Abstract
Malignant gliomas are uniformly lethal tumors whose morbidity is mediated in large part by the angiogenic response of the brain to the invading tumor. This profound angiogenic response leads to aggressive tumor invasion and destruction of surrounding brain tissue as well as blood-brain barrier breakdown and life-threatening cerebral edema. To investigate the molecular mechanisms governing the proliferation of abnormal microvasculature in malignant brain tumor patients, we have undertaken a cell-specific transcriptome analysis from surgically harvested nonneoplastic and tumor-associated endothelial cells. SAGE-derived endothelial cell gene expression patterns from glioma and nonneoplastic brain tissue reveal distinct gene expression patterns and consistent up-regulation of certain glioma endothelial marker genes across patient samples. We define the G-protein-coupled receptor RDC1 as a tumor endothelial marker whose expression is distinctly induced in tumor endothelial cells of both brain and peripheral vasculature. Further, we demonstrate that the glioma-induced gene, PV1, shows expression both restricted to endothelial cells and coincident with endothelial cell tube formation. As PV1 provides a framework for endothelial cell caveolar diaphragms, this protein may serve to enhance glioma-induced disruption of the blood-brain barrier and transendothelial exchange. Additional characterization of this extensive brain endothelial cell gene expression database will provide unique molecular insights into vascular gene expression.
Malignant brain tumors (gliomas) represent a deadly form of cancer that has seen little survival improvement throughout the last 30 years.1 Despite advances in surgical technique, chemotherapy regimens, and radiation protocols, the median survival for the most malignant and common form of these tumors, glioblastoma multiforme, remains less than 1 year from initial diagnosis. Tumor growth and proliferation is associated with a profound angiogenic response that leads to aggressive invasion and destruction of surrounding brain tissue as well as blood-brain barrier breakdown and life-threatening cerebral edema.1 The vascular microenvironment within gliomas has been studied previously through morphological, circulatory, and perfusion-based experiments.2,3 Increased fenestrations, malperfusion, hyperpermeability, and reduced leukocyte-endothelial cell (EC) interaction are all phenotypic changes that have been demonstrated within glioma microvasculature.4–6 The molecular characterization of glioma ECs has been primarily limited to the evaluation of common growth factor/receptor pathways, previously defined brain EC transporters, or select endothelial markers.7–9 We hypothesized that a comprehensive comparison of gene expression patterns in microvascular brain ECs between nonneoplastic brain and glial tumors would yield new insights into astrocyte-regulated cross talk with brain ECs, brain-specific endothelial differentiation, and the mechanisms by which these systems are disrupted to promote central nervous system neoplasia.
Materials and Methods
Sample Preparation
Five separate brain tissue samples were resected and immediately subjected to EC isolation with slight modifications to the protocol described previously.10 Briefly, samples were surgically excised and submerged in Dulbecco’s modified Eagle’s medium. The samples were minced into 2-cm cubes and subjected to tissue digestion with a collagenase cocktail. Samples were mixed at 37°C until dissolved. Cells were centrifuged and washed two times with phosphate-buffered saline (PBS)/bovine serum albumin (BSA) and filtered through successive nylon mesh filters of 250, 100, and 40 μm. Samples were resuspended in PBS/BSA and applied to a 30% Percoll gradient centrifuging for 15 minutes at 800 × g. Five ml of the top layer of the Percoll gradient was diluted in 50 ml of Dulbecco’s modified Eagle’s medium, and cells were centrifuged, washed with PBS, and resuspended in 3 ml of PBS/BSA. Cells were filtered, centrifuged, and resuspended in 1 ml of PBS/BSA. One hundred μl of prewashed anti-CD45 magnetic beads (Dynal, Brown Deer, WI) were added and the solution allowed to gently mix for 10 minutes. Bead-bound cells were discarded and the supernatant transferred to a fresh microcentrifuge tube. Ten μl of P1H12 mAb (1:100) (brain N1, T1, and T2 samples) or UEA-I lectin (brain N2 and T3 samples) was added and the samples were mixed gently at 4°C for 45 minutes. ECs for samples N1, T1, and T2 were harvested from single cell tumor digests using immunomagnetic beads coated with the EC-specific monoclonal antibody (P1H12), whereas samples N2 and T3 were prepared using beads coated with the endothelial-specific lectin, UEA-I. Dual selection methods were used to ensure that any observed gene expression changes were generalizable across subsets of microvascular ECs and not an artifact of the purification method. Cells were centrifuged and washed three times in PBS/BSA and resuspended in 500 μl of PBS/BSA. Prewashed goat anti-mouse M450 Dynabeads were added to each tube and allowed to mix for 15 minutes at 4°C. Bead-bound cells were washed eight times with PBS/BSA and resuspended in a final volume of 500 μl of PBS. Cells were counted and frozen at −70°C before RNA extraction.
Data Analysis
Differential expression for brain endothelial tags was evaluated as follows: for tumor/normal induction, the median of three tumor samples versus the maximum of two normal samples were compared. For normal/tumor induction ratios, the maximum of three tumor samples versus the minimum of two normal samples were compared. In practice, our data-sorting filters select for two of three tumors showing high expression relative to each of the two normal samples [glioma endothelial markers (GEMs)], or conserved repression in each of the two normal samples relative to each of the three tumor samples (glioma-repressed genes). A Bayesian statistical analysis was used to assess the probability that the observed counts for a given tag represent at least a twofold induction in expression of the corresponding gene.11 To facilitate quantitative comparison with colon endothelial and other SAGE tag data, 17-bp long tags derived from brain endothelial samples were truncated to 10-bp short tags. Multiple long tags corresponding to the same truncated tag were aggregated. Full long tags were used for mapping of differentially expressed tags to reference transcript and genomic sequences. Reference long SAGE tags for known genes were derived from a set of UniGene sequences annotated as 3′ or mRNA, and screened for low confidence sequences.
Microarray Analysis
Custom 50 nucleotide oligomer arrays were constructed containing 606 unique gene elements. The 606 genes were derived from tumor and normal induced genes from both colon and brain data (328 genes), as well as 278 genes from both literature reviews and housekeeping genes. Arrays were interrogated with Cy3 and Cy5 dye-swapped labeled aRNA samples comparing human microvascular endothelial cells (HMVECs) grown on plastic, collagen, fibrin, or Matrigel.
In Situ Hybridizations and Immunohistochemistry
In situ hybridizations for RDC1, PV1, VEGFR2, and von Willebrand factor (vWF) were performed as described previously.10 Co-staining of PV1 and CD31 was performed as follows: four 500 nucleotide riboprobe fragments specific for PV1 were transcribed and used to probe formalin-fixed 5-μm tissue sections. Final detection of the bound riboprobes was delayed until after the CD31 immunohistochemical staining. After PV1 hybridization and washing, tissue sections were fixed for 20 minutes in 4% formaldehyde. After a brief rinse in Tris-buffered saline, antigen retrieval was performed using DAKO target retrieval solution (catalog no. S1699; DAKO, Carpinteria, CA) according to the manufacturer’s instructions. After a 5-minute wash in Tris-buffered saline, slides were digested with Proteinase K at 20 ng/ml in Tris-buffered saline for 20 minutes at 37°C, then blocked for 20 minutes at room temperature in block (10% goat serum/0.5% casein/0.05% Tween-20/PBS). Slides were incubated with DAKO CD31 (catalog no. M0823) at a final concentration of 1 μg/slide in block solution, for 60 minutes at room temperature. After two 5-minute TBST (catalog no. S3306, DAKO) washes at room temperature, PV1 riboprobe and CD31 antibody were detected with streptavidin-Cy2 (catalog no. 016-220-084; Jackson ImmunoResearch, West Grove, PA) at 5 μg/slide for the PV1 riboprobe, and goat anti-mouse-Cy3 (catalog no. 115-165-146, Jackson ImmunoResearch) at 2.5 μg/slide for CD31, for 60 minutes at room temperature. After three 5-minute washes in TBST, the slides were mounted with anti-fade medium containing 4,6-diamidino-2-phenylindole nuclear counterstain, coverslipped and stored at −20°C until viewing. Single images of 4,6-diamidino-2-phenylindole, Cy2, and Cy3 images were acquired separately on a Zeiss Axioplan at ×40 with a Hammamatsu camera, then merged together to form a composite image using universal imaging metamorph software, and stored at −20°C until viewing.
Results
Sample Characterization
Five independent EC populations were purified from glioma tumor tissue (three samples) or nonneoplastic brain tissue (two samples) removed surgically in the course of routine patient care. Whole tumor bulk tissue was used to derive EC populations thus minimizing any contribution from vasculature that may have been within nonhyperplastic regions of the tissue mass. The normal samples used were nonangiogenic EC populations derived from the middle temporal gyrus of patients undergoing temporal lobectomy for intractable epilepsy. Each temporal lobe specimen was radiographically normal based on magnetic resonance imaging and demonstrated no evidence of increased vascularity/angiogenesis on pathological sectioning with hematoxylin and eosin. We subsequently refer to these nonangiogenic samples as normal to reflect the nonneoplastic nature of the tissue. The samples are summarized in Table 1. ECs for samples N1, T1, and T2 were harvested from single cell tumor digests using immunomagnetic beads coated with the EC-specific monoclonal antibody (P1H12), whereas samples N2 and T3 were prepared using beads coated with the endothelial-specific lectin, UEA-I. Dual selection methods were used to ensure that any observed gene expression changes were generalizable across subsets of microvascular ECs and not an artifact of the purification method. Before SAGE analysis, each sample was assessed by reverse transcriptase-polymerase chain reaction (RT-PCR) for the relative mRNA abundance of vWF, glial fibrillary acidic protein, and EF1. Abundant levels of vWF and the control housekeeper EF1, and low levels of the glial cell-specific gene glial fibrillary acidic protein suggested that the cell populations were primarily endothelial (data not shown). The purity of the cell population was further confirmed by the SAGE output, which demonstrated that expression levels of recognized EC marker genes were consistent with SAGE data from clinical colonic ECs, the only other organ system in which comprehensive endothelial gene expression has been examined (Table 2).10 Additionally, markers specific for epithelial, hematopoietic, or glial cells showed limited or no expression in the brain endothelial libraries confirming little contamination from non-EC populations (Table 2). Finally, seven genes previously determined to be EC-specific are represented in each of our brain EC libraries providing further evidence of near pure EC populations (see Huminiecki and Bicknell12 and supporting on-line material, Supplement A at http://ajp.amjpathol.org).
Table 1.
SAGE Libraries of Normal (N) ECs or Tumor ECs (T) Were Derived from Fresh Surgical Samples of Temporal Lobe Cortex or Glioma, Respectively
| Sample | Description | Tags generated | EC selection |
|---|---|---|---|
| Brain N1 | Normal temporal lobectomy ECs | 43,000 | P1H12 |
| Brain N2 | Normal temporal lobectomy ECs | 49,000 | UEA-I |
| Brain T1 | Grade IV glioma ECs | 46,000 | P1H12 |
| Brain T2 | Grade III glioma ECs | 50,000 | P1H12 |
| Brain T3 | Grade IV glioma ECs | 58,000 | UEA-I |
ECs were separated using magnetic beads coated with either an endothelial specific monoclonal antibody, P1H12, or an endothelial specific lectin, UEA-I, as indicated. Sequencing of each library was performed to the indicated tag depth.
Table 2.
Normalized SAGE Tag Frequencies Per 50,000 Total Tags for Selected Cell-Type-Specific Genes
| Gene | Specificity | Brain N1 | Brain N2 | Brain T1 | Brain T2 | Brain T3 | Colon N | Colon T |
|---|---|---|---|---|---|---|---|---|
| Hevin | EC | 51 | 99 | 223 | 121 | 48 | 161 | 69 |
| VWF | EC | 12 | 53 | 37 | 51 | 110 | 35 | 33 |
| Tie2/Tek | EC | 2 | 4 | 1 | 4 | 3 | 4 | 2 |
| CD34 | EC | 3 | 10 | 12 | 4 | 11 | 5 | 2 |
| CD14 | Hematopoeitic | 1 | 2 | 0 | 0 | 1 | 1 | 1 |
| CK8 | Epithelial | 0 | 0 | 2 | 1 | 1 | 1 | 2 |
| GLUT1 | Brain EC | 8 | 37 | 2 | 25 | 8 | 0 | 1 |
| GFAP | Glial | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Previously published SAGE tag counts for isolated colonic EC libraries are provided for comparison.
Expression Profiling
The Long SAGE variant of the SAGE protocol was used to determine gene expression profiles for each EC library.13 Long SAGE uses a novel type II restriction enzyme as the tagging enzyme under the SAGE protocol to yield a 17-bp tag compared to the original 10-bp tag generated by traditional SAGE. The Long SAGE variant, therefore, improves confidence in tag to gene assignment compared to traditional SAGE methods. Because the data from our brain analysis will be used for comparison to traditional SAGE data, the long tags were aggregated to short tags for consistency within the comparative analyses. Sequencing of each library was performed to a depth of ∼50,000 tags per sample representing 72,954 unique tags overall (Table 1). For data analysis, each SAGE profile was normalized to exactly 50,000 tags. Pair-wise comparisons between expression data derived from tumor samples selected with P1H12 or UEA-I showed correlation coefficients at 80%, slightly higher than a comparison between two tumor samples selected with P1H12. This suggests that selecting ECs with either P1H12 or UEA-I results in highly similar cell populations. Moreover, nearly half of the tumor-specific markers revealed in this study are induced fourfold relative to each of the normal samples used, suggesting that normal samples are similar populations as well. Thus, we felt that combining data for the two normal samples and for the three tumor samples was appropriate, and that the SAGE profiles represented gene expression across the central nervous system microvasculature as a whole.
Glioma Endothelial Markers (GEMS)
GEMS expressed differentially in glioma-derived ECs with respect to normal brain ECs are potentially involved in regulating angiogenesis-dependent tumor growth and blood-brain barrier abnormalities. To identify such genes, we calculated fold-ratios and differential probabilities for the normalized tag abundances. Application of conservative threshold values revealed a subset of glioma-induced tags, which were subsequently mapped to known gene sequences. Overall, a bulk comparative analysis between glioma samples and normal brain samples yielded an 85% correlation coefficient suggesting that the vast majority of genes are expressed to similar levels. Tags that found matches to Unigene include 122 genes induced in the glioma ECs based on a fourfold induction ratio (supporting on-line material, Supplement B at http://ajp.amjpathol.org). The field of 122 glioma-induced genes was narrowed to 14 GEMS by applying additional statistical filters (Table 3). In this case, a β distribution was used to establish a 90% probability of observing at least a twofold change in expression.11
Table 3.
Fourteen Glioma-Induced Genes Meeting Both Fold Induction (T/N) and Statistical Filters (T/N prob.)
| T/N | T/N prob. | SAGE tag | UG ID | UG description | Localization |
|---|---|---|---|---|---|
| 17 | 95 | GTCTCAGTGC | 118893 | Melanoma-associated gene MG50 | Surface/secreted |
| 14 | 90 | CTTATGCTGC | 82002 | Endothelin receptor type B | Surface |
| 13 | 99 | CCACCCTCAC | 211573 | HSPG2 Perlecan | Extracellular |
| 13 | 94 | GTGCTACTTC | 119129 | Collagen, type IV, alpha 1 | Extracellular |
| 12 | 98 | GAGTGAGACC | 345643 | Thy-1 cell surface antigen | Surface |
| 10 | 94 | ATGGCAACAG | 149609 | ITGA5 integrin alpha 5 (Fn receptor) receptor | Surface |
| 9 | 91 | TCACACAGTG | 23016 | G protein-coupled receptor RDC-1 | Surface |
| 8 | 100 | GACCGCAGG | 119129 | Collagen, type IV, alpha 1 | Extracellular |
| 8 | 97 | GGGAGGGGTG | 2399 | Matrix metalloproteinase 14 (membrane-inserted) | Extracellular |
| 7 | 99 | CCCTACCCTG | 75736 | Apolipoprotein D | Extracellular |
| 8 | 97 | TTCTCCCAAA | 75617 | Collagen, type IV, alpha 2 | Extracellular |
| 8 | 98 | GGATGCGCAG | 302741 | Homo sapiens mRNA full length insert cDNA clone EU | |
| 5 | 98 | GTGCTAAGCG | 159263 | Collagen, type VI, alpha 2 Exon 1 | Extracellular |
| 4 | 93 | CCCAGGACAC | 110443 | Homo sapiens cDNA: FLJ22215 fis, clone HRC01580 |
Among the gene products identified as GEMs by this study are several genes that regulate tumor endothelium extracellular matrix architecture, including HSPG2 (perlecan), several type IV collagen transcript variants, and matrix metalloprotease 14 (MMP14). Interestingly, other genes that play roles in either cellular signaling or cell-cell communication are also highly expressed exclusively in glioma-associated ECs. Melanoma-associated antigen (MG50), endothelin receptor type B, the G-protein coupled receptor RDC-1, and integrin αV are all cell surface proteins previously demonstrated to play a role in signaling cascades.14–16 Although the endothelin receptor B, RDC-1, and integrin αV have suggested roles in angiogenesis, MG50 does not have a known association with angiogenesis. Moreover, MG50 has been previously linked with several types of tumor cells but its function is yet to be defined.16 In total, of the 14 GEMs listed, 12 are known to be present on the cell surface or secreted. The localization of the remaining two gene products has yet to be determined because these genes remain uncharacterized. Finally, it is noteworthy that only a select few genes from this list show significant (>2 occurrences per 50,000 tags) expression in either a fetal brain library or in a fetal kidney library in which angiogenesis is expected to be robust (data not shown).
Glioma-Repressed Genes
In contrast to the highly biased localization of glioma-induced EC gene products defined above, genes that are induced in normal brain ECs relative to glioma ECs encode proteins with a radically different cellular distribution. Twenty-one genes are induced fourfold or greater in the normal brain ECs (glioma-repressed genes, supporting on-line materials, Supplement C at http://ajp.amjpathol.org). A twofold β distribution filter refines this list to 14 genes (Table 4). Protein products predicted for these 14 genes show a range of cellular localizations including four intracellular proteins, five integral membrane proteins, three extracellular proteins, and one each either secreted, on the cell surface or a nuclear membrane receptor. Several of these genes have functions consistent with either tumor suppressor or anti-angiogenic functions. Such anti-proliferative functions have been ascribed to the early growth response gene 1 (EGR1), BTG2, Kruppel-like factor 4 (KLF4), and the serine protease inhibitor SPINT2, although associations with angiogenesis are limited to SPINT2.17–20 Although previous work on genes such as EGR1 typically suggest a progrowth function, recent results demonstrate an anti-proliferative function for EGR1 associated with its regulation of p53.21 The down-regulation of these genes in each of the three gliomas suggests that these genes may function to encode proteins with anti-angiogenic properties or properties associated with endothelial differentiation and blood-brain barrier development. Both SPINT2 and BTG2 are secreted and may act via paracrine mechanisms. SPINT2 serves to reduce the effective concentration of hepatocyte growth factor, a known brain EC mitogen.20,22,23 Interestingly, EGR1 and KLF4 encode transcription factors suggesting that some part of the anti-angiogenic pathway may be initiated by these gene products.
Table 4.
Fourteen Glioma-Repressed Genes Meeting Both Fold Induction (T/N) and Statistical Filters (T/N prob.)
| Brain N/T | Brain N/T prob. | SAGE tag | UG ID | UG description | Localization |
|---|---|---|---|---|---|
| 9 | 72 | TAGTTGGAAA | 1119 | Nuclear receptor subfamily 4, group A, member 1 NR4A1 | Nuclear membrane |
| 9 | 72 | AAGGGCGCGG | 1378 | Annexin A3 ANXA3 | Membrane |
| 9 | 72 | AGCTGTGCCA | 348254 | Metallothionein 1A (functional) MT1A | Extracellular |
| 7 | 60 | ACAAAATCAA | 110613 | Nuclear pore complex interacting protein SMG-1 | Membrane |
| 6 | 68 | GCCTGCAGTC | 31439 | Serine protease inhibitor, Kunitz type, 2 SPINT2 | Extracellular |
| 6 | 52 | ACCAGGTCCA | 5167 334549 | Solute carrier family 5 (sodium-dependent vitamin) | Membrane |
| 6 | 52 | GGCTAATTAT | 34114 | ATPase, Na+/K+ transporting, alpha 2 (+) | Membrane |
| 6 | 75 | TTTAAATAGC | 7934 | KLF4 Kruppel-like factor 4 (gut) | Intracellular |
| 5 | 81 | CAGTTCATTA | 326035 | Early growth response 1 EGR1 | Intracellular |
| 5 | 61 | CTGCCGTGAC | 75462 | BTG family, member 2 BTG2 | Extracellular |
| 5 | 65 | TTTTAACTTA | 160483 | Erythrocyte membrane protein band 7.2 (stomatin) | Membrane |
| 4 | 77 | TAGAAACCGG | 8997 | Heat shock 70-kd protein 1A HSP70 | Intracellular |
| 4 | 77 | CTTCTTGCC | 272572 347939 | Hemoglobin, alpha 2 | Intracellular |
| 4 | 53 | TAGAAAAAAT | 8906 | Syntaxin 7 | Surface |
Comparison with Peripherally Derived Tumor ECs
The specificity of gene expression for tumor EC subtypes is important to define and can be addressed by comparing the glioma EC data with data obtained previously for peripherally derived colon EC populations.10 Only the MT1A gene is tumor-repressed to fourfold or greater levels in both the colon and brain samples, suggesting a correlation with cell proliferation and protection from apoptosis (data not shown). In contrast, 16 genes were induced at least fourfold in both colon and glioma tumor EC fractions (supporting on-line material, Supplement D at http://ajp.amjpathol.org). Twelve of these genes also meet the applied twofold β distribution filter (Table 5). The majority of these genes (seven) are collagen transcripts, however, tumor endothelial marker 1 (TEM1), THY1, and RDC1 also show consistent induction in both glioma and colon tumor EC cells. This limited conservation of tumor-induced EC expression suggests highly specific EC expression profiles dependent on the tissue source. More specifically, brain tissue may contain an endothelium that is transcriptionally unique relative to endothelium in peripheral vasculature.
Table 5.
Common Tumor-Induced EC Genes between Colon Tumor and Glioma Samples Exhibiting a Minimal Fourfold Induction when Compared to Normal EC Samples
| Brain T/N | Colon T/N | SAGE tag | UG ID | UG description | Localization |
|---|---|---|---|---|---|
| 13 | 4 | GTGCTACTTC | 119129 | Collagen, type IV, alpha 1 | Extracellular |
| 12 | 16 | GAGTGAGACC | 125359 | Thy-1 cell surface antigen | Surface |
| 9 | 4 | TCACACAGTG | 23016 | G protein-coupled receptor RDC-1 | Surface |
| 8 | 6 | GACCGCAGGA | 119129 | Collagen, type IV, alpha 1 | Extracellular |
| 8 | 13 | GGGAGGGGTG | 2399 | Matrix metalloproteinase 14 (membrane-inserted) | Extracellular |
| 7 | 14 | GGGGCTGCCC | 195727 | Tumor endothelial marker 1 precursor | Surface |
| 6 | 4 | TTCTCCCAAA | 75617 | Collagen, type IV, alpha 2 | Extracellular |
| 6 | 18 | CCACAGGGGA | 119571 | Collagen, type III, alpha 1 (Ehlers-Danlos syndrome) | Extracellular |
| 6 | 9 | TCAAGTTCAC | 351928 | Homo sapiens mRNA full length insert cDNA Euroimage 1977059 | |
| 5 | 10 | ACCAAAAACC | 172928 | Collagen, type I, alpha 1 | Extracellular |
| 4 | 7 | GATCAGGCCA | 119571 | Collagen, type III, alpha 1 | Extracellular |
| 4 | 4 | AGAAACCACG | 119129 | Collagen, type IV, alpha 1 | Extracellular |
RDC1 Localization to Tumor Endothelium
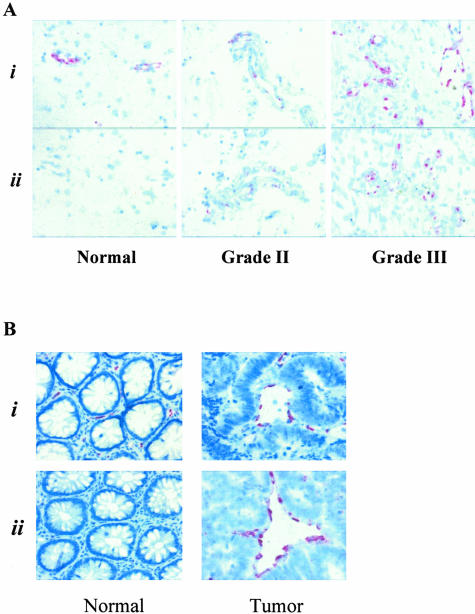
Because G protein-coupled receptors (GPCRs) are intriguing molecules involved in signal transduction as well as being amenable to therapeutic intervention, we chose to evaluate the specificity of RDC1 expression by in situ hybridization. RDC1 mRNA was localized exclusively to vascular regions within glioma samples showing near coincident expression with the vascular-specific marker vWF (Figure 1A). Moreover, RDC1 was observed to be expressed only in the glioma samples and was not observed in normal brain cortex sections. Because RDC1 was also demonstrated to be induced in colon cancer via SAGE, we localized the expression of RDC1 in both disease-free colon and tumor-bearing colon samples. As observed for the glioma samples, RDC1 showed both tumor and vascular specificity in colon samples (Figure 1B). We were also able to confirm the vascular-specific expression of another conserved tumor EC marker, TEM1, in gliomas (data not shown). Previous results confirmed the vascular-specific expression of TEM1 in colon tumor samples.10
Figure 1.
RDC1 expression. Representative tissue in situ hybridizations demonstrating RDC1 mRNA localization in gliomas (A) and colon tumors (B). Hybridizations were performed as described previously10 using tissue microarrays (Novagen) for brain sections or colon tissue specimens (CHTN). i, vWF; ii, RDC1.
EC-Restricted GEMs
Defining the specificity of gene expression to particular cell types can assist in determining function and designing therapeutics. Our non-EC SAGE database currently contains 76 libraries encoding 255,000 unique SAGE tags. The epithelial cell lines derive from normal and diseased lung, ovary, kidney, prostate, breast, colon, and pancreas. Additional nonepithelial sources include cardiomyocytes, melanocytes, glioblastoma tumor cells, and monocytes. Genes that show induction in glioma ECs and that also demonstrate a restricted expression in non-ECs may be ideal targets for anti-angiogenic therapies. Allowing for one or fewer tags in any non-EC library and at least a fourfold induction in glioma ECs yielded only five genes that were differentially expressed (Table 6). Some of these genes may prove not to be EC-specific because the majority of the data derives from a limited, epithelial-biased database. However, both PV1 and Plexin A2 (PLXNA2) were identified as EC-specific and are interesting genes with potential functional relevance to angiogenesis.24,25 We chose to further investigate the expression of the transmembrane protein PV1 in additional tissue samples and evaluate the functional significance of its expression.
Table 6.
Five Genes Showing a Minimal Fourfold Induction in Glioma Versus Normal Brain Samples and Showing a Restricted Expression in Non-EC Samples (1 or Fewer Tags in 76 Non-EC SAGE Libraries)
| Brain T/N | Brain T/N prob. | Short tag | Non-EC count | UG ID | UG description | Localization |
|---|---|---|---|---|---|---|
| 9 | 83 | AAGGTTCTTC | 1 | 89695 | Insulin receptor | Surface |
| 7 | 74 | CCCTTTCACA | 1 | 107125 | PV1 | Surface |
| 6 | 75 | AGACTAGGGG | 1 | 350065 | Plexin A2 | Surface |
| 4 | 69 | CATAAACGGG | 1 | 69954 | Laminin, gamma 3 | Extracellular |
| 4 | 53 | GGCCAACATT | 1 | 36353 | Homo sapiens mRNA full length insert cDNA clone EU |
PV1 Characterization
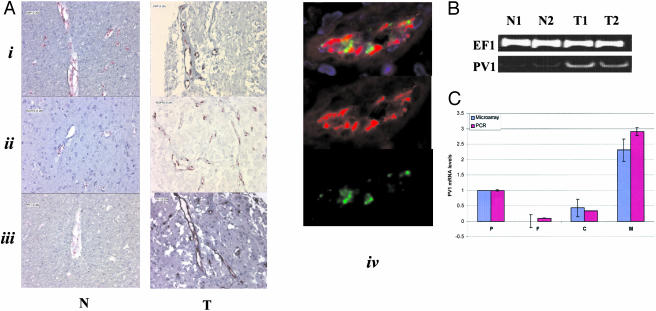
PV1 is normally expressed at low or undetectable levels within the normal adult brain cortex.24 Our SAGE data showed 7, 3, and 12 tags in the tumor samples and no tags in either normal sample, suggesting a moderately expressed level of the PV1 transcript in tumor ECs. In situ localization and PCR experiments demonstrate a significant increase in PV1 expression in astrocytoma and glioblastoma samples (Figure 2, A and B). Both SAGE data and in situ hybridization results demonstrate comparable levels of vWF and VEGFR2 in normal and tumor samples as well as a co-localization of PV1 with the EC-specific protein CD31 in glioblastoma tissue (Figure 2A). Moreover, in situ evaluation of PV1 expression in an additional eight tumor samples, including three metastatic adenocarcinomas, showed consistent up-regulation of PV1 similar to the results presented here. The induction of PV1 expression in metastases to the brain demonstrates that the vascular-specific expression of PV1 is not limited to primary brain tumors.
Figure 2.
PV1 expression. A: Representative tissue in situ hybridizations demonstrating PV1 localization in glioma EC. Hybridizations were performed as described previously10 using tissue microarrays (Novagen). N, Normal brain cortex tissue sections; T, glioma tissue sections from an astrocytoma (i to iii) or glioblastoma (iv). Probes for vWF and VEGFR2 were hybridized in sequential sections to confirm expression of these EC markers and to define vascular structure. i, vWF; ii, VEGFR2; iii, PV1. iv: Co-localization of PV1 (Cy3) and CD31 (FITC) in glioblastoma vasculature. Top: A composite of single channel labeling for PV1 (middle), and CD31 (bottom). B: RT-PCR analysis of four of the brain EC samples used for SAGE comparing normal (N) versus tumor (T) expression of PV1 and EF1. C: PV1 induction in HMVECs induced to form tubes on Matrigel. Single donor, neonatal dermal HMVEC IF1645 (Clonetics) were grown to confluency on plastic followed by splitting cultures onto plastic, fibrin, collagen, or Matrigel. Cells were allowed to grow for 24 hours at which time HMVEC networks formed for all cultures excluding cells grown on plastic. RNA was harvested and subjected to real-time PCR analysis or custom microarray interrogation. PV1 expression is relative to expression on plastic (arbitrarily set to 1) for real-time PCR (red) or microarray results (blue). Microarray results are log2 ratios. Real-time PCR values represent an average of two replicates and the microarray results are an average from quadruplicate PV1 spots. P, Plastic; F, fibrin; C, collagen; M, Matrigel.
Certain growth matrices are known to induce angiogenic phenotypes in ECs.26 To better assess the correlation between PV1 expression and angiogenesis, we evaluated PV1 expression in HMVECs induced to form tubes on Matrigel. A custom oligonucleotide microarray composed of both colon- and brain-specific EC markers, as well as several hundred genes previously recognized to play a role in angiogenesis was interrogated with RNA probes derived from HMVECs grown on plastic, fibrin, collagen, or Matrigel. Although PV1 is not induced to a significant degree in HMVECs grown in fibrin or collagen, it is significantly induced in HMVECs grown in Matrigel when compared to ECs grown on plastic (Figure 2C). Of the 606 genes contained on our custom array, PV1 was the most highly induced gene in cells grown on Matrigel versus plastic. Real-time PCR analysis confirmed the induced expression of PV1 in these cultures (Figure 2C). Although growth of ECs on fibrin or collagen does induce networks of ECs, true tube-like structures seem to be confined to Matrigel-induced differentiation.
Discussion
To identify key regulators of the tumor-promoting angiogenic process in brain, we have defined the transcriptomes for both normal and malignant brain. Our transcript profiling analysis has revealed numerous genes previously unrecognized to play a role in brain angiogenesis. Moreover, comparative analysis with colon-derived EC transcriptomes allowed us to define conserved tumor vascular genes in addition to sets defining brain-specific genes and genes derived from peripheral vasculature. We have confirmed the tumor and vascular specificity for two genes that are likely localized to the cell surface.
The PV1 gene is significantly induced in each of the three glioma samples studied as compared to normal brain ECs. The gene is expressed at substantial levels in colon ECs but is not expressed differentially between normal and tumor colon ECs. PV1 is also expressed in normal fenestrated endothelium exemplified by kidney peritubular capillaries.24 Previous studies using a PV1-specific antibody suggested that this protein is severely down-regulated on differentiation of the blood brain barrier.27 The up-regulation of PV1 in brain tumors is consistent with a dedifferentiated vasculature lacking the blood-brain barrier phenotype. Besides caveolae, PV1 was previously shown to be localized to diaphragms within fenestrae, transendothelial channels, and vesiculo-vacuolar organelles (VVOs).24 Moreover, previous work has demonstrated a coincident induction of fenestrated endothelium during tumor growth, consistent with an increase in diaphragm structures.28,29 SAGE analysis has also confirmed both a threefold and eightfold induction of PV1 in two individual breast ductal carcinoma EC samples as compared to normal breast EC samples (unpublished results). Indeed, using real-time PCR we observed a 60% up-regulation of PV1 (twofold or greater) within the vasculature of invasive ductal carcinomas of the breast relative to normal breast tissue vasculature (n = 10 normal and 30 tumor samples) (unpublished results). By its localization, PV1 may function as a regulator of transendothelial exchange. Therefore, our results suggest the PV1 protein may prove to be a mediator of nutrient flow to tumor cells. We did not observe the up-regulation of any caveolin in our analysis suggesting our observation of PV1 regulation is not simply an indirect effect of increased numbers of EC caveolae.
Genes that show conserved tumor-specific EC expression across several tumor types may play key roles in regulating global neo-angiogenesis. We demonstrate here that the orphan GPCR RDC1 is induced in both glioma and colon tumor vasculature relative to normal tissue vasculature. RDC1 may indeed represent a vascular marker definitive for active angiogenesis because the derived SAGE data showed an eightfold induction of RDC1 in grade IV tumors versus normal. No SAGE tags for RDC1 were observed in the normal colon data set. Interestingly, although by SAGE the RDC1 levels for the single grade III glioma were not significantly differential from the normal samples, the localization results demonstrate distinct expression of RDC1 in both grade II and grade III gliomas suggesting that RDC1 expression may not be restricted to later stage gliomas.
The data presented here suggests that additional genes identified in this extensive brain microvascular transcriptome analysis may also be directly affecting angiogenesis. Importantly, one conclusion from our study is that the vasculature from different tumor types shows limited conservation in gene expression with respect to both tumor-induced and tumor-repressed markers. Overall, however, the transcriptomes from diverse sources are remarkably similar. The implications of our transcriptome analysis could extend beyond helping to define the biology of glioma tumors because many of the genes identified in this study may also prove to be important regulators in nonglioma brain tumors and tumors that metastasize to the brain. Additional analysis of the brain EC data discussed here, as well as the elucidation of further tumor EC transcriptomes, will assist in characterizing the function of tumor-specific EC genes and may provide valuable targets for tumor therapies.
Supplementary Material
Acknowledgments
We thank Kenneth W. Kinzler (Johns Hopkins University) and Brad St. Croix (National Cancer Institute) for sharing colon gene expression data.
Footnotes
Address reprint requests to Stephen Madden, Genetics and Genomics, 5 Mountain Rd., Framingham, MA 01701. E-mail: steve.madden@genzyme.com.
Supported in part by the National Institutes of Health (grant RO1 NS32148 to J.L.).
Supplemental information can be found at http://ajp.amjpathol.org.
References
- Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN, Cavenee WK, DePinho RA. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- Vajkoczy P, Menger MD. Vascular microenvironment in gliomas. J Neurooncol. 2000;50:99–108. doi: 10.1023/a:1006474832189. [DOI] [PubMed] [Google Scholar]
- Bart J, Groen HJ, Hendrikse NH, van der Graaf WT, Vaalburg W, de Vries EG. The blood-brain barrier and oncology: new insights into function and modulation. Cancer Treat Rev. 2000;26:449–462. doi: 10.1053/ctrv.2000.0194. [DOI] [PubMed] [Google Scholar]
- Bernsen HJ, Rijken PF, Oostendorp T, van der Kogel AJ. Vascularity and perfusion of human gliomas xenografted in the athymic nude mouse. Br J Cancer. 1995;71:721–726. doi: 10.1038/bjc.1995.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick NA, Bigner DD. Microvascular abnormalities in virally-induced canine brain tumors. Structural bases for altered blood-brain barrier function. J Neurol Sci. 1972;17:29–39. doi: 10.1016/0022-510x(72)90019-6. [DOI] [PubMed] [Google Scholar]
- Hobbs SK, Monsky WL, Yuan F, Roberts WG, Griffith L, Torchilin VP, Jain RK. Regulation of transport pathways in tumor vessels: role of tumor type and microenvironment. Proc Natl Acad Sci USA. 1998;95:4607–4612. doi: 10.1073/pnas.95.8.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holash J, Maisonpierre PC, Compton D, Boland P, Alexander CR, Zagzag D, Yancopoulos GD, Wiegand SJ. Vessel cooption, regression, and growth in tumors mediated by angiopoietins and VEGF. Science. 1999;284:1994–1998. doi: 10.1126/science.284.5422.1994. [DOI] [PubMed] [Google Scholar]
- Guerin C, Wolff JE, Laterra J, Drewes LR, Brem H, Goldstein GW. Vascular differentiation and glucose transporter expression in rat gliomas: effects of steroids. Ann Neurol. 1992;31:481–487. doi: 10.1002/ana.410310504. [DOI] [PubMed] [Google Scholar]
- Unger RE, Oltrogge JB, von Briesen H, Engelhardt B, Woelki U, Schlote W, Lorenz R, Bratzke H, Kirkpatrick CJ. Isolation and molecular characterization of brain microvascular endothelial cells from human brain tumors. In Vitro Cell Dev Biol Anim. 2002;38:273–281. doi: 10.1290/1071-2690(2002)038<0273:IAMCOB>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- St. Croix B, Rago C, Velculescu V, Traverso G, Romans KE, Montgomery E, Lal A, Riggins GJ, Lengauer C, Vogelstein B, Kinzler KW. Genes expressed in human tumor endothelium. Science. 2000;289:1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- Chen H, Centola M, Altschul SF, Metzger H. Characterization of gene expression in resting and activated mast cells. J Exp Med. 1998;188:1657–1668. doi: 10.1084/jem.188.9.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huminiecki L, Bicknell R. In silico cloning of novel endothelial-specific genes. Genome Res. 2000;10:1796–1806. doi: 10.1101/gr.150700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Sparks AB, Rago C, Akmaev V, Wang CJ, Vogelstein B, Kinzler KW, Velculescu VE. Using the transcriptome to annotate the genome. Nat Biotechnol. 2002;20:508–512. doi: 10.1038/nbt0502-508. [DOI] [PubMed] [Google Scholar]
- Wang XQ, Sun P, Al-Qamari A, Paller AS. Gangliosides block keratinocyte binding to fibronectin through carbohydrate-carbohydrate binding to the alpha5 subunit of alpha5beta1. J Invest Dermatol. 2000;115:333. doi: 10.1046/j.1523-1747.2000.115002333.x. [DOI] [PubMed] [Google Scholar]
- Libert F, Parmentier M, Lefort A, Dumont JE, Vassart G. Complete nucleotide sequence of a putative G protein coupled receptor: rDC1. Nucleic Acids Res. 1990;18:1917. doi: 10.1093/nar/18.7.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell MS, Kan-Mitchell J, Minev B, Edman C, Deans RJ. A novel melanoma gene (MG50) encoding the interleukin 1 receptor antagonist and six epitopes recognized by human cytolytic T lymphocytes. Cancer Res. 2000;60:6448–6456. [PubMed] [Google Scholar]
- Liu C, Rangnekar VM, Adamson E, Mercola D. Suppression of growth and transformation and induction of apoptosis by EGR-1. Cancer Gene Ther. 1998;5:3–28. [PubMed] [Google Scholar]
- Rouault JP, Falette N, Guehenneux F, Guillot C, Rimokh R, Wang Q, Berthet C, Moyret-Lalle C, Savatier P, Pain B, Shaw P, Berger R, Samarut J, Magaud JP, Ozturk M, Samarut C, Puisieux A. Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat Genet. 1996;14:482–486. doi: 10.1038/ng1296-482. [DOI] [PubMed] [Google Scholar]
- Shields JM, Christy RJ, Yang VW. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J Biol Chem. 1996;271:20009–20017. doi: 10.1074/jbc.271.33.20009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi T, Qin L, Shimomura T, Kondo J, Matsumoto K, Denda K, Kitamura N. Purification and cloning of hepatocyte growth factor activator inhibitor type 2, a Kunitz-type serine protease inhibitor. J Biol Chem. 1997;272:27558–27564. doi: 10.1074/jbc.272.44.27558. [DOI] [PubMed] [Google Scholar]
- Krones-Herzig A, Adamson E, Mercola D. Early growth response 1 protein, an upstream gatekeeper of the p53 tumor suppressor, controls replicative senescence. Proc Natl Acad Sci USA. 2003;100:3233–3238. doi: 10.1073/pnas.2628034100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abounader R, Lal B, Luddy C, Koe G, Davidson B, Rosen EM, Laterra J. In vivo targeting of SF/HGF and c-met expression via U1snRNA/ribozymes inhibits glioma growth and angiogenesis and promotes apoptosis. FASEB J. 2002;16:108–110. doi: 10.1096/fj.01-0421fje. [DOI] [PubMed] [Google Scholar]
- Rosen EM, Laterra J, Joseph A, Jin L, Fuchs A, Way D, Witte M, Weinand M, Goldberg ID. Scatter factor expression and regulation in human glial tumors. Int J Cancer. 1996;67:248–255. doi: 10.1002/(SICI)1097-0215(19960717)67:2<248::AID-IJC16>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Stan RV, Kubitza M, Palade GE. PV-1 is a component of the fenestral and stomatal diaphragms in fenestrated endothelia. Proc Natl Acad Sci USA. 1999;96:13203–13207. doi: 10.1073/pnas.96.23.13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagnone L, Artigiani S, Chen H, He Z, Ming GI, Song H, Chedotal A, Winberg ML, Goodman CS, Poo M, Tessier-Lavigne M, Comoglio PM. Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell. 1999;99:71–80. doi: 10.1016/s0092-8674(00)80063-x. [DOI] [PubMed] [Google Scholar]
- Unger RE, Krump-Konvalinkova V, Peters K, Kirkpatrick CJ. In vitro expression of the endothelial phenotype: comparative study of primary isolated cells and cell lines, including the novel cell line HPMEC-ST16R. Microvasc Res. 2002;64:384–397. doi: 10.1006/mvre.2002.2434. [DOI] [PubMed] [Google Scholar]
- Hallman R, Mayer DN, Berg EL, Broermann R, Butcher EC. Novel mouse endothelial cell surface marker is suppressed during differentiation of the blood brain barrier. Dev Dyn. 1995;202:325–332. doi: 10.1002/aja.1002020402. [DOI] [PubMed] [Google Scholar]
- Roberts WG, Delaat J, Nagane M, Huang S, Cavenee WK, Palade GE. Host microvasculature influence on tumor vascular morphology and endothelial gene expression. Am J Pathol. 1998;153:1239–1248. doi: 10.1016/s0002-9440(10)65668-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts WG, Palade GE. Neovasculature induced by vascular endothelial growth factor is fenestrated. Cancer Res. 1997;57:765–772. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.