Abstract
Deregulation of Notch signaling, which normally affects a broad spectrum of cell fates, has been implicated in various neoplastic conditions. Here we describe a transgenic mouse model, which demonstrates that expression of a constitutively active form of the Notch1 receptor in the mammary epithelium induces the rapid development of pregnancy/lactation-dependent neoplasms that consistently exhibit a characteristic histopathological pattern. These signature tumors retain the ability to respond to apoptotic stimuli and regress on initiation of mammary gland involution, but eventually appear to progress in subsequent pregnancies to nonregressing malignant adenocarcinomas. Additionally, we present evidence indicating that cyclin D1 is an in vivo target of Notch signals in the mammary glands and demonstrate that we can effectively inhibit Hras1-driven, cyclin D1-dependent mammary oncogenesis by transgenic expression of the Notch antagonist Deltex.
The Notch locus encodes a large transmembrane receptor,1,2 which is the central element of an evolutionarily conserved signaling pathway controlling a broad spectrum of cell fate decisions during metazoan development. Signals through Notch couple cell-fate acquisition by an individual cell to the cell fate choices made by its immediate neighbors.3–7 Modulation of Notch signaling is known to affect developmental programs of proliferation, differentiation, and apoptosis in many different cell types. Thus, aberrant Notch signaling through any one of the four mammalian receptor paralogues (Notch1 through Notch4) can deregulate the balance between these processes and can lead to tumorigenesis.8–14
The binding of transmembrane ligands expressed in one cell to the Notch1 receptor expressed on a neighboring cell is thought to release, through a series of proteolytic steps, the intracellular domain of the receptor (N1IC), which is then translocated into the nucleus and acts as a transcriptional regulator together with other factors. In view of the potential involvement of Notch signaling in human breast cancer,15 and considering that truncated forms of Notch1 consisting only of N1IC are constitutively active,14,16–24 we sought to examine the consequences of hN1IC expression in the mammary glands of transgenic mice. In parallel, for further analysis of Notch-related activities in breast tissue, we pursued the study of a transgene encoding human Deltex (hDTX1),25 a modulator of the Notch signaling pathway that acts as an inhibitor of Notch-dependent transcription in mammalian cells.26,27
For our purposes, we used the promoter/enhancer of the mouse mammary tumor virus (MMTV) long terminal repeat (LTR)25,28 to express in breast epithelial cells human transgenes encoding hN1IC or hDTX1. We are assuming that the human and mouse homologues, which display 88% and 95% amino acid identities, respectively, are functionally interchangeable.
Here we show that transgenic activation of Notch1 signaling in mammary glands leads to the development of lactation-dependent tumors that, however, regress at weaning, because the neoplastic cells apparently retain the ability to respond to apoptotic cues encountered during the process of mammary involution. Eventually, these regressing neoplasms apparently evolve into nonregressing adenocarcinomas. Moreover, by exploring the phenotypic consequences of antagonizing the endogenous Notch activity in the mammary epithelium through the transgenic expression of Deltex, we provide evidence indicating that cyclin D1 is an in vivo target of Notch signals. Consistent with this observation, we show that Hras1-induced tumorigenesis, which depends on the presence of cyclin D1 activity, can be suppressed by expressing Deltex.
Materials and Methods
Mice
To generate genetically modified mice, cDNAs encoding hN1IC (human Notch1 intracellular sequence, amino acids 1758 to 2556; UniGene Hs.129053) and hDTX1 (human Deltex 1 sequence, amino acids 1 to 621; Hs.124024) were first cloned into the pMMTV/SV40 vector.28 Transgenic animals were then generated by standard procedures, after injecting into fertilized eggs (derived from C3H × C57BL6 F2 mice; JacksonLaboratories, Bar Harbor, ME) SpeI/SalI DNA fragments excised from the constructs. The donor animals bearing C3H background, which is known to host MMTV, were free of endogenous milk-borne virus according to the supplier [confirmed by our own polymerase chain reaction (PCR) analysis].
MMTV-hN1IC and MMTV-hDTX1 transgenic lines, maintained in a mixed C3H × C57BL6 × FVB genetic background, were established from four and three founder animals, respectively. Animals of different lines carrying the same transgene exhibited similar phenotypes. Mice of inbred strains for the generation and breeding of founders, and also MMTV/v-Ha-ras and MMTV/c-myc animals,28 were obtained from Charles River Laboratories (Wilmington, MA) and Jackson Laboratories. For all experiments, littermates and/or isogenic animals were used as controls.
Molecular Analyses
For genotyping by PCR, we used tail DNA and the pairs of oligonucleotide primers 5′-AAGGCACGGAGGAAGAAGTC-3′ and 5′-CGCATTGACCATTCAAACTG-3′, which amplify a 397-bp fragment of the MMTV-hN1IC transgene, and 5′-CTGGTCACAGCATCAGGCTA-3′ and 5′-GGTCTTGTGGTGGATCTCGT-3′, which amplify a 501-bp fragment of the MMTV-hDTX1 transgene. The same primers, in addition to the pairs 5′-CACCACCTTCTCCACCAACT-3′ and 5′-TTGTCCACAGAATTCGCAAG-3′ (401-bp product) for CK18, were used for reverse transcriptase (RT)-PCR of total RNA extracted from mammary tissue with TRIzol reagent (Invitrogen, Carlsbad, CA). Western analysis was performed according to standard protocols using primary antibodies against cyclin D1 (CC12; Oncogene Research Products, San Diego, CA) and β-actin (A4700; Sigma, St. Louis, MO).
To perform luciferase reporter assays, we used the following procedure. First, a primary culture of mammary epithelial cells from a breast tumor developed in a MMTV c-myc animal was established in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. Cells were then seeded in a 24-well plate at a density of 7.0 × 104 cells/well and, after 24 hours, co-transfected using SuperFect reagent (Qiagen, Valencia, CA) with 0.4 μg of the TP1-luciferase plasmid, 2.0 ng of the Renilla luciferase control plasmid (Promega, Madison, WI), and variable quantities of pCDNA3 vector, hN1iC, and hDTX1. The TP1-luciferase reporter consists of hexamerized 50-bp Epstein-Barr virus nuclear antigen 2 (EBNA2) response element of the TP1 promoter in front of a minimal β-globin promoter driving the luciferase gene. Each EBNA2 response element contains two CBF1 binding sites.29 NotchIC binding converts the CBF transcriptional repressor to an activator. Firefly and Renilla luciferase activities were determined in whole cell extracts 24 hours after transfection using the Dual Luciferase Assay kit (Promega) and a Turner esigns TD200 dual luminometer.
Histological Analyses
Dissected inguinal mammary glands were used for whole mounts or were fixed for 24 hours in 4% neutralized buffered formalin, dehydrated, and embedded in paraffin. Paraffin blocks were sectioned and stained with hematoxylin and eosin. For whole mount analyses, tissues were fixed for 4 hours in Carnoy’s Fix containing ethanol, chloroform, and acetic acid (6:3:1 v:v:v), hydrated, and stained with carmine alum. Immunohistochemical analysis was performed by standard procedures using an anti-p63 primary antibody (559951; BD Biosciences Pharmingen, San Diego, CA). Terminal dUTP nick-end labeling (TUNEL) analysis was performed by using the in situ cell death detection kit (Roche Applied Science, Indianapolis, IN) according to the manufacturer’s instructions. For cell proliferation studies, animals 1.5 hours before sacrifice were injected with BrdU (Sigma) at 200 μg/g body weight. Sections (4 μm) were deparaffinized, and BrdU-positive cells were detected by immunocytochemistry using a mouse monoclonal anti-BrdU antibody (B2531; Sigma) according to the manufacturer’s instructions. Before observation, a weak counter stain with hematoxylin was performed, and the ratio of the epithelial cells that had incorporated BrdU was evaluated in three to four ducts per mouse in at least three mice per genotype. Hes1 in situ hybridization was performed as described in Chatzistamou and colleagues30 using 5-UCGUUCAUGCACUCGCUGAA-3 (anti-sense) and 5-UCAGCGAGUGCAUGAACGA-3 (sense) oligonucleotide riboprobes.
Tumorigenicity Assays
Tumors were allowed to develop in MMTV-hN1IC animals and, after histological examination, were aseptically dissected and mechanically minced. Approximately 3-mm3 pieces were transplanted subcutaneously by trocar needle into the right flanks of five female nude mice (Nu/Nu; Charles River Laboratories). Animals were subsequently observed for the development of palpable tumors for a period of 5 weeks. Tumors from MMTV-c-myc animals (n = 5) were used as positive controls (all transplantations resulted in the development of tumors within ∼2 weeks).
Results
MMTV-hN1IC and MMTV-hDTX1 Transgenic Lines
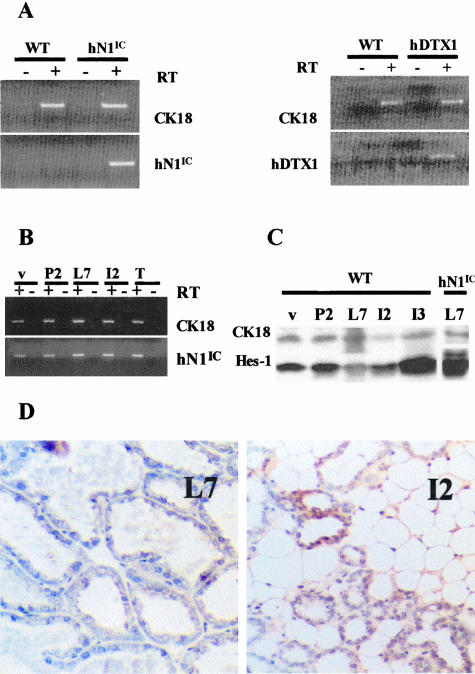
To investigate the consequences of modulating Notch activity in the murine mammary epithelium, we generated mouse lines carrying MMTV LTR-driven transgenes encoding a constitutively activated form of the human Notch1 receptor (hN1IC) or full-length human Deltex (hDTX1), a negative effector of Notch signals.26,31 Expression of each of these transgenes in the mammary glands of virgin females was demonstrated by RT-PCR analysis, using specifically designed oligonucleotide pairs that could discriminate between the endogenous mouse transcripts and the human transgenic sequences [assays for transcripts encoding the epithelial marker cytokeratin 18 (CK18) served as controls; Figure 1A. In the case of the hN1IC transgene, additional RT-PCR assays showed that it is also expressed in pregnant females during all phases of the mammary gland cycle (pregnancy, lactation, and involution; Figure 1B).
Figure 1.
Generation of MMTV-hN1IC and MMTV-hDTX1 transgenic mice. The preparations shown in all panels involve the fourth inguinal mammary gland. A: RT-PCR analysis of mammary gland tissue isolated from MMTV-hN1IC and MMTV-hDTX1 transgenic animals. The panel on the left depicts the RT-PCR amplification products with or without reverse transcriptase (RT+ and RT−) in samples extracted from a wild-type (WT) or hN1IC transgenic animal whereas the right shows a similar analysis for the hDTX1 transgenic mice. In both cases the epithelial marker CK18 was used as a control. B: hN1IC expression during various stages of mammary gland development and hN1IC tumors as revealed by RT-PCR analysis: 5-week virgin (v), pregnant day 2 (P2), lactating at day 7 (L7), after 2 (I2) days of involution and hN1IC tumors (T). The epithelial marker CK18 was used as a control. C: Western analysis of Hes-1 expression during wild-type mammary gland development. Protein extracts are from 5-week virgins (v), pregnant day 2 (P2), lactation day 7 (L7), and after 2 (I2) and 3 (I3) days of involution. The up-regulation of Hes1 in lactating MMTV-hN1IC animals is also shown. The epithelial marker CK18 was used as a control. D: In situ hybridization for Hes-1 in wild-type animals during lactation (L7) and after 2 days of involution (I2). Consistent with the Western blot analysis in C, stage I2 appears to express higher levels of Hes1 than stage L7. Note that the Hes1-positive cells are epithelial.
To determine whether the expression of the truncated Notch1 receptor (hN1IC) is capable of activating the Notch signaling cascade in the transgenic animals, we examined the expression of Hes1, a transcriptional target of Notch signals.24 For this evaluation, we used Western analysis and took into consideration that in wild-type controls the level of Hes1 is not constant during different developmental stages of the mammary glands (Figure 1C). Thus, in comparison with the CK18 control,32 the lowest amount of immunoreactive Hes1 was detected during lactation (day 7, L7). The immunoblotting results demonstrated that the level of Hes1 in the lactating (L7) glands of transgenic animals is very high (Figure 1C, lane 6) indicating that the ectopic expression of the hN1IC transgene activates the Notch pathway in the mammary epithelium. This was confirmed by in situ hybridization analysis that detected higher Hes1 levels during the second day of involution (I2) than during L7 (Figure 1D), showing also that Hes1 is an appropriate marker for our analysis because it is expressed in mammary epithelial cells rather than in other elements of the gland, such as adipose tissue.
MMTV-hN1IC and MMTV-hDTX1 Expression in Virgin Animals
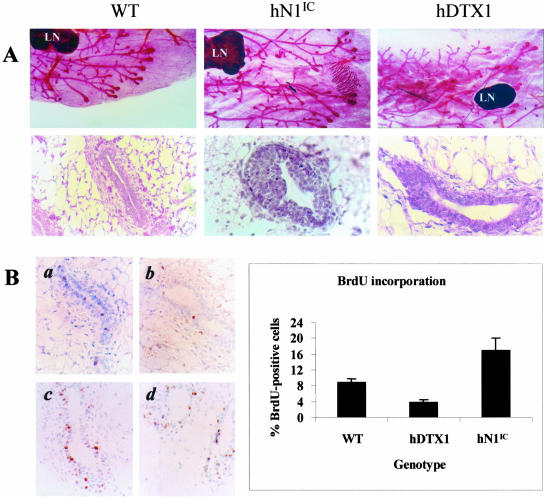
Whole mount analyses of mammary glands from 5-week-old MMTV-hN1IC females revealed the presence of elongated ductal structures and reduced side branching in comparison with the controls (Figure 2A, top panel). In histological sections (Figure 2B, bottom panel), the mammary glands of hN1IC transgenic animals at puberty exhibited variable hyperplasia ranging from normal to atypical, a phenotype associated with nonuniform but consistently increased levels of BrdU incorporation as compared to wild-type littermates (Figure 2B; compare a with c and d). Notably, in two of four MMTV hN1IC virgin mammary glands, we observed the presence of a ductal carcinoma in situ (DCIS; Figure 2A, middle) in the examined sections (the analysis of these specimens was not exhaustive). As seen in the figure, this neoplasm consisted of multiple layers of medium size cells that were mitotically active and contained uniform to mildly pleiomorphic nuclei with definitive nuclear borders. Although a small number of MMTV-hN1IC virgin females (n = 6) monitored for a period of ∼6 months remained free of palpable tumors, evolution of in situ carcinomas to full-fledged malignancy in virgin transgenic mice after a long latency cannot be formally excluded. Nevertheless, the hN1IC-induced, regressing, and nonregressing mammary tumors that we have studied (see below) appear to be primarily, if not exclusively, pregnancy/lactation-dependent neoplasms.
Figure 2.
Characterization of MMTV-hN1IC and MMTV-hDTX1 transgenic mice. A: Morphology of mammary glands from 5-week-old wild-type (WT), MMTV-hDTX1, and MMTV-hN1IC female mice. The top panel shows whole mounts stained with carmine alum and the bottom panel shows histological sections stained with H&E. Note the hyperplastic epithelium composed of atypical cells (ductal carcinoma in situ) in a duct of an MMTV-hN1IC animal. The lymph node (LN) is also shown. B: BrdU incorporation in the mammary gland of 5-week-old wild-type (WT) (a) compared to the mammary gland of an MMTV-hDTX1 transgenic (b) and MMTV-hN1IC (c and d) female mice. Average values of the ratio of BrdU-positive cells, obtained from three to four animals (three to four ducts each), from each genotype are shown in the diagram. We note that despite the high heterogeneity noted in the mammary glands of the MMTV-hN1IC animals, expression of hDTX1 led to reduced BrdU incorporation whereas expression of hN1IC led overall to an increase in the BrdU incorporation index.
In contrast to the consequences of activation of Notch signals in the mammary epithelium, the expression of Deltex in MMTV-hDTX1 animals leaves the ductal tree unperturbed, except for the occasional appearance of mild atrophy relative to the wild type. This atrophic phenotype was evident in only 5 of 10 cases examined and was consistent with the somewhat reduced levels of BrdU incorporation in MMTV-hDTX1 animals (Figure 2B, b). The MMTV-hDTX1 lines did not exhibit any additional phenotype after more that 1 year of observation.
Notch-Induced Regressing Tumors
A striking phenotype that was manifested with extraordinary rapidity as a consequence of Notch1 activation in MMTV-hN1IC female mice was the development of pregnancy-dependent palpable mammary tumors. These neoplasms appeared simultaneously at multiple mammary glands during lactation and before weaning of the pups (Figure 3A). Monitoring of a cohort of 34 transgenic females showed that 22 of them developed tumors after the first pregnancy and 8 after the second pregnancy, whereas 4 animals remained free of palpable tumors even after three pregnancies. It remains to be seen whether this high, but apparently incomplete penetrance can be attributed to segregation of modifier loci because of the mixed genetic background of these animals (see Materials and Methods).
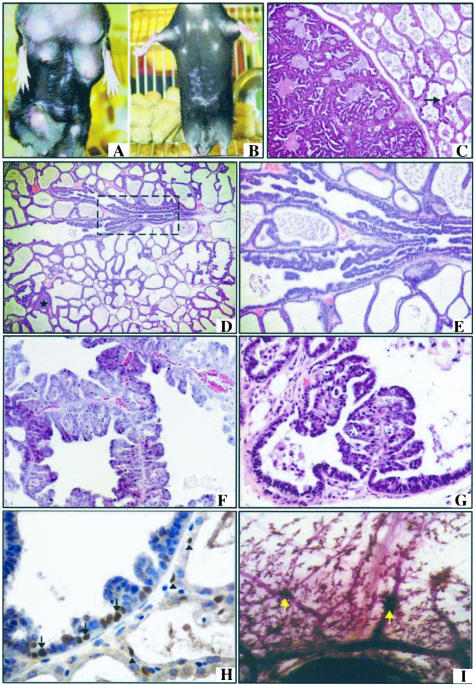
Figure 3.
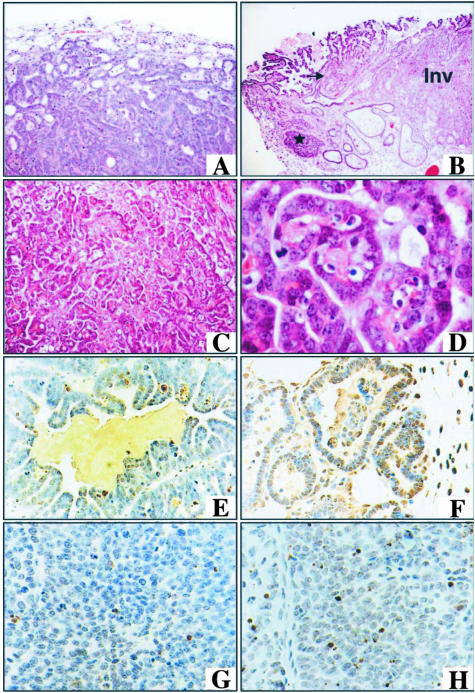
Lactation-dependent regressing mammary neoplasms induced by Notch1 activation. Mammary tumors develop in multiple glands of MMTV-hN1IC transgenic animals during lactation (A) and regress after 3 weeks of involution (B). Typical tumors seen in MMTV-hN1IC transgenic animals exhibit a characteristic signature papillary morphology and involve the entire lobular alveolar unit (C–G). Tumors were observed in ductules and alveoli [C, D (asterisk), and F] and in terminal ducts (D, E, and G; E is an enlargement of the area indicated by a rectangle in D). Architecturally, the tumors have the appearance of evenly spaced papillary fronds. In C, incipient lesions arising in alveoli (arrow) were observed adjacent to a fully developed regressing papillary tumor (left). Such lesions were surrounded by a layer of myoepithelial cells highlighted by nuclear staining with an anti-p63 antibody (H, brown color). Note that the tumor tissue on the left and the normal epithelium on the right do not differ in the pattern of p63 staining (arrows and arrowheads, respectively, indicate some of the positive nuclei). Whole mount of a mammary gland of the animal in B is shown in I. The arrows indicate focal nodules, which are apparently remnants of the regressed tumor. Original magnifications: ×4 (D); ×10 (C, E); ×20 (F, G, H).
Histopathological analysis indicated that multiple, noninvasive neoplasms had arisen in a background of lactational hyperplasia (Figure 3, C and D). Further detailed examination of six tumors from four animals indicated that these neoplasms represented in situ carcinomas because they were found to grow invariably within recognizable anatomical structures. Thus, they predominantly involved lactating alveoli (Figure 3, C and F), but also exhibited extensions into terminal breast ducts that were detectable in four of six cases (Figure 3; D, E, and G). The strictly intraglandular presence of tumor cells and the absence of invasion was demonstrated by immunostaining (Figure 3H) using an antibody against p63 (a p53 homologue, Mm.20894), a marker of breast myoepithelial cells, which are lost in malignancy.33 Whereas a lack of myoepithelial cells is noted even in microinvasive foci on commencement of infiltration, positive staining with the anti-p63 antibody demonstrated that, in the tumors induced by Notch1 in lactating glands, the myoepithelial layer was preserved.
These regressible neoplasms, which were not tumorigenic when inoculated into nude mice (n = 5), exhibited reproducibly a characteristic histopathological pattern (signature tumors). Thus, architecturally, the neoplastic cells formed papillary structures ranging from papillary tufts and evenly spaced delicate papillary fronds in alveoli to bulky papillae with prominent fibrovascular cores in terminal breast ducts (Figure 3, F and G). Cytologically, the tumors were composed of cuboidal cells with abundant cytoplasm and hyperchromatic, mildly pleiomorphic nuclei with a coarse chromatin pattern (not shown).
The tumorigenic effect of the MMTV-hN1IC transgenic activity appeared to be dependent on lactation after pregnancy, suggesting that maintenance of the lactogenic differentiation status of the mammary epithelium in a favorable hormonal milieu was a prerequisite for manifestation of the phenotype. Thus, blocking lactation by separating dams (n = 5) from their litters shortly after parturition prevented the appearance of palpable regressing tumors.
It is notable that removal of the suckling stimulus by forced weaning, experimental block of milk release, or impairment of milk ejection by genetic ablation of oxytocin, triggers, within hours, a process of involution in the mammary gland, ie, apoptosis and remodeling of lobuloalveolar structures.34,35 Although removal of the pups from the mother immediately after birth prevented MMTV-hN1IC-dependent tumor development, natural weaning and initiation of involution caused tumor regression almost invariably (Figure 3, compare B with A). Some of the transgenic animals in which the lactation-dependent tumors had regressed were sacrificed 3 weeks after weaning. Histological analysis revealed persistence of scattered microscopic papillary lesions in breast ducts possibly representing remnants of previous tumors (three of seven cases) (Figure 3I).
Notch-Induced Nonregressing Tumors
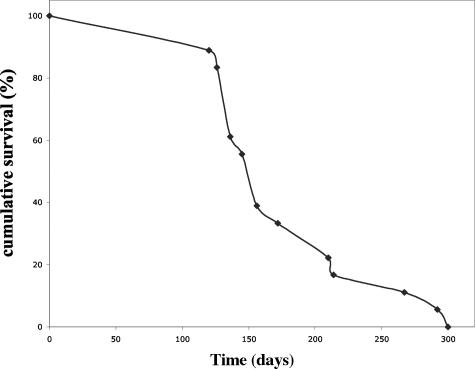
A cohort of MMTV-hN1IC females (n = 18) monitored for a period of ∼1 year developed focal nonregressing tumors (one to three per animal, occasionally in the same breast). In most cases, these neoplasms appeared after three or four pregnancies and repeated rounds of regressible neoplastic formation (the time for 50% tumor-free survival, T50, was 5.2 months; Figure 4). Tumors were also detected in two animals that underwent two pregnancies and in two animals older than 8 months, which by chance were not mated beyond their first pregnancy.
Figure 4.
Nonregressing Notch tumor latency. Eighteen MMTV-hN1IC animals were subjected to consecutive rounds of pregnancy/lactation and scored for the appearance of palpable nonregressing hN1IC-induced tumors.
Detailed histopathological analysis of 14 nonregressing tumors from eight animals (overall 25 specimens from the entire cohort of 18 mice were examined) showed that all of these neoplasms were invasive. Infiltration of adjacent adipose tissue (Figure 5A) and occasionally of skeletal muscle was further demonstrated by the absence of a myoepithelial layer after immunostaining with the anti-p63 antibody (not shown). In at least six of the cases in which breast tissue adjacent to the invading neoplasms was present, we detected an in situ component in terminal ducts (Figure 5B) invariably exhibiting the same papillary signature as that characterizing the regressible papillary tumors of lactating glands. Although the smaller, early invading carcinomas (∼0.4 cm in diameter; see Figure 5C) had retained an evenly spaced papillary architecture, the larger invasive cancers (0.9 to 1.2 cm) exhibited irregular, blunt papillae that were lined with up to 14 layers of malignant cells. In tumors that had reached this advanced stage, we observed complex arborization with secondary and tertiary branching and also merging of papillary fronds that gave rise to various secondary patterns, including trabecular and solid configurations (not shown). Moreover, numerous cancer cells in the outer layers had become necrotic, apparently by outgrowing their vascular supply. It is notable that the malignant cells had retained the low-grade cytological appearance found in in situ lesions. Their size was small to intermediate and they contained uniform or slightly pleiomorphic dark round to ovoid nuclei with a sharp nuclear border (Figure 5D). These invasive murine papillary carcinomas are comparable to well-differentiated, low-grade human papillary breast carcinomas,36 which are a distinct clinicopathological entity comprising up to 2% of all female breast cancers and tend to appear at a later age than the usual NOS (not otherwise specified) type of ductal carcinomas
Figure 5.
Nonregressing hN1IC-dependent tumors. Nonregressing hN1IC-dependent papillary invasive adenocarcinomas with low-grade nuclei and necrotic areas. A: Invasion of adipose tissue by a papillary adenocarcinoma. B: An invasive tumor (Inv) is found adjacent to a benign papillary lesion in a terminal breast duct (arrow). DCIS is also visible in this section and is indicated by an asterisk. C: Invasive tumor with evident papillary structure composed of (enlargement in D) a uniform population of cancer cells with low-grade nuclei, sharp nuclear borders, and inconspicuous nucleoli. Regressing papillary hN1IC neoplasias are more sensitive to apoptosis than the invasive tumors. E and F: Apoptotic cells identified by TUNEL analysis (brown staining) in hN1IC lactation-dependent tumors from animals sacrificed either during lactation (E) or during involution (F). Note the increased number of the TUNEL-positive cells in F, as compared to E, consistent with the notion that tumors induced during lactation retain the ability to respond to apoptotic signals during involution. G and H: TUNEL analysis in nonregressing tumors during lactation and involution, respectively. The scarcity of apoptotic nuclei, as compared to that of the regressing tumors shown in E and F indicates that these lesions are resistant to the apoptotic stimuli encountered during involution. Original magnifications: ×4 (B); ×10 (A, C); ×60 (D).
Notch-Induced Tumors and Apoptotic Processes
Assuming that focal nonregressing tumors evolve through the occurrence of secondary events from remnants of the N1IC-induced polyclonal neoplasms, an expectation was that these two lesion forms would exhibit a difference in apoptotic levels, if apoptosis during involution is indeed associated with tumor regression. TUNEL analysis showed that this is indeed the case, as abundant apoptotic cells were detected in regressing tumors during involution (Figure 5, E and F), in numbers comparable to those seen in wild-type animals. Thus, as far as we can judge by this analysis, Notch activation does not appreciably interfere with the apoptotic machinery. In contrast, the invasive nonregressing adenocarcinomas (Figure 5, G and H) exhibited only a few apoptotic figures, and this pattern was indistinguishable between tumors examined during lactation or involution (Figure 5, G and H).
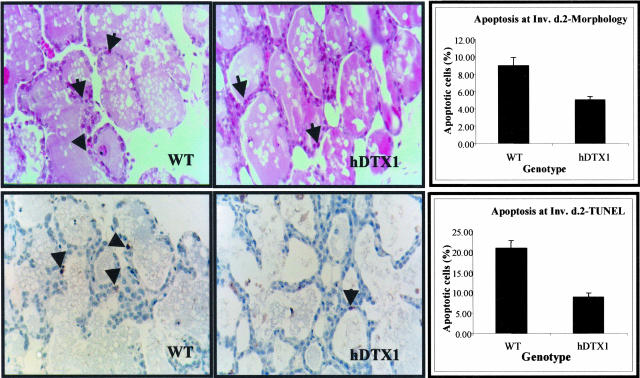
According to our observations, Notch overexpression resulting in signaling that exceeds a normal threshold seems to deregulate proliferation without interfering with apoptosis. On the other hand, reduction of the normal level of Notch signaling appears to impair the operation of the apoptotic program. Thus, when the endogenous Notch signals were inhibited in the mammary glands of MMTV-hDTX1 transgenic females by expression of the Deltex antagonist, the occurrence of involution was delayed, apparently because of suppression of apoptosis (Figure 6). However, terminal differentiation of the mammary epithelium during lactation could proceed normally.
Figure 6.
The expression of the Notch signal antagonist Deltex inhibits apoptosis during involution. The ratio of apoptotic versus total number of cells was evaluated by hematoxylin staining and TUNEL in animals at day 2 of involution. Cells were counted in groups of three to five ducts with each group deriving from an individual gland. A total of three glands, each from a different mouse, are included in this analysis. Top panels show an analysis based on the identification of apoptotic figures (examples are indicated by arrows) revealed by hematoxylin staining, and the bottom panels identify apoptosis by TUNEL (examples are indicated by arrowheads) in wild-type (WT) and MMTV-hDTX1 (hDTX1) animals at day 2 of involution. Both methods indicate that the expression of the Notch antagonist Deltex inhibits apoptosis during involution, indicating an opposite phenotype seen in MMTV-hN1IC(hN1IC) in which involution-dependent apoptosis seems intact and may even be elevated. The average values of the apoptotic cells determined by the two methods are also shown in the graphs.
Notch Signal Modulation Inhibits Hras1- but Not c-Myc-Induced Mammary Tumors
Considering the central role that Notch signaling plays in cell fate control and the dramatic tumor phenotypes associated with the expression of a constitutively active Notch1 transgene in the mammary epithelium, we sought to also examine whether the modulation of the normal endogenous Notch pathway can influence the action of other oncogenic stimuli. Thus, we asked whether Notch antagonism by transgenic expression of Deltex could affect Hras1-induced mammary tumorigenesis. For this purpose, we crossed MMTV-hDTX1 and MMTV/v-Ha-ras mice and monitored the bitransgenic progeny for tumor development.
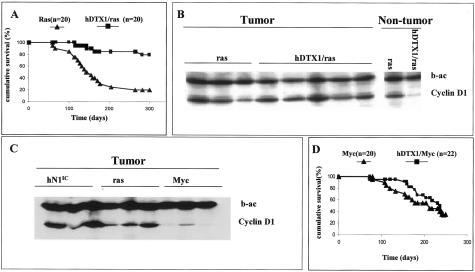
As shown in a Kaplan-Meier plot (Figure 7A), expression of Deltex inhibited strongly the oncogenic effects of Hras1 expression in the mammary glands. Thus, in the majority of bitransgenic animals, palpable mammary tumors were not detected, even after allowing for a relatively long period of latency. In fact, at ∼10 months of age, when 80% of the mice carrying only the Hras1 transgene had developed malignancies, only 20% of the bitransgenic females exhibited palpable tumors. This suppression of tumor development cannot be simply attributed to a reduction in oncogene transcription because the mammary glands of both of these animal groups expressed the transgenic Hras1 at indistinguishable levels (data not shown). We note that, the suppressing activity of Deltex exhibited tissue specificity for the mammary epithelium because salivary gland tumors, known to appear in mice carrying an MMTV-Hras1 transgene,28 were still detectable in the Ras/hDTX1 bitransgenic mice.
Figure 7.
Cyclin D1 expression and its oncogenic action can be modulated by the expression of Deltex. A: Occurrence of palpable breast cancers in MMTV-v-Ha-ras in the presence and absence of MMTV-hDTX1 transgene. Bitransgenic animals show that the expression of the Notch antagonist Deltex inhibits Ras-driven oncogenesis. Tumor incidence was recorded at least twice weekly by palpation until the experiment was terminated for the period indicated in the graph. B: In the left panel, we compare the expression of cyclin D1 in tumors from MMTV-v-Ha-ras mice in the presence or absence of the MMTV-hDTX1 transgene. For this analysis, protein extracts are shown from three independent breast tumors deriving from MMTV-v-Ha-ras animals (ras) and from all of the five instances in which mammary glands from MMTV-hDTX1/MMTV-v-Ha-ras (hDTX1/ras) mice escaped Deltex inhibition and developed tumors. In the right panel, protein extracts from nontumoral epithelium were pooled from the mammary glands of three 12-week-old virgin, tumor-free MMTV-Ha-ras (ras) and MMTV-hDTX1/MMTV-v-Ha-ras (hDTX1/ras) females, respectively. b-ac indicates the levels of β-actin, the marker used here as loading control. C: Expression of cyclin D1 in mammary tumors developed in transgenic mice carrying the MMTV-hN1IC, MMTV-v-Ha-ras, or MMTV-c-Myc transgene. Three anatomically independent tumors from each genotype were analyzed. Tumors from MMTV-hN1IC mice exhibited cyclin D1 expression levels comparable to those deriving from MMTV-v-Ha-ras animals whereas tumors from MMTV-c-Myc animals do not show cyclin D1 up-regulation. D: Occurrence of palpable breast cancers in MMTV-c-Myc mice in the presence and absence of MMTV-hDTX1 transgene. Bitransgenic animals show that the expression of the Notch antagonist Deltex does not have a clearly appreciable effect on Myc-driven oncogenesis. Tumor incidence was recorded at least twice weekly by palpation until the experiment was terminated for the period indicated in the graph.
Given that MMTV-Hras1-induced mammary tumorigenesis is inhibited in a cyclin D1-null background, while the development of salivary gland tumors is not affected,37 we asked, whether the similar phenotype of the Hras1/hDTX1 bitransgenic animals could be correlated with reduced cyclin D1 levels. To address this question, we used Western analysis and compared the amounts of cyclin D1 present in protein extracts prepared from tumor-free mammary glands dissected from 3-month-old virgin females carrying the Hras1 oncogene either alone or in combination with the hDTX1 transgene (Figure 7B). The results indicated that in MMTV-Hras1 animals, which are presumably destined to develop mammary tumors, there is Hras1-induced up-regulation of cyclin D1 expression even in a precancerous state.37–40 In contrast, in the bitransgenic mice, there was a significant reduction in the amount of cyclin D1 compared to the levels detected in the MMTV-Hras1 mice (Figure 7B). Significantly, we found that all bitransgenic animals (n = 4), which escaped the inhibition of the Hras1 oncogenic action by hDTX1 and developed mammary tumors (n = 5), displayed an up-regulated expression pattern of cyclin D1, similar to what is seen in animals expressing only Hras1 (Figure 7B).
The correlation between the action of Deltex, the Notch signaling antagonist, and low levels of cyclin D1 in mammary glands is further consistent with a reciprocal up-regulation of cyclin D1 expression in hN1IC-induced regressible tumors. In fact, as shown by Western analysis, the amounts of cyclin D1 in hN1IC- and Hras1-induced mammary tumors are comparable and much higher than those detected in tumors induced by c-myc (Figure 7C).
These observations suggest that cyclin D1 is a target of Notch signals in the mammary epithelium. This notion is consistent with the finding that the expression of hN1IC activates cyclin D1 gene transcription in cultured cells.41 Nevertheless, evidence that, beyond being a marker, the up-regulation of cyclin D1 plays a role in the tumorigenic action of hN1IC is currently lacking. This question would be directly addressed by examining the action of the hN1IC transgene in cyclin D1-null background. In analogous genetic experiments, for example, it was shown that mice lacking cyclin D1 were resistant to the development of mammary tumors induced by Hras1 or Neu, but continued to be sensitive to the action of c-Myc and Wnt1.37 Unfortunately, because of the lactation dependence of Notch1-induced tumors, such a critical experiment could not be performed in our case because the cyclin D1-null females do not lactate.42
Nevertheless, the hypothesis that the Deltex inhibitory effect on Hras1-induced mammary tumorigenesis rests on its ability to down-regulate cyclin D1 is viable. In this regard, it is noteworthy that the development of breast carcinomas remained unaffected in females carrying both MMTV-c-Myc and MMTV-hDTX1 transgenes (Figure 7D). Similarly, c-Myc induced oncogenesis in mouse mammary glands is not inhibited in a genetic background lacking cyclin D1.37
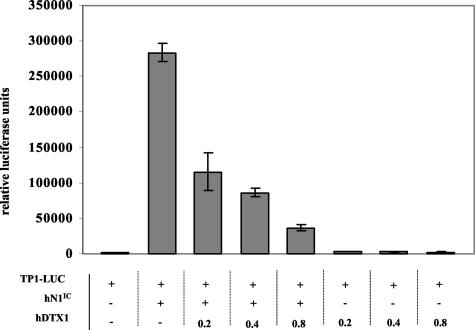
The inability of Deltex to inhibit the formation of c-Myc tumors cannot be interpreted as reflecting a failure of Deltex antagonistic activity in those tumors because, in a reporter assay using a primary culture of cells derived from a c-Myc tumor explant, Deltex was still able to inhibit the transcriptional activity of hN1IC in a dose-dependent manner (Figure 8).
Figure 8.
hDTX1 inhibits hNotch1IC-dependent transcription in MMTV c-Myc cell explants. 537M cells, derived from a c-Myc tumor explant, were transiently transfected with 0.4 μg of the TP1 reporter plasmid (see text) alone or in combination with 0.4 μg hN1IC alone or 0.4μg hN1IC together with increasing concentrations of hDTX1 (0.2, 0.4, 0.8 μg) and finally with hDTX1 alone in increasing concentrations (0.2, 0.4, 0.8 μg). The total amount of DNA transfected under each condition was normalized with the empty vector pCDNA3. After 24 hours, the cultures were lysed and assayed for luciferase activity. The histogram represents three independent luciferase assays.
Discussion
Extensive studies throughout the past 2 decades established that the developmental action of Notch signaling is highly dependent on cellular context. It is known, in addition, that regardless of context, ectopic Notch signals can be powerful modulators of cell fates during development.3 The animal model we presented here extends these concepts in the context of mammary tumorigenesis. We observed that the mammary gland is particularly sensitive to the oncogenic action of Notch1 overexpression during lactation, demonstrating that the outcome of Notch signaling in this tissue depends on the developmental stage. To our knowledge, this is the first description of a lactation-dependent breast tumor. In addition, we have demonstrated that modulation of Notch activity in the mammary gland can alter the manifestation of breast cancer driven by mutated Hras1, indicating that Notch signals have the ability to influence the oncogenic fate guided by specific genetic lesions.
The regressing tumors we observed in the lactating gland provide a model in which deregulation of proliferation appears to precede a derangement of apoptosis during tumor evolution. The cells in these tumors retain their ability to respond to apoptotic signals during involution and consequently the tumors regress, unless secondary mutagenic events drive them toward full-fledged malignancy. The exceedingly short latency in the appearance of the regressing in situ tumors, their signature nature, and their dependence on lactation does not conform with a multihit mutational model of stepwise combinatorial engagement of deranged signaling pathways involved in proliferation and apoptosis control. In fact, the incidence of somatic mutations, if any, appears to be low because we have failed to identify allelic imbalances or instability (assayed by using 10 microsatellite markers) in 12 independent papillary tumors (data not shown). Moreover, these neoplasms were not tumorigenic when transplanted into nude mice. We posit, therefore, that, in a lactation-dependent favorable background, neoplasms with a signature morphological pattern appear deterministically as a consequence of the aberrant expression of activated Notch1, which has the ability to affect and derange diverse signaling programs in a single step without a need for mutagenic events.
On the basis of these considerations, the regressing lesions can be thought of as representing a premalignant state, in which the performance of the apoptotic machinery remains intact. In fact, our model differs from other transgenic models in which tumor regression follows the withdrawal of the oncogenic stimulus.43–46 In the case of the MMTV-hN1IC animals, the continuous expression of the transgene is insufficient to maintain the neoplasms when the mammary epithelium undergoes involution. Thus, the premalignant state that we have identified provides a basis to search in the nonregressing tumors for mutational events that together with activated Notch are responsible for dissociating the reception and/or interpretation of proliferative and apoptotic cues in the mammary epithelium.
We note that in an analogous experimental design, Notch4IC transgenes driven either by the MMTV LTR10 or the whey acidic protein (WAP) gene promoter12 induced mammary adenocarcinomas in both virgin and parous mice. The ductal epithelium developed normally only in WAP-Notch4IC virgins, but both models exhibited a nonlactational phenotype, in contrast to the MMTV-Notch1IC females described here. In WAP-Notch4IC transgenic animals, the dysplastic lesions appeared throughout the mammary glands and, in contrast to our model, they did not regress after weaning. Instead, they developed into Dunn type B adenocarcinomas (sheets or cords of poorly differentiated cells), which are histologically different from the histopathological pattern of the nonregressing tumors that we have seen.
The reasons for the differences between the Notch1 and Notch4 oncogenic phenotypes are not clear.10,12 Excluding the trivial explanation that the different manifestations are because of dissimilarity in genetic backgrounds, it is reasonable to consider that the signaling cascades of the two receptors either use some nonoverlapping downstream targets or, perhaps more likely, exhibit quantitative differences in signal intensity, which in turn are manifested in distinct developmental outputs. It is notable, in this regard, that gene dosage studies in both invertebrate and vertebrate systems have indicated that normal development can be exquisitely sensitive to the quantity of Notch signals.3 Thus, until we have rigorously examined the qualitative and quantitative aspects of Notch1 and Notch 4 signaling in the mammary gland, it cannot be assumed that the Notch1 and Notch 4 animal models described are directly comparable.
Regardless of the specific molecular mechanisms underlying the formation of either the Notch1 tumors we report here or the Notch4 tumors previously described,10,12 these studies indicate that the mammary gland is sensitive to Notch signal modulation. However, although expression of all four murine Notch genes in the mammary glands of virgin, pregnant, and lactating mice has been reported,23 the exact role of Notch signaling in normal mammary gland development remains unknown.
The demonstration that the expression of Deltex interferes with Ras-driven oncogenesis in the mammary gland is also an indication, albeit indirect, that the mammary tissue is sensitive to Notch signals and consequently suggests the existence of a crosstalk between Ras and Notch signals in this tissue via cyclin D1. It must be noted however that, not withstanding the documented physical interactions between the intracellular segment of the Notch receptor and hDTX25,47 and its well-established role in Notch signal down-regulation in mammalian cells26,27 (Figure 8), we understand neither the mechanism of Deltex action nor its specificity. Thus, we cannot exclude formally that the transgenic expression of Deltex could have some effects unrelated to Notch antagonism.
Despite these caveats, an interaction between the Notch and Ras pathways in breast epithelial cells would not be surprising, because such signaling cross-talk has been documented in diverse tissues and experimental systems, including Drosophila, Caenorhabditis elegans, and mammalian cultured cells.46,48–51 Despite extensive genetic analyses in Drosophila and C. elegans, which are amenable to explorations of epigenetic relationships between the two pathways using double-mutant genetic backgrounds affecting both Notch and Ras signaling, it is still not possible to formulate a unifying interaction model.46,48–51
Although the possibility that cyclin D1 may define a node of interaction between Notch and Ras is an appealing working hypothesis, the mode of such an interplay and its consequences may vary depending on developmental context.46 The difference in tumor development between mammary and salivary glands in the Hras1/hDTX1 bitransgenic mice that we have described emphasizes this point. However, an epistatic relationship with Notch acting downstream of Ras is currently compatible not only with our data but also with experiments involving Ras-transformed human fibroblasts and epithelial cultured cells.52 As shown in the latter study, oncogenic Ras up-regulate the intracellular Notch1 activity, whereas the Ras-dependent transformed phenotype was suppressed by inhibition of Notch1 expression. On the other hand, the complexities in signaling relationships can be exemplified by the observation that Notch4-induced cellular transformation depends on parallel activation of the Ras pathway.53
Given the potency of Notch signaling in altering cell fate, it is likely, as we have argued in the past,54 that Notch signal modulation could provide a relatively general means of manipulating the fate of malignant cells, thus modifying or inhibiting their malignant character. Certainly, the inhibition of Ras-driven oncogenesis by the antagonism of Notch signaling would suggest that in principle, this hypothesis may be correct. Thus, the notion that Notch signal modulation may serve as a more general cancer therapeutic target seems to merit further consideration. Moreover, we hope that the mouse model described here could assist in the unraveling of mechanistic details related to the complex interplay between aberrant Notch signals and a variety of effectors in other pathways, especially providing a basis to search for mutational events that collaborate with activated Notch1 to trigger the onset of nonregressing tumors.
Acknowledgments
We thank Lin Wu for help with the generation of transgenic animals and our colleague Dr. Andi McClatchey for numerous helpful discussions throughout the course of this work.
Footnotes
Address reprint requests to Spyros Artavanis-Tsakonas, Massachusetts General Hospital Center for Cancer Research, Department of Cell Biology, Harvard Medical School, 13th St., Bldg. 149, Charlestown, MA 02129. E-mail: tsakonas@helix.mgh.harvard.edu.
Supported by the National Institutes of Health [grants NS26084, GM62931, and CA098402 to S.A.-T.; CA97403 (project 2) to A.E.; predoctoral fellowship provided by cancer biology training grant T32 CA09503 to K.P.] and the Herbert Irving Comprehensive Cancer Center of the Columbia Presbyterian Medical Center from the Avon Products Foundation Breast Cancer Research and Care Program (to A.E.).
References
- Wharton KA, Johansen KM, Xu T, Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985;43:567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- Kidd S, Kelley MR, Young MW. Sequence of the notch locus of Drosophila melanogaster: relationship of the encoded protein to mammalian clotting and growth factors. Mol Cell Biol. 1986;6:3094–3108. doi: 10.1128/mcb.6.9.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Greenwald I. LIN-12/Notch signaling: lessons from worms and flies. Genes Dev. 1998;12:1751–1762. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- Egan SE, St-Pierre B, Leow CC. Notch receptors, partners and regulators: from conserved domains to powerful functions. Curr Top Microbiol Immunol. 1998;228:273–324. doi: 10.1007/978-3-642-80481-6_11. [DOI] [PubMed] [Google Scholar]
- Kopan R, Cagan R. Notch on the cutting edge. Trends Genet. 1997;13:465–467. doi: 10.1016/s0168-9525(97)01318-8. [DOI] [PubMed] [Google Scholar]
- Baron M, Aslam H, Flasza M, Fostier M, Higgs JE, Mazaleyrat SL, Wilkin MB. Multiple levels of Notch signal regulation (review). Mol Membr Biol. 2002;19:27–38. doi: 10.1080/09687680110112929. [DOI] [PubMed] [Google Scholar]
- Bellavia D, Campese AF, Alesse E, Vacca A, Felli MP, Balestri A, Stoppacciaro A, Tiveron C, Tatangelo L, Giovarelli M, Gaetano C, Ruco L, Hoffman ES, Hayday AC, Lendahl U, Frati L, Gulino A, Screpanti I. Constitutive activation of NF-kappaB and T-cell leukemia/lymphoma in Notch3 transgenic mice. EMBO J. 2000;19:3337–3348. doi: 10.1093/emboj/19.13.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dievart A, Beaulieu N, Jolicoeur P. Involvement of Notch1 in the development of mouse mammary tumors. Oncogene. 1999;18:5973–5981. doi: 10.1038/sj.onc.1202991. [DOI] [PubMed] [Google Scholar]
- Jhappan C, Gallahan D, Stahle C, Chu E, Smith GH, Merlino G, Callahan R. Expression of an activated Notch-related int-3 transgene interferes with cell differentiation and induces neoplastic transformation in mammary and salivary glands. Genes Dev. 1992;6:345–355. doi: 10.1101/gad.6.3.345. [DOI] [PubMed] [Google Scholar]
- Gallahan D, Callahan R. The mouse mammary tumor associated gene INT3 is a unique member of the NOTCH gene family (NOTCH4). Oncogene. 1997;14:1883–1890. doi: 10.1038/sj.onc.1201035. [DOI] [PubMed] [Google Scholar]
- Gallahan D, Jhappan C, Robinson G, Hennighausen L, Sharp R, Kordon E, Callahan R, Merlino G, Smith GH. Expression of a truncated Int3 gene in developing secretory mammary epithelium specifically retards lobular differentiation resulting in tumorigenesis. Cancer Res. 1996;56:1775–1785. [PubMed] [Google Scholar]
- Hubmann R, Schwarzmeier JD, Shehata M, Hilgarth M, Duechler M, Dettke M, Berger R. Notch2 is involved in the overexpression of CD23 in B-cell chronic lymphocytic leukemia. Blood. 2002;99:3742–3747. doi: 10.1182/blood.v99.10.3742. [DOI] [PubMed] [Google Scholar]
- Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- Brennan K, Brown AM. Is there a role for Notch signalling in human breast cancer? Breast Cancer Res. 2003;5:69–75. doi: 10.1186/bcr559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- Struhl G, Fitzgerald K, Greenwald I. Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell. 1993;74:331–345. doi: 10.1016/0092-8674(93)90424-o. [DOI] [PubMed] [Google Scholar]
- Capobianco AJ, Zagouras P, Blaumueller CM, Artavanis-Tsakonas S, Bishop JM. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol Cell Biol. 1997;17:6265–6273. doi: 10.1128/mcb.17.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J, Baltimore D. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohn JL, Lauring AS, Linenberger ML, Overbaugh J. Transduction of Notch2 in feline leukemia virus-induced thymic lymphoma. J Virol. 1996;70:8071–8080. doi: 10.1128/jvi.70.11.8071-8080.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano JV, Uyttendaele H, Kitajewski J, Montesano R. Transduction of Notch2 in feline leukemia virus-induced thymic lymphoma. Int J Cancer. 2000;86:652–659. doi: 10.1002/(sici)1097-0215(20000601)86:5<652::aid-ijc8>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Soriano JV, Uyttendaele H, Kitajewski J, Montesano R. Expression of an activated Notch4(int-3) oncoprotein disrupts morphogenesis and induces an invasive phenotype in mammary epithelial cells in vitro. Int J Cancer. 2000;86:652–659. doi: 10.1002/(sici)1097-0215(20000601)86:5<652::aid-ijc8>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Callahan R, Raafat A. Notch signaling in mammary gland tumorigenesis. J Mammary Gland Biol Neoplasia. 2001;6:23–36. doi: 10.1023/a:1009512414430. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Matsuno K, Eastman D, Mitsiades T, Quinn AM, Carcanciu ML, Ordentlich P, Kadesch T, Artavanis-Tsakonas S. Human deltex is a conserved regulator of Notch signalling. Nat Genet. 1998;19:74–78. doi: 10.1038/ng0598-74. [DOI] [PubMed] [Google Scholar]
- Sestan N, Artavanis-Tsakonas S, Rakic P. Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science. 1999;286:741–746. doi: 10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- Izon DJ, Aster JC, He Y, Weng A, Karnell FG, Patriub V, Xu L, Bakkour S, Rodriguez C, Allman D, Pear WS. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity. 2002;16:231–243. doi: 10.1016/s1074-7613(02)00271-6. [DOI] [PubMed] [Google Scholar]
- Sinn E, Muller W, Pattengale P, Tepler I, Wallace R, Leder P. Coexpression of MMTV/v-Ha-ras and MMTV/c-myc genes in transgenic mice: synergistic action of oncogenes in vivo. Cell. 1987;49:465–475. doi: 10.1016/0092-8674(87)90449-1. [DOI] [PubMed] [Google Scholar]
- Strobl LJ, Hofelmayr H, Stein C, Marschall G, Brielmeier M, Laux G, Bornkamm GW, Zimber-Strobl U. Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch1 transactivate genes by interacting with the cellular protein RBP-J kappa. Immunobiology. 1997;198:299–306. doi: 10.1016/s0171-2985(97)80050-2. [DOI] [PubMed] [Google Scholar]
- Chatzistamou I, Schally AV, Pafiti A, Kiaris H, Koutselini H. Expression of growth hormone-releasing hormone in human primary endometrial carcinomas. Eur J Endocrinol. 2002;147:381–386. doi: 10.1530/eje.0.1470381. [DOI] [PubMed] [Google Scholar]
- Yun K, Fischman S, Johnson J, Hrabe de Angelis M, Weinmaster G, Rubenstein JL. Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development. 2002;129:5029–5040. doi: 10.1242/dev.129.21.5029. [DOI] [PubMed] [Google Scholar]
- Ehmann UK, DeVries JT, Chen MS, Adamos AA, Guzman RC, Omary MB. An in vitro model of epithelial cell growth stimulation in the rodent mammary gland. Cell Prolif. 2003;36:177–190. doi: 10.1046/j.1365-2184.2003.00265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbareschi M, Pecciarini L, Cangi MG, Macri E, Rizzo A, Viale G, Doglioni C. p63, a p53 homologue, is a selective nuclear marker of myoepithelial cells of the human breast. Am J Surg Pathol. 2001;25:1054–1060. doi: 10.1097/00000478-200108000-00010. [DOI] [PubMed] [Google Scholar]
- Quarrie LH, Addey CV, Wilde CJ. Programmed cell death during mammary tissue involution induced by weaning, litter removal, and milk stasis. J Cell Physiol. 1996;168:559–569. doi: 10.1002/(SICI)1097-4652(199609)168:3<559::AID-JCP8>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Li M, Liu X, Robinson G, Bar-Peled U, Wagner KU, Young WS, Hennighausen L, Furth PA. Mammary-derived signals activate programmed cell death during the first stage of mammary gland involution. Proc Natl Acad Sci USA. 1997;94:3425–3430. doi: 10.1073/pnas.94.7.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansidhar BJ, Garguilo GA. Papillary carcinoma of the breast: characteristics and classification. Am Surg. 2003;69:400–403. [PubMed] [Google Scholar]
- Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411:1017–1021. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- Filmus J, Robles AI, Shi W, Wong MJ, Colombo LL, Conti CJ. Induction of cyclin D1 overexpression by activated ras. Oncogene. 1994;9:3627–3633. [PubMed] [Google Scholar]
- Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell RG. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem. 1995;270:23589–23597. doi: 10.1074/jbc.270.40.23589. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Chao JR, Jiang MC, Ng SY, Yen JJ, Yang-Yen HF. Ras transformation results in an elevated level of cyclin D1 and acceleration of G1 progression in NIH 3T3 cells. Mol Cell Biol. 1995;15:3654–3663. doi: 10.1128/mcb.15.7.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchini C, Capobianco AJ. Induction of cyclin D1 transcription and CDK2 activity by Notch(ic): implication for cell cycle disruption in transformation by Notch(ic). Mol Cell Biol. 2001;21:5925–5934. doi: 10.1128/MCB.21.17.5925-5934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicinski P, Donaher JL, Parker SB, Li T, Fazeli A, Gardner H, Haslam SZ, Bronson RT, Elledge SJ, Weinberg RA. Cyclin D1 provides a link between development and oncogenesis in the retina and breast. Cell. 1995;82:621–630. doi: 10.1016/0092-8674(95)90034-9. [DOI] [PubMed] [Google Scholar]
- Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N, Shen Q, O’Hagan R, Pantginis J, Zhou H, Horner JW, II, Cordon-Cardo C, Yancopoulos GD, DePinho RA. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400:468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- D’Cruz CM, Gunther EJ, Boxer RB, Hartman JL, Sintasath L, Moody SE, Cox JD, Ha SI, Belka GK, Golant A, Cardiff RD, Chodosh LA. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nat Med. 2001;7:235–239. doi: 10.1038/84691. [DOI] [PubMed] [Google Scholar]
- Gunther EJ, Moody SE, Belka GK, Hahn KT, Innocent N, Dugan KD, Cardiff RD, Chodosh LA. Impact of p53 loss on reversal and recurrence of conditional Wnt-induced tumorigenesis. Genes Dev. 2003;17:488–501. doi: 10.1101/gad.1051603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena A, Buff E, Halfon MS, Gisselbrecht S, Jimenez F, Baylies MK, Michelson AM. Reciprocal regulatory interactions between the Notch and Ras signaling pathways in the Drosophila embryonic mesoderm. Dev Biol. 2002;244:226–242. doi: 10.1006/dbio.2002.0606. [DOI] [PubMed] [Google Scholar]
- Matsuno K, Diederich RJ, Go MJ, Blaumueller CM, Artavanis-Tsakonas S. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development. 1995;121:2633–2644. doi: 10.1242/dev.121.8.2633. [DOI] [PubMed] [Google Scholar]
- Fortini ME, Rebay I, Caron LA, Artavanis-Tsakonas S. An activated Notch receptor blocks cell-fate commitment in the developing Drosophila eye. Nature. 1993;365:555–557. doi: 10.1038/365555a0. [DOI] [PubMed] [Google Scholar]
- Karim FD, Chang HC, Therrien M, Wassarman DA, Laverty T, Rubin GM. A screen for genes that function downstream of Ras1 during Drosophila eye development. Genetics. 1996;143:315–329. doi: 10.1093/genetics/143.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyen EM, Purcell KJ, Fortini ME, Artavanis-Tsakonas S. Analysis of dominant enhancers and suppressors of activated Notch in Drosophila. Genetics. 1996;144:1127–1141. doi: 10.1093/genetics/144.3.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaye DD, Greenwald I. Endocytosis-mediated downregulation of LIN-12/Notch upon Ras activation in Caenorhabditis elegans. Nature. 2002;420:686–690. doi: 10.1038/nature01234. [DOI] [PubMed] [Google Scholar]
- Weijzen S, Rizzo P, Braid M, Vaishnav R, Jonkheer SM, Zlobin A, Osborne BA, Gottipati S, Aster JC, Hahn WC, Rudolf M, Siziopikou K, Kast WM, Miele L. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat Med. 2002;8:979–986. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- Fitzgerald K, Harrington A, Leder P. Ras pathway signals are required for notch-mediated oncogenesis. Oncogene. 2000;19:4191–4198. doi: 10.1038/sj.onc.1203766. [DOI] [PubMed] [Google Scholar]
- Zagouras P, Stifani S, Blaumueller CM, Carcangiu ML, Artavanis-Tsakonas S. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proc Natl Acad Sci USA. 1995;92:6414–6418. doi: 10.1073/pnas.92.14.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]