 66 genes had previously been reported to be up-regulated in other types of cancer. The 66 genes identified in this study represented a variety of proteins, including 11 cell adhesion molecules/receptors, 11 enzymes, 10 transcription factors, 7 cell-signalingproteins, 6 cell cycle/cell proliferation proteins, 6 ion transport proteins, 4 ligand-binding proteins, 3 accessory proteins, 3 tumor antigens, 2 cytokines, 1 scavenger receptor, 1 histone, and 1 unknown. Interestingly, the cellular localization of the gene products of these 66 genes was rather equally divided between the membrane, nucleus, and secretory (∼30% in each group) while fewer of the gene products were localized to the cytoplasm (∼15%).
The design of the current study has several advantages in identifying potential ovarian carcinoma tumor markers compared to many of the earlier ovarian cancer gene expression studies. First, a relatively large number of ovarian tissues were used for the microarray analyses (50 normal ovaries, 20 serous papillary ovarian tumors, and 17 ovarian tumors metastatic to the omentum). By analyzing a large number of tissues, a more accurate picture of ovarian carcinoma gene profiles can be obtained. Earlier studies used fewer ovarian carcinoma tissues or cell lines in their large-scale analyses; the resulting gene expression profiles may have been skewed because of the expression of genes that were altered during the perpetuation of the cell lines.10,20,21
A second advantage of this study was that protein expression was verified by using a relatively large number of ovarian tissues samples (15 normal ovaries, 15 ovarian carcinoma tumors, and 15 ovarian carcinomas metastatic to the omentum). In one earlier study, validation by immunohistochemistry was performed on 13 ovarian tumors, but no normal ovary tissues were similarly screened.6
A third advantage of this study is that 321 tissue samples from 24 other sites were analyzed, including: endometrial adenocarcinoma, lung adenocarcinoma, kidney carcinoma, squamous cell carcinoma of the cervix, squamous cell carcinoma of the lung, and colon adenocarcinoma. By comparison, in one earlier study, only seven tissue samples from other sites were included in the cDNA hybridization analysis.9 However, the majority of previously published ovarian carcinoma gene expression studies did not analyze any other type of tissue except for ovarian tissues and/or cell lines.14–18 Others have noted that a key step in determining the diagnostic potential of gene expression profiling is to compare the gene expression of a variety of tumors derived from many different organs.40
One potential shortcoming of gene expression studies on ovarian carcinoma, the present study included, is the limited quantity of normal ovarian surface epithelium available for microarray and immunohistochemical analysis. Although controversial, it is widely accepted that epithelial ovarian carcinomas arise from the thin layer of epithelial cells surrounding the ovary.23 Not surprisingly, it is difficult to obtain sufficient quantities of surface epithelial cells for further analysis. Therefore, the surface epithelial cells represented a very low percentage of the total normal ovary cells that are included in the microarray analysis. Some groups have circumvented this problem by enriching for the surface ovarian epithelial cells by creating a short-term ovarian surface epithelium cell culture.6,10,11 Others have immortalized normal ovarian surface epithelial cells with SV40 large T-antigen41 or telomerase.42 Zorn and colleagues42 have recently shown that the development and maintenance of normal ovarian surface epithelial cell lines alter the gene expression pattern when compared to whole normal ovaries or brushings taken from the surface of fresh normal ovaries. For these reasons, in this study we used more than 300 other tissues to determine the specificity of the up-regulated genes to ovarian carcinoma, and we verified our findings by immunohistochemistry. It should be noted that benign ovarian epithelial tumors10 or laser capture microdissection followed by amplification may provide alternative sources of RNA for gene expression analysis.43,44
In our study, seven genes selected for further analysis were: the β8 integrin subunit, BMP-7, claudin-4, COL IX α2, CRABP-1, FOX J1, and S100A1. Immunohistochemical staining of 45 ovarian tissues for the presence and localization of the proteins corresponding to each of the genes, followed by statistical analysis, revealed that the β8 integrin subunit, claudin-4, and S100A1 are the most promising candidate ovarian carcinoma tumor markers.
The β8 integrin subunit is a member of the family of integrins that mediates cell-cell and cell-ECM interactions.45 The β8 integrin subunit protein has been reported in mice and rat neural synapses, suggesting its potential role in synaptic function.45 A recent study by Mu and colleagues46 revealed that the β8 integrin subunit binds the cytokine transforming growth factor-β, leading to changes in cell growth and matrix production, and thus regulating epithelial cell homeostasis. Another study revealed that the αVβ8 integrin may complex with the ECM components laminin and fibronectin, and that these interactions may play a role in human glial cell invasion.47 Our study is the first to report and validate the expression of the β8 integrin subunit mRNA and protein in ovarian carcinoma. Another study reported up-regulation of β8 integrin subunit mRNA in highly differentiated serous ovarian adenocarcinomas compared to benign serous adenocarcinomas, but this was not verified by a second technique.7 It is possible that overexpression of the β8 integrin subunit in ovarian carcinoma may enhance tumor cell adhesion and stabilize contacts between the epithelial tumor cells, thus enabling further progression of the disease.
Claudin-4, a member of the claudin family of tight junction proteins,35 also showed promise as a candidate biomarker of ovarian carcinoma. Overexpression of claudin-4 mRNA has been demonstrated in several types of cancer including pancreatic48 and prostate cancer.49 In addition, claudin-4 has recently been reported to be overexpressed in ovarian carcinoma by two other groups.6,13 Based on its role as a tight junction protein, it is possible that overexpression of claudin-4 in ovarian tumor cells may enhance and stabilize tumor cell connections, and could contribute to increased growth at secondary sites.
S100A1 is 1 of 19 members that make up the S100 protein family.50 S100 proteins are localized in the cytoplasm of a variety of cells, and are involved in cell cycle progression and differentiation.50 Several studies have shown that the S100A1 protein is highly expressed in the heart, and that the protein plays a key role in a variety of myocardial functions.51,52 S100A1 proteins are also involved in the assembly and disassembly of microtubules and intermediate filaments.53 Several members of the S100 protein family have been shown to promote invasion and metastasis of many human cancers.50 S100A1, S100A2, and S100B proteins have been detected in epithelial skin tumors.54 Expression of S100A4 has been demonstrated in many different human cancers, including pancreatic cancer, gastric adenocarcinoma, breast carcinomas, and colorectal cancer.50,55 A yeast two-hybrid system has demonstrated a strong interaction between S100A4 and S100A1, suggesting that S100A1 may mediate the metastatic capabilities of S100A4.56 In addition, the S100A2 gene was reported and confirmed to be up-regulated in ovarian carcinoma tissues.5 Our study confirms that of Su and colleagues22 in reporting that S100A1 mRNA is overexpressed in ovarian carcinoma tissues. However, our study is the first to show that S100A1 protein expression is up-regulated in ovarian carcinoma tissues compared to normal ovaries. Our statistical analyses revealed that S100A1 protein expression could be used to distinguish ovarian carcinoma tissues from normal ovary tissues.
A fourth protein that we studied, BMP-7, is a member of the transforming growth factor-β cytokine family.34 Bone morphogenetic proteins are involved in tissue differentiation, development, and remodeling.57 BMP-7 is expressed in articular cartilage,58 where it induces cartilage and bone formation.59 BMP-7 is also expressed in the kidney and may induce kidney epithelial cell differentiation.60,61 Several bone morphogenetic proteins have been implicated in various forms of cancer. BMP-4 mRNA was overexpressed in poorly differentiated gastric cancer cell lines,62 and BMP-4, -5, and -6 mRNA were overexpressed in colon cancer cells.63,64 BMP-7 mRNA has been shown to be up-regulated in osteosarcoma and in some breast and prostate tumors.65–67 However, the exact role of bone morphogenetic proteins in various cancers has yet to be elucidated. This study is the first to report that BMP-7 mRNA is up-regulated in ovarian carcinoma tissue samples, but we were unable to verify the presence of the corresponding protein by immunohistochemistry. Among the few ovarian carcinoma tissues in which the BMP-7 protein was detected, it was occasionally found in the stromal cells surrounding the tumor nests. A possible explanation for this finding is that ovarian carcinoma tumor cells may induce the expression of various factors in the surrounding stromal cells, thus forcing the stromal cells to participate in tumor invasion and metastasis.68
Another protein that we observed to be up-regulated in ovarian carcinoma was COL IX α2, one of three different α chains that combine to form the heterotrimer type IX collagen.36 Type IX collagen is an ECM protein and is a major component of hyaline cartilage. Type IX collagen forms cross-links between type II collagen and other type IX collagen molecules.69 Several studies have identified COL IX α2 mutations that give rise to multiple epiphyseal dysplasia.70 Collagens type I, III, and IV have all been implicated in ovarian cancer.71–73 Interestingly, one study postulated that type IV collagen and BMP-2 may play a role in ovarian cancer.74 The accelerated synthesis and breakdown of type I and type III collagen was shown to be characteristic of ovarian cancer.72 Moser and colleagues75 reported that ovarian epithelial carcinoma cells exhibit preferential adhesion to type I collagen, and that this interaction may stimulate the production of other factors that promote the dissemination of ovarian cancer. Perhaps in a similar manner, COL IX α2 may interact with ovarian carcinoma cells in such a way to promote ovarian tumorigenesis. Alternatively, changes in expression of the COL IX α2 protein may lead to the disruption of the ECM, enabling enhanced tumor cell migration and invasion. Although this study is the first to report the up-regulation of COL IX α2 mRNA in ovarian carcinoma compared to normal ovaries, attempts to confirm this specificity by immunohistochemistry were not possible because of a high degree of nonspecific staining. Because of a lack of additional commercially available antibodies against COL IX α2 or COL IX, it was not possible to verify our gene expression data. Clearly, additional studies are necessary in assessing the role of COL IX α2 in ovarian carcinoma.
CRABP-1, a carrier protein known to mediate the transport and biological activity of retinoic acid,37 also showed some promise as an ovarian carcinoma marker. Recent studies have revealed CRABP-1 mRNA expression in mouse cerebellum and rat lung,76,77 and CRAPB-1 protein expression in chick retina.78 However, few studies have examined CRABP-1 expression in normal human tissues. The expression of cellular retinoic acid-binding proteins has been evaluated in several human cancers. One study reported that changes in CRABP-2 gene expression affected retinoic acid-mediated target gene response, resulting in phenotypic alterations in various squamous carcinoma cells.79 Other studies have detected CRABP-1 protein in human cervical carcinoma tissues80 and large bowel cancer.81 Ono and colleagues12 reported the presence of the CRABP-1 gene in ovarian carcinoma tissues, but this finding was not confirmed by a second method. In our study, we only detected the CRABP-1 protein in a few of the ovarian carcinoma tissues by immunohistochemistry. It is possible that the CRABP-1 mRNA may not be translated into a protein product. Alternatively, if the cells have a high protein turnover rate, then the protein product may not be detected despite its continual production. Whether CRABP-1 plays a role in ovarian carcinoma remains to be determined.
We also studied FOX J1, a transcription factor that belongs to the winged helix/forkhead gene family.38 Members of this family are thought to be involved in cell-specific differentiation.38 FOX J1 is present in ciliated cells of the human lung, oviduct, testis, and brain cortex, suggesting a possible role for FOX J1 in regulating axonemal structural proteins.38 FOX J1 may also play a role in the determination of left-right asymmetry,82 ciliated cell development,82 liver metabolism in humans,83 lung morphogenesis,84 and lung epithelial cell differentiation in mice.84 Other studies have demonstrated the expression of FOX J1 in lung epithelial cells84 and hepatocellular carcinoma.83,85 This study is the first to report the overexpression of FOX J1 mRNA in ovarian carcinoma compared with normal ovaries. Interestingly, FOX J1 was the only gene of the seven we studied that was found to be significantly up-regulated (over threefold) in ovarian carcinoma compared to ovarian carcinoma metastatic to the omentum. Contrary to the RNA expression data, immunohistochemical analysis revealed that the FOX J1 protein was found in the majority of the ovarian tissues examined, including a majority of the normal ovaries, suggesting nonspecific staining. Because of the lack of an additional commercially available Ab against FOX J1, we were not able to further test this finding.
Further studies are needed to assess the ability to use the β8 integrin subunit, claudin-4, and S100A1 as tumor markers alone, or in combination with other markers such as CA-1252,3 in the detection of patients with ovarian carcinoma. It is possible these three proteins may be secreted by ovarian carcinoma cells, and that the proteins may be soluble in bodily fluids. In particular, two of the three proteins that proved by immunohistochemistry to be overexpressed in ovarian carcinoma tissues compared to normal ovaries (β8 integrin and claudin-4) are cell surface proteins. These two proteins are involved in cell-cell or cell-extracellular matrix interactions. Based on the examples of CA-1252,3 and mesothelin,86 these proteins may be cleaved from the surface of ovarian carcinoma cells and be present in the serum of patients. The third protein that proved by immunohistochemistry to be overexpressed in ovarian carcinoma tissues compared to normal ovaries, S100A1, is a cytoplasmic protein that is involved in the cell cycle. By immunohistochemistry, we observed that S100A1 was also present in membranous areas of ovarian carcinoma tissues, not just cytoplasmic areas. Thus, S100A1 may also be secreted by ovarian carcinoma cells. Accessible fluids such as serum and urine, as well as ascites fluid, will be analyzed in future studies for the presence of these three proteins.
The results of this study emphasize the usefulness of microarray analysis in elucidating the genetic profiles of ovarian carcinoma. By comparing the gene expression profiles of ovarian carcinoma tissues to those of a variety of other normal and malignant tissues, genes that are unique and specific to ovarian carcinoma may be identified. These genes may be further analyzed in subsequent studies in an attempt to obtain new and biologically relevant information about the molecular mechanisms involved in ovarian carcinogenesis. In addition, some of the proteins whose presence was confirmed in ovarian carcinoma samples may be studied as potential ovarian carcinoma tumor markers and may contribute to the diagnosis and/or treatment of ovarian carcinoma.
Acknowledgments
We thank Diane Rauch and Sarah Bowell of the University of Minnesota Cancer Center’s Tissue Procurement Facility for assistance in collecting and processing the human tissue samples; the staff of Gene Logic Inc., Gaithersburg, MD, for performing the gene expression experiments with the human tissue samples; Dr. Stephen Nishimura, University of California, San Francisco, CA, for generously providing the mAb against the β8 integrin subunit; Dr. Leo Furcht for providing the mAb P5D2 against the β1 integrin subunit; and the University of Minnesota Supercomputing Institute for providing computing resources.
Footnotes
Address reprint requests to Amy P.N. Skubitz, Ph.D., Department of Laboratory Medicine and Pathology, University of Minnesota, MMC 609, 420 Delaware St. S.E., Minneapolis, MN 55455. E-mail: skubi002@umn.edu.
Supported by grants from the Department of the Army (DA/DAMD17-99-1-9564), the National Institutes of Health/National Cancer Institute (CA0913825), the Minnesota Medical Foundation, and the Minnesota Ovarian Cancer Alliance.
The content of the information presented in this manuscript does not necessarily reflect the position of the United States Government.
This work was presented, in part, at the Marsha Rivkin Center for Ovarian Cancer Research “Fourth Biennial Ovarian Carcinoma Research Symposium: Integration of Research and Treatment” held September 19 to 20, 2002, in Seattle, WA. Portions of this manuscript were submitted as a thesis in partial fulfillment of the requirements for the degree of Master of Science (K.H.) and a Master of Science joint degree in Law, Health, and the Life Sciences (J.J.T).
References
- Ahmedin J, Murray T, Samuels A, Ghafoor A, Ward E, Thun MJ. Cancer statistics. CA Cancer J Clin. 2003;53:5–26. doi: 10.3322/canjclin.53.1.5. [DOI] [PubMed] [Google Scholar]
- Verheijen RHM, von Mensdorff-Pouilly S, van Kamp GJ, Kenemans P. CA 125: fundamental and clinical aspects. Cancer Biol. 1999;9:117–124. doi: 10.1006/scbi.1998.0114. [DOI] [PubMed] [Google Scholar]
- Bast Jr RC, Urban N, Shridhar V, Smith D, Zhang Z, Skates S, Lu K, Liu J, Fishman D, Mills G: Early detection of ovarian cancer: promise and reality. Ovarian Cancer. Edited by Stack MS, Fishman DA. Cancer Treatment and Research, vol 107 Edited by Rosen ST (Series Editor). Boston, Kluwer Academic Publishers, 2002, pp 61–97 [DOI] [PubMed] [Google Scholar]
- Welsh JB, Zarrinkar PP, Sapinoso LM, Kern SG, Behling CA, Monk BJ, Lockhart DJ, Burger RA, Hampton GM. Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc Natl Acad Sci USA. 2001;98:1176–1181. doi: 10.1073/pnas.98.3.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough CD, Cho KR, Zonderman AB, Schwartz DR, Morin PJ. Coordinately up-regulated genes in ovarian cancer. Cancer Res. 2001;61:3869–3876. [PubMed] [Google Scholar]
- Hough CD, Sherman-Baust CA, Pizer ES, Montz FJ, Im DD, Rosenshein NB, Cho KR, Riggins GJ, Morin PJ. Large-scale serial analysis of gene expression reveals genes differentially expressed in ovarian cancer. Cancer Res. 2000;60:6281–6287. [PubMed] [Google Scholar]
- Tapper J, Kettunen E, El-Rifai W, Seppala M, Andersson L, Knuutila S. Changes in gene expression during progression of ovarian carcinoma. Cancer Genet Cytogenet. 2001;128:1–6. doi: 10.1016/s0165-4608(01)00386-7. [DOI] [PubMed] [Google Scholar]
- Wang K, Gan L, Jeffery E, Gayle M, Gown AM, Skelly M, Nelson PS, Ng WV, Schummer M, Hood L, Mulligan J. Monitoring gene expression profile changes in ovarian carcinomas using cDNA microarray. Gene. 1999;229:101–108. doi: 10.1016/s0378-1119(99)00035-9. [DOI] [PubMed] [Google Scholar]
- Schummer M, Ng WV, Bumgarner RE, Nelson PS, Schummer B, Bednarski DW, Hassell L, Baldwin RL, Karlan BY, Hood L. Comparative hybridization of an array of 21,500 ovarian cDNA’s for the discovery of genes overexpressed in ovarian carcinomas. Gene. 1999;238:375–385. doi: 10.1016/s0378-1119(99)00342-x. [DOI] [PubMed] [Google Scholar]
- Ismail RS, Baldwin RL, Fang J, Browning D, Karlan BY, Gasson JC, Chang DD. Differential gene expression between normal and tumor-derived ovarian epithelial cells. Cancer Res. 2000;60:6744–6749. [PubMed] [Google Scholar]
- Mok SC, Chao J, Skates S, Wong K, Yiu GK, Muto MG, Berkowitz RS, Cramer DW. Prostatin, a potential serum marker for ovarian cancer: identification through microarray technology. J Natl Cancer Inst. 2001;93:1458–1464. doi: 10.1093/jnci/93.19.1458. [DOI] [PubMed] [Google Scholar]
- Ono K, Tanaka T, Tsunoda T, Kitahara O, Kihara C, Okamoto A, Ochiai K, Takagi T, Nakamura Y. Identification by cDNA microarray of genes involved in ovarian carcinogenesis. Cancer Res. 2000;60:5007–5011. [PubMed] [Google Scholar]
- Shridhar V, Lee J, Pandita A, Iturria S, Avula R, Staub J, Morrissey M, Calhoun E, Sen A, Kalli K, Keeney G, Roche P, Cliby W, Lu K, Schmandt R, Mills GB, Bast RC, Jr, James D, Couch FJ, Hartmann LC, Lillie J, Smith DL. Genetic analysis of early- versus late-stage ovarian tumors. Cancer Res. 2001;61:5895–5904. [PubMed] [Google Scholar]
- Bayani J, Brenton JD, Macgregor PF, Beheshti B, Albert M, Nallainathan D, Karaskova J, Rosen B, Murphy J, Laframboise S, Zanke B, Squire JA. Parallel analysis of sporadic primary ovarian carcinomas by spectral karyotyping, comparative genomic hybridization, and expression microarrays. Cancer Res. 2002;62:3466–3476. [PubMed] [Google Scholar]
- Martoglio A, Tom BDM, Starkey M, Corps AN, Charnock-Jones DS, Smith SK. Changes in tumorigenesis and angiogenesis-related gene transcript abundance profiles in ovarian cancer detected by tailored high density cDNA arrays. Mol Med. 2000;6:750–765. [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Moore DH, Ginzinger DG, Godfrey TE, Barclay J, Powell B, Pinkel D, Zaloudek C, Lu K, Mills G, Berchuck A, Gray JW. An approach to analysis of large-scale correlations between genome changes and clinical endpoints in ovarian cancer. Cancer Res. 2000;60:5382–5385. [PubMed] [Google Scholar]
- Van Niekerk CC, Boerman OC, Ramaekers FCS, Poels LG. Marker profile of different phases in the transition of normal human ovarian epithelium to ovarian carcinomas. Am J Pathol. 1991;138:455–463. [PMC free article] [PubMed] [Google Scholar]
- Van Niekerk CC, Ramaekers FCS, Hanselaar GJM, Aldeweireldt J, Poels LG. Changes in expression of differentiation markers between normal ovarian cells and derived tumors. Am J Pathol. 1993;142:157–177. [PMC free article] [PubMed] [Google Scholar]
- Shvartsman HS, Lu KH, Lee J, Lillie J, Deavers MT, Clifford SL, Wolf J, Mills GB, Bast RC, Gershenson DM, Schmandt RE: Over-expression of NES-1 in epithelial ovarian carcinoma. Society of Gynecology 32nd Annual Meeting, Abstract p. 103 [Google Scholar]
- Tonin PN, Hudsno TJ, Rodier F, Bossolasco M, Lee PD, Novak J, Mandreson EN, Provencher D, Mes-Masson A. Microarray analysis of gene expression mirrors the biology of ovarian cancer model. Oncogene. 2001;20:6617–6626. doi: 10.1038/sj.onc.1204804. [DOI] [PubMed] [Google Scholar]
- Wong KK, Cheng RS, Mok SC. Identification of differentially expressed genes from ovarian cancer cells by MICROMAX cDNA microarray system. Biotechniques. 2001;30:670–675. doi: 10.2144/01303dd05. [DOI] [PubMed] [Google Scholar]
- Su AI, Welsh JB, Sapinoso LM, Kern SG, Dimitrov P, Lapp H, Schultz PG, Powell SM, Moskaluk CA, Frierson HF, Jr, Hampton GM. Molecular classification of human carcinomas by use of gene expression signatures. Cancer Res. 2001;61:7388–7393. [PubMed] [Google Scholar]
- Dubeau L. The cell of origin of ovarian epithelial tumors and the ovarian surface epithelium dogma: does the emperor have no clothes? Gynecol Oncol. 1999;72:437–442. doi: 10.1006/gyno.1998.5275. [DOI] [PubMed] [Google Scholar]
- Feng H, Ghazizadeh M, Konishi H, Araki T. Expression of MUC1 and MUC2 mucin gene products in human ovarian carcinomas. Jpn J Clin Oncol. 2002;32:525–529. doi: 10.1093/jjco/hyf111. [DOI] [PubMed] [Google Scholar]
- Alaiya AA, Franzen B, Hagman A, Silfversward C, Moberger B, Linder S, Auer G. Classification of human ovarian tumors using multivariate data analysis of polypeptide expression patterns. Int J Cancer. 2000;86:731–736. doi: 10.1002/(sici)1097-0215(20000601)86:5<731::aid-ijc20>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, Mills GB, Simone C, Fishman DA, Kohn EC, Liotta LA. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- Skubitz KM, Skubitz APN. Differential gene expression in renal-cell cancer. J Lab Clin Med. 2002;140:52–64. doi: 10.1067/mlc.2002.125213. [DOI] [PubMed] [Google Scholar]
- Skubitz KM, Skubitz APN. Differential gene expression in uterine leiomyoma. J Lab Clin Med. 2003;141:297–308. doi: 10.1016/S0022-2143(03)00007-6. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skubitz APN, Bast RC, Jr, Wayner EA, Letourneau PC, Wilke MS. Expression of α6 and β4 integrins in serous ovarian carcinoma correlates with expression of the basement membrane protein laminin. Am J Pathol. 1996;148:1445–1461. [PMC free article] [PubMed] [Google Scholar]
- Casey RC, Burleson KM, Skubitz KM, Pambuccian SE, Oegema TR, Jr, Ruff LE, Skubitz APN. β1-integrins regulate the formation and adhesion of ovarian carcinoma multicellular spheroids. Am J Pathol. 2001;159:2071–2080. doi: 10.1016/s0002-9440(10)63058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galmozzi E, Tomassetti A, Sforzini S, Mangiarotti F, Mazzi M, Nachmanoff K, Elwood PC, Canevari S. Exon 3 of the α folate receptor gene contains a 5′ splice site which confers enhanced ovarian carcinoma specific expression. FEBS Lett. 2001;502:31–34. doi: 10.1016/s0014-5793(01)02659-x. [DOI] [PubMed] [Google Scholar]
- Moyle M, Napier MA, McLean JW. Cloning and expression of a divergent integrin subunit β8. J Biol Chem. 1991;266:19650–19658. [PubMed] [Google Scholar]
- Ozkaynak E, Rueger DC, Drier EA, Corbett C, Ridge RJ, Sampath TK, Oppermann H. OP-1 cDNA encodes an osteogenic protein in the TGF-beta family. EMBO J. 1990;9:2085–2093. doi: 10.1002/j.1460-2075.1990.tb07376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K, Furuse M, Fujimoto K, Tsukita S. Claudin multigene family encoding four-transmembrane domain protein components of tight junction strands. Proc Natl Acad Sci USA. 1999;96:511–516. doi: 10.1073/pnas.96.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perala M, Hanninen M, Hastbacka J, Elima K, Vuorio E. Molecular cloning of the human alpha-2 (IX) collagen cDNA and assignment of the human COL9A2 gene to chromosome 1. FEBS Lett. 1993;319:177–180. doi: 10.1016/0014-5793(93)80062-y. [DOI] [PubMed] [Google Scholar]
- Cornic M, Guidez F, Delva L, Agadir A, Degos L, Chomienne C. Mechanism of action of retinoids in a new therapeutic approach to acute promyelocytic leukemia. Bull Cancer. 1992;79:697–704. [PubMed] [Google Scholar]
- Murphy DB, Seeman S, Wiese S, Kirschner R, Grzeschik KH, Thies U. The human hepatocyte nuclear factor 3/fork head gene FKHL13: genomic structure and pattern of expression. Genomics. 1997;40:462–469. doi: 10.1006/geno.1996.4587. [DOI] [PubMed] [Google Scholar]
- Schafer BW, Wicki R, Engelkamp D, Mattei M-G, Heizmann CW. Isolation of a YAC clone covering a cluster of nine S100 genes on human chromosome 1q21: rationale for a new nomenclature of the S100 calcium-binding protein family. Genomics. 1995;25:638–643. doi: 10.1016/0888-7543(95)80005-7. [DOI] [PubMed] [Google Scholar]
- Giordano TJ, Shedden KA, Schwartz DR, Kuick R, Taylor JMG, Lee N, Misek DE, Greenson JK, Kardia SLR, Beer DG, Rennert G, Cho KR, Gruber SB, Fearon ER, Hanash S. Organ-specific molecular classification of primary lung, colon, and ovarian adenocarcinomas using gene expression profiles. Am J Pathol. 2001;159:1231–1238. doi: 10.1016/S0002-9440(10)62509-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazaeri AA, Yee CJ, Sotiriou C, Brantley KR, Boyd J, Liu ET. Gene expression profiles of BRCA1-linked, BRCA2-linked, and sporadic ovarian cancers. J Natl Cancer Inst. 2002;94:990–1000. doi: 10.1093/jnci/94.13.990. [DOI] [PubMed] [Google Scholar]
- Zorn KK, Jazaeri AA, Awtrey CS, Gardner GJ, Mok SC, Boyd J, Birrer MJ. Choice of normal ovarian control influences determination of differentially expressed genes in ovarian cancer expression profiling studies. Clin Cancer Res. 2003;9:4811–4818. [PubMed] [Google Scholar]
- Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- Luo L, Salunga RC, Guo H, Bittner A, Joy KC, Galindo JE, Xiao H, Rogers KE, Wan JS, Jackson MR, Erlander MG. Gene expression profiles of laser-captured adjacent neuronal subtypes. Nat Med. 1999;5:117–122. doi: 10.1038/4806. [DOI] [PubMed] [Google Scholar]
- Nishimura SL, Boylen KP, Einheber S, Milner TA, Ramos DM, Pytela R. Synaptic and glial localization of the integrin αvβ8 in mouse and rat brain. Brain Res. 1998;791:271–282. doi: 10.1016/s0006-8993(98)00118-8. [DOI] [PubMed] [Google Scholar]
- Mu D, Cambier S, Fjellbirkeland L, Baron JL, Munger JS, Kawakatsu H, Sheppard D, Broaddus VC, Nishimura SL. The integrin αvβ8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-β1. J Cell Biol. 2002;157:493–507. doi: 10.1083/jcb.200109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belot N, Rorive S, Doyen I, LeFranc F, Bruyneel E, Dedecker R, Micik S, Brotchi J, Decaestecker C, Salmon I, Kiss R, Camby I. Molecular characterization of cell substratum attachments in human glial tumors relates to prognostic features. Glia. 2001;36:375–390. doi: 10.1002/glia.1124. [DOI] [PubMed] [Google Scholar]
- Terris B, Blaveri E, Crnogorac-Jurcevic T, Jones M, Missiaglia E, Ruszniewski P, Sauvanet A, Lemoine NR. Characterization of gene expression profiles in intraductal papillary-mucinous tumors of the pancreas. Am J Pathol. 2002;160:1745–1754. doi: 10.1016/S0002-9440(10)61121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H, Crean CD, Lee WH, Cummings OW, Gabig TG. Expression of Clostridium perfringens enterotoxin receptors claudin-3 and claudin-4 in prostate cancer epithelium. Cancer Res. 2001;61:7878–7881. [PubMed] [Google Scholar]
- Mazzucchelli L. Protein S100A4: too long overlooked by pathologists? Am J Pathol. 2002;160:7–13. doi: 10.1016/S0002-9440(10)64342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett W, Mandinova A, Remppis A, Sauder U, Ruter F, Heizmann CW, Aebi U, Zerkowski HR. Translocation of S100A1(1) calcium binding protein during heart surgery. Biochem Biophys Res Comm. 2001;284:698–703. doi: 10.1006/bbrc.2001.4996. [DOI] [PubMed] [Google Scholar]
- Most P, Bernotat J, Ehlermann P, Pleger ST, Reppel M, Borries M, Niroomand F, Pieske B, Janssen PML, Eschenhagen T, Karczewski P, Smith GL, Koch WJ, Katus HA, Remppis A. S100A1: a regulator of myocardial contractility. Proc Natl Acad Sci USA. 2001;98:13889–13894. doi: 10.1073/pnas.241393598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorci G, Agneletti AL, Donato R. Effects of S100A1 and S100B on microtubule stability. An in vitro study using Triton-cytoskeletons from astrocyte and myoblast cell lines. Neuroscience. 2000;99:773–783. doi: 10.1016/s0306-4522(00)00238-4. [DOI] [PubMed] [Google Scholar]
- Shrestha P, Muramatsu Y, Kudeken W, Mori M, Takai Y, Ilg EC, Schafer BW, Heizmann CW. Localization of Ca(2+)-binding S100 proteins in epithelial tumours of the skin. Virchows Arch. 1998;432:53–59. doi: 10.1007/s004280050134. [DOI] [PubMed] [Google Scholar]
- Rosty C, Ueki T, Argani P, Jansen M, Yeo CJ, Cameron JL, Hruban RH, Goggins M. Overexpression of S100A4 in pancreatic ductal adenocarcinomas is associated with poor differentiation and DNA hypomethylation. Am J Pathol. 2002;160:45–50. doi: 10.1016/S0002-9440(10)64347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Rudland PS, Whites MR, Barraclough R. Interaction in vivo and in vitro of the metastatic-inducing S100 protein, S100A4 (p9Ka) with S100A1. J Biol Chem. 2000;275:11141–11146. doi: 10.1074/jbc.275.15.11141. [DOI] [PubMed] [Google Scholar]
- Yamamoto T, Saatcioglu F, Matsuda T. Cross-talk between bone morphogenic proteins and estrogen receptor signaling. Endocrinology. 2002;143:2635–2642. doi: 10.1210/endo.143.7.8877. [DOI] [PubMed] [Google Scholar]
- Rajshankar D, McCulloch CA, Tenenbaum HC, Lekic PC. Osteogenic inhibition by rat periodontal ligament cells: modulation of bone morphogenic protein-7 activity in vivo. Cell Tissue Res. 1998;294:475–483. doi: 10.1007/s004410051199. [DOI] [PubMed] [Google Scholar]
- Hidaka C, Quitoriano M, Warren RF, Crystal RG. Enhanced matrix synthesis and in vitro formation of cartilage-like tissue by genetically modified chondrocytes expressing BMP-7. J Orthopaedic Res. 2001;19:751–758. doi: 10.1016/S0736-0266(01)00019-5. [DOI] [PubMed] [Google Scholar]
- Simon M, Maresh JG, Harris SE, Hernandez JD, Arar M, Olson MS, Abboud HE. Expression of bone morphogenetic protein-7 mRNA in normal and ischemic adult rat kidney. Am J Physiol. 1999;276:F382–F389. doi: 10.1152/ajprenal.1999.276.3.F382. [DOI] [PubMed] [Google Scholar]
- Ishibashi K, Sasaki S, Akiba T, Marumo F. Expression of bone morphogenic protein 7 mRNA in MDCK cells. Biochem Biophys Res Comm. 1993;193:235–239. doi: 10.1006/bbrc.1993.1614. [DOI] [PubMed] [Google Scholar]
- Katoh M, Terada M. Overexpression of bone morphogenic protein (BMP)-4 mRNA in gastric cancer cell lines of poorly differentiated type. J Gastroenterol. 1996;31:137–139. doi: 10.1007/BF01211202. [DOI] [PubMed] [Google Scholar]
- Kim JS, Crooks H, Dracheva T, Nishanian TG, Singh B, Jen J, Waldman T. Oncogenic beta-catenin is required for bone morphogenetic protein 4 expression in human cancer cells. Cancer Res. 2002;62:2744–2748. [PubMed] [Google Scholar]
- Imai N, Iwai A, Hatakeyama S, Masuzaki K, Kitagawa Y, Kato S, Hokari R, Kawaguchi A, Nagao S, Miyahara T, Itoh K, Miura S. Expression of bone morphogenetic proteins in colon carcinoma with heterotopic ossification. Pathol Int. 2001;51:643–648. doi: 10.1046/j.1440-1827.2001.01243.x. [DOI] [PubMed] [Google Scholar]
- Sulzbacher I, Birner P, Trieb K, Pichlbauer E, Lang S. The expression of bone morphogenetic proteins in osteosarcoma and its relevance as a prognostic parameter. J Clin Pathol. 2002;55:381–385. doi: 10.1136/jcp.55.5.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber KL, Bolander ME, Rock MG, Pritchard D, Sarkar G. Evidence for the upregulation of osteogenic protein-1 mRNA expression in musculoskeletal neoplasms. J Orthopaedic Res. 1998;16:8–14. doi: 10.1002/jor.1100160103. [DOI] [PubMed] [Google Scholar]
- Lin DL, Tarnowski CP, Zhang J, Dai J, Rohn E, Patel AH, Morris MD, Keller ET. Bone metastatic LNCaP-derivative C4–2B prostate cancer cell line mineralizes in vitro. Prostate. 2001;47:212–221. doi: 10.1002/pros.1065. [DOI] [PubMed] [Google Scholar]
- Weber BL. Cancer genomics. Cancer Cell. 2002;1:37–47. doi: 10.1016/s1535-6108(02)00026-0. [DOI] [PubMed] [Google Scholar]
- Diab M, Wu JJ, Eyre DR. Collagen type IX from human cartilage: a structural profile of intermolecular cross-linking sites. Biochem J. 1996;314:327–332. doi: 10.1042/bj3140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden P, Canty EG, Mortier GR, Zabel B, Spranger J, Carr A, Grant ME, Loughlin JA, Briggs MD. Identification of novel pro-alpha-2(IX) collagen gene mutations in two families with distinctive oligo-epiphyseal forms of multiple epiphyseal dysplasia. Am J Hum Genet. 1999;65:31–38. doi: 10.1086/302440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolanska M, Sobolewski K, Drozdzewicz M. Extracellular matrix components in ovarian tumors. Ginekologia Polska. 1999;70:179–185. [PubMed] [Google Scholar]
- Santala M, Risteli J, Risteli L, Puistola U, Kacinski BM, Stanley ER, Kauppila A. Synthesis and breakdown of fibrillar collagens: concomitant phenomena in ovarian cancer. Br J Cancer. 1998;77:1825–1831. doi: 10.1038/bjc.1998.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu GG, Risteli J, Puistola U, Kauppila A, Ristela L. Progressive ovarian carcinoma induces synthesis of type I and type III procollagens in the tumor tissue and peritoneal cavity. Cancer Res. 1993;53:5028–5032. [PubMed] [Google Scholar]
- Kiyozuka Y, Nakagawa H, Senzaki H, Uemura Y, Adachi S, Teramoto Y, Matsuyama T, Bessho K, Tsubura A. Bone morphogenetic protein-2 and type IV collagen expression in psammoma body forming ovarian cancer. Anticancer Res. 2001;21:1723–1730. [PubMed] [Google Scholar]
- Moser TL, Pizzo SV, Bafetti LM, Fishman DA, Stack MS. Evidence for preferential adhesion of ovarian epithelial carcinoma cells to type I collagen mediated by the alpha2beta1 integrin. Int J Cancer. 1996;67:695–701. doi: 10.1002/(SICI)1097-0215(19960904)67:5<695::AID-IJC18>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Parenti R, Wassef M, Cicirata F. Expression of CRABP I mRNA in fastigial cells of the developing cerebellum. Eur J Neurosci. 2002;15:211–215. doi: 10.1046/j.0953-816x.2001.01853.x. [DOI] [PubMed] [Google Scholar]
- Whitney D, Massaro GD, Massaro D, Clerch LB. Gene expression of cellular retinoid-binding proteins: modulation by retinoic acid and dexamethasone in postnatal rat lung. Pediatr Res. 1999;45:2–7. doi: 10.1203/00006450-199901000-00002. [DOI] [PubMed] [Google Scholar]
- Mey J, McCaffery P, Klemeit M. Sources and sink of retinoic acid in the embryonic chick retina: distribution of aldehyde dehydrogenase activities, CRABP-I, and sites of retinoic acid inactivation. Brain Res Dev Brain Res. 2001;127:135–148. doi: 10.1016/s0165-3806(01)00127-4. [DOI] [PubMed] [Google Scholar]
- Vo HP, Crowe DL. Transcriptional regulation of retinoic acid responsive genes by cellular retinoic acid binding protein-II modulates RA mediated tumor cell proliferation and invasion. Anticancer Res. 1998;18:217–224. [PubMed] [Google Scholar]
- Hillemanns P, Tannous-Khuri L, Koulos JP, Talmage D, Wright TC., Jr Localization of cellular retinoid-binding proteins in human cervical intraepithelial neoplasia and invasive carcinoma. Am J Pathol. 1992;141:973–980. [PMC free article] [PubMed] [Google Scholar]
- Di Fronzo G, Cappelletti V, Miodini P, Bertario L, Ravasi G. Role of cellular retinoic acid binding protein (cRABP) in patients with large bowel cancer. Cancer Detect Prevent. 1987;10:327–333. [PubMed] [Google Scholar]
- Chen J, Knowles HJ, Hebert JL, Hackett BP. Mutation of the mouse hepatocyte nuclear factor/forkhead homologue 4 gene results in an absence of cilia and random left-right asymmetry. J Clin Invest. 1998;102:1077–1082. doi: 10.1172/JCI4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naiki T, Nagaki M, Shidoji Y, Kojima H, Imose M, Kato T, Ohishi N, Yagi K, Moriwaki H. Analysis of gene expression profile induced by hepatocyte nuclear factor 4 alpha in hepatoma cells using an oligonucleotide microarray. J Biol Chem. 2002;277:14011–14019. doi: 10.1074/jbc.M105403200. [DOI] [PubMed] [Google Scholar]
- Tichelaar JW, Lim L, Costa RH, Whitsett JA. HNF-3/forkhead homologue-4 influences lung morphogenesis and respiratory epithelial cell differentiation in vivo. Dev Biol. 1999;213:405–417. doi: 10.1006/dbio.1999.9380. [DOI] [PubMed] [Google Scholar]
- Xu L, Hui L, Wang S, Gong J, Jin Y, Wang Y, Ji Y, Wu X, Han Z, Hu G. Expression profiling suggested a regulatory role of liver-enriched transcription factors in human hepatocellular carcinoma. Cancer Res. 2001;61:3176–3181. [PubMed] [Google Scholar]
- Scholler N, Fu N, Yang Y, Ye Z, Goodman GE, Hellstrom KE, Hellstrom I. Soluble member(s) of the mesothelin/megakaryocyte potentiating factor family are detectable in sera from patients with ovarian carcinoma. Proc Natl Acad Sci USA. 1999;96:11531–11536. doi: 10.1073/pnas.96.20.11531. [DOI] [PMC free article] [PubMed] [Google Scholar]