Abstract
Human papilloma virus (HPV) causes focal infections of epithelial layers in skin and mucosa. HIV-infected patients on highly active antiretroviral therapy (HAART) appear to be at increased risk of developing HPV-induced oral warts. To identify the mechanisms that allow long-term infection of oral epithelial cells in these patients, we used a combination of laser-dissection microscopy (LDM) and highly sensitive and quantitative, non-biased, two-step multiplex real-time RT-PCR to study pathogen-induced alterations of specific tissue subcompartments. Expression of 166 genes was compared in three distinct epithelial and subepithelial compartments isolated from biopsies of normal mucosa from HIV-infected and non-infected patients and of HPV32-induced oral warts from HIV-infected patients. In contrast to the underlying HIV infection and/or HAART, which did not significantly elaborate tissue substructure-specific effects, changes in oral warts were strongly tissue substructure-specific. HPV 32 seems to establish infection by selectively enhancing epithelial cell growth and differentiation in the stratum spinosum and to evade the immune system by actively suppressing inflammatory responses in adjacent underlying tissues. With this highly sensitive and quantitative method tissue-specific expression of hundreds of genes can be studied simultaneously in a few cells. Because of its large dynamic measurement range it could also become a method of choice to confirm and better quantify results obtained by microarray analysis.
Human papilloma virus (HPV)-induced pre-neoplastic lesions can precede cancerous transformation of skin and mucosal surface epithelium.1,2 Innate and/or adaptive immunity seem to provide some control of HPV infection and/or replication, since HPV-induced oral warts are more common in HIV-infected patients and in immunocompromised renal transplant recipients compared to fully immunocompetent persons.3,4 Interestingly, the incidences of oral warts seem to increase in HIV+ patients on highly active anti-retroviral therapy (HAART),5,6 a treatment that leads to the reconstitution of the peripheral immune system.7,8 This finding together with the fact that peripheral immunity to HPV seems poorly induced,9,10 points to a crucial role for local, oral mucosa-specific immune functions in controlling HPV infection. This control mechanism might not be completely restored by HAART.
Laser-dissection microscopy (LDM) can facilitate the in situ study of biological processes in solid tissues and organs by precisely isolating cells and tissue substructures for further analysis. Individual gene expression studies by quantitative single-gene RT-PCR analysis or multi-gene microarray technologies from cells isolated by LDM have been reported.11–16 Single-gene analysis has obvious limitations and although microarray analyses provide the most comprehensive approach to date for gene expression studies, they require large amounts of starting RNA and/or extensive pre-amplification with complex quality control issues because of possible bias introduced by non-linear amplification.11–14 Importantly, microarray-based hybridization methods have significantly lower sensitivities and dynamic ranges compared to PCR-based methodologies.17 Furthermore, technologies are lacking that allow the independent verification of multi-gene changes indicated by microarray analyses. To overcome these limitations we have developed a novel, highly sensitive and quantitative approach of laser-dissection microscopy (LDM)-assisted multiplex nested real-time RT-PCR that allows gene expression studies for hundreds of genes in situ from distinct tissue subcompartments isolated from biopsy materials. We show the great potential of this approach by demonstrating highly compartmentalized focal effects of HPV32 infection on a single epithelial layer (stratum spinosum) of the oral mucosa in HIV-infected patients.
Materials and Methods
Patients and Biopsy Material
Five-mm punch biopsies, snap-frozen in liquid nitrogen and stored until sectioned, were taken at the University of California, San Francisco (UCSF) Oral AIDS Center Clinic from oral warts of six HIV+ male and one HIV+ female patient (46 ± 7.8 years) and from non-diseased buccal mucosa of three male and five female HIV+ patients (46.0 ± 6.1 years) and from seven male and five female (31.7 ± 6.9 years) HIV− patients. Non-diseased buccal mucosa was taken from patients with no oral lesions at the time of biopsy. Overall CD4 counts were similar between patients that presented with oral warts and those that presented without lesions (385 ± 217 and 354 ± 185 CD4+ cells/μl blood, respectively). Viral loads in both patient groups varied from undetectable to 95,000 copies/L plasma with no statistical significant differences between the groups (P = 0.934). All HIV+ patients were on HAART. All procedures followed protocols approved by the UCSF Internal Review Board. The Tissue Core, UCSF Comprehensive Cancer Center, provided tonsil materials.
Diagnosis of HPV-Induced Oral Warts and HPV-Typing
Clinical diagnosis of oral warts as single or multiple mucosal nodules, usually with multiple papillomatous surface projections was confirmed microscopically on H&E-stained paraffin-embedded tissue sections as mucosal nodules with verruciform projections, hyper- or hyperpara-keratosis and koilocytosis. Part of the OCT-embedded biopsy underwent DNA extraction and PCR analysis for HPV typing.18,19 All oral wart biopsies were identified as HPV consensus sequence positive, HPV 32 positive, and negative for all other HPV types tested, confirming previous reports on the prevalence of HPV32 in oral warts.20
Laser-Dissection Microscopy
Seven μm cryosections from freshly frozen tonsils and 10 μm sections from oral biopsies were placed on uncoated slides and fixed for 1 minute in 70% ethanol. The sections were then subjected to the following series of reagents, each for 30 seconds (5 seconds for oral biopsies): water, Mayer’s hematoxylin, water, bluing reagent, 70% ethanol, 95% ethanol, eosin (alcoholic with phloxine), 70% ethanol, 95% ethanol, and 100% ethanol. For capturing with the Arcturus PixCell II system (Arcturus, Moutain View, CA), sections were incubated twice for 5 minutes in xylene then allowed to air dry for 20 minutes before proceeding to microdissection. Microdissection was performed using the Arcturus PixCell II system with high sensitivity caps (Arcturus), or a P.A.L.M system (P.A.L.M. Microlaser Technologie GmbH, Bernried, Germany), using polyethylene naphthalene membrane-covered slides. For the Arcturus system, loosely adherent tissue was removed first using “LCM prep strips” (Arcturus). Cell populations were captured using 75 mW, 1 msec pulses, and lysed in RLT buffer (Qiagen, Valencia, CA) containing linear acrylamide (20 μg/ml).
RNA Purification
Depending on the experiments conducted, different amounts of tissue were collected by LDM for RNA purification. For the analysis of oral warts and normal oral mucosa close to the entire tissue substructure (stratum spinosum, basal layer or superficial connective tissue) from several 5 mm × 10 μm 127 section was isolated. The number of sections from which tissue was collected had to be adjusted from biopsy to biopsy depending on the way the biopsy was cut and the depths of the biopsy. The goal was to keep the overall captured surface area similar for all biopsies, as evaluated by studying the raw GAPDH values obtained (see below). Successful amplification, however, was achieved with as little as 5 to 6 cells captured by LDM (data not shown, see also discussion). Larger amounts of tissue were captured mainly to avoid sampling bias. RNA was purified using the RNeasy system (Qiagen) following manufacturer’s protocol. RNA was eluted with RNase-free water (Sigma-Aldrich, St. Louis, MO), subjected to RQ1 RNase-free DNase (5 U, Promega, Madison, WI) for 1 hour at 37°C and re-purified following the “RNA cleanup” protocol (Qiagen) and eluted into 30-μl storage buffer (Ambion, Austin, TX) in the presence of linear acrylamide. Total RNA from entire sections of snap-frozen tonsils before and following staining were assessed with a Bioanalyser (Agilent Technologies, Palo Alto, CA) to determine RNA quality. Although staining did affect RNA quality slightly (ratio of 28S to 18S rRNA: 1.67 and 2.14, respectively) as also noted by others14 the overall RNA quality was very good (data not shown).
RT-Multiplexed-Nested-Real Time-PCR
Purified RNA (one-third of total obtained) was reverse transcribed in 20-μl volume following the manufacturer’s instructions (80 U RNase H- Superscript II (Invitrogen, Carlsbad, CA), 250 ng random hexamers (Invitrogen), 20 nmoles dNTPs each (10 nm for oral biopsies; Clontech, BD Biosciences, Palo Alto, CA), 20 U RNase inhibitor (Superase In, Ambion)). Following reverse transcription, two separate amplification steps were performed. First, half of the cDNA obtained from the reverse-transcription step was used for a multiplex pre-amplification step in a volume of 50 μl. Pre-amplification was done using the Advantage 2 cDNA polymerase system (Clontech) with a mixture of all “outer” primers (10 pmoles each, dNTPs at 10 nmoles each) for the genes to be analyzed, including the housekeeping genes. This cDNA/outer primer mix was amplified in a thermocycler (Perkin Elmer Applied Biosystems, Foster City, CA): 1 cycle at 94°C for 1 minute, 25 cycles at 94°C for 15 seconds, 55°C for 15 seconds, 70°C for 40 seconds, and 1 cycle at 70°C for 5 minutes.
Second, individual real-time RT-PCR reactions were set up using individual sets of “inner” primers. This included separate reactions for the housekeeping genes. Preliminary experiments established that for the amount of RNA obtained by LDM in the experiments involving oral biopsies, addition of between 0.1 to 0.5 μl of pre-amplification material to 25 μl total real-time PCR mix (for oral biopsies 10 μl reactions) containing universal master mix (Applied Biosystems), primers (300 nmol each), and a 6-FAM and BHQ-1-labeled probe (100 nmol, Biosearch Technologies, Novato, CA) would result in linear second-round amplification (see Figure 2 and data not shown). Amplification occurred on a 7700 or 7900 Sequence Detection System (Applied Biosystems): 1 cycle at 50°C for 2 minutes, 1 cycle at 95°C for 10 minutes, 40 cycles at 95°C for 15 seconds, 60°C for 1 minute. Great care was given to the analysis of all raw Ct values obtained from these amplifications. Amplifications resulting in Ct values <6 (threshold 0.04) were repeated with smaller amounts of input pre-amplification cDNA to ensure that amplification plateaus were not reached. Outer and inner primers were designed with the “Primer Express” software (Applied Biosystems). Amplicon lengths for outer primers was kept <250 bp. Second round amplicon lengths were between 75 to 120 bp. Because of the large number of primers needed, further primer optimization was done only when primers designed by the software did not provide amplification with commercial available positive bulk control RNA. Primer sequences are provided on request.
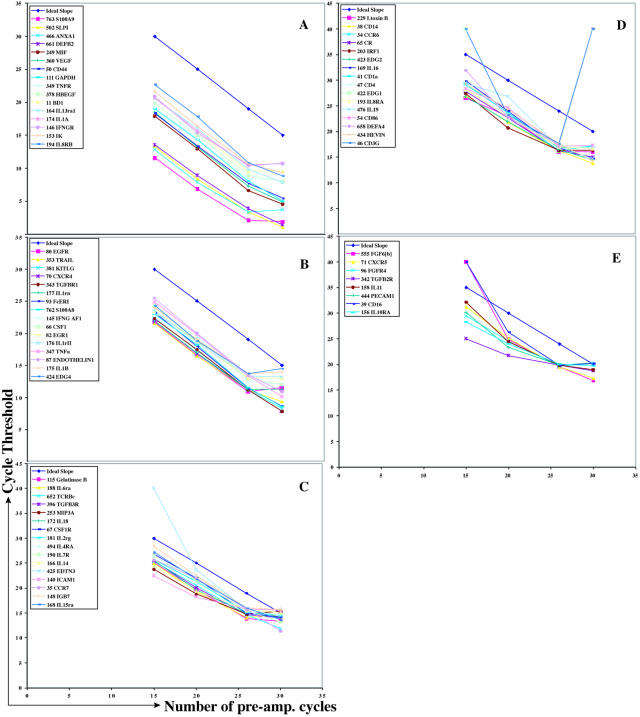
Figure 2.
Multiplex pre-amplification yields linear amplification over a wide range of pre-amplification cycles. A–E: Results from real-time RT-PCR analysis of 88 genes (A, strongest expressers; E, weakest expressers) quantified in LDM-isolated tonsil material following multiplex RT-PCR pre-amplification when varying 15 to 30 pre-amplification cycles were used. Note the good correlation between Ct values obtained with real-time RT-PCR and the number of amplification cycles used during the first (multiplex) amplification step between 20 and <30 amplification cycles. Ct value of 40, 0 copies of RNA.
Cloning of PCR Products
PCR products for 11 randomly chosen genes were amplified with outer primer sets under pre-amplification conditions (see above). Purified products were cloned into the pCRII-TOPO plasmid vector (Invitrogen) and sequenced (UC Davis Sequencing Facility). RNA was generated using AmpliScribe T7polymerase (Epicenter, Madison, WI) according to the manufacturer’s instructions and quantified by spectrophotometry.
Data Normalization and Statistical Analysis
To identify a robustly expressed control gene, eight genes (glucoronidase β, glyceraldehydes-3-phosphate dehydrogenase (GAPDH), peptidylprolyl isomerase A, phosphoglycerate kinase 1, polymerase II polypeptide G, protein phosphatase 2, transferring receptor, and ubiquitin) were compared by geometric averaging their mean Ct values, obtained from 10 oral epithelium and 10 basal layer samples of 10 HIV+ patients with or without oral warts, using the GeNorm software (www.wzw.tum.de/gene-quantification/) as outlined by others.17 GAPDH was consistently identified as one of the most stably expressed genes (data not shown) and was used for data normalization. All eukaryotes express GAPDH and contamination with for example, yeast, could affect those values. Therefore, we ensured by BLAST search (data not shown) that the primer and probe sets do not amplify known sequences of GAPDH from species other than human.
Ct values were normalized using the raw GAPDH values obtained for each of two separate collections of tonsil epithelium and follicles of the same patient. Statistical evaluation was conducted using the rank test. Statistical analysis for oral warts was conducted on the gene expression profiles of stratum spinosum, basal plus superbasal layers, and superficial connective tissue isolated by LDM from oral warts and normal buccal mucosa, respectively. After Ct values were normalized using the raw GAPDH values, log Ct values were obtained for all 167 genes because of the logarithmic correlation between Ct value and gene expression levels. The genes were classified into six gene families (chemokines, chemokine-receptors, cytokine/growth factor receptors, cytokines/growth factors, effector molecules, and surface markers). An analysis of variance model was used to determine the effects of the tissue substructure (epithelium versus basal layer/superbasal layer versus superficial connective tissue), individual genes, gene family, and diagnosis (oral wart versus normal mucosa) on gene expression profiles. Least squares means comparisons were performed to determine statistical significance of each of those comparisons. A separate analyses of male- and female-derived samples revealed no significant gender differences (P = 0.37) and data from combined analysis are therefore presented.
Results
Gene Expression Profiles from Tissue Substructures Using an Assay with a Large Dynamic Range and a Low Threshold of Detection
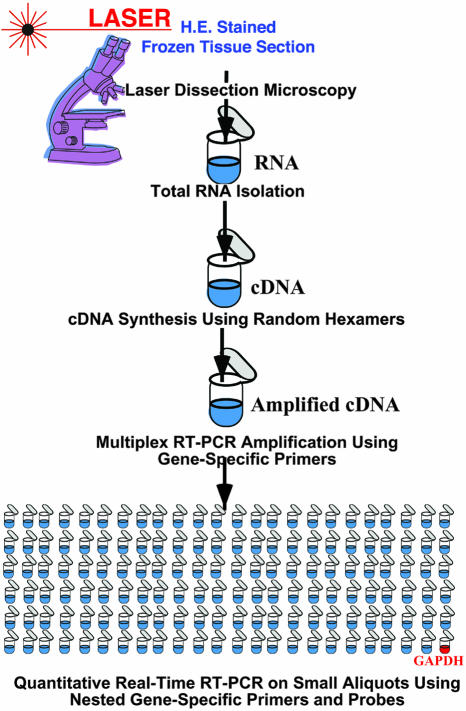
To obtain unbiased quantitative gene expression profiles from small numbers of cells while retaining information on their tissue location, we have combined LDM with nested multiplex real-time RT-PCR. The major steps in this approach include the isolation of total RNA from cells isolated by LDM followed by bulk reverse transcription then pre-amplification of cDNA by multiplex RT-PCR using a cocktail of specific primers. From this step, sufficient material is obtained to perform hundreds of nested real-time RT-PCR reactions with single gene-specific (inner) primers and probes to create expression profiles of the isolated cells (Figure 1).
Figure 1.
Schematic outline of LDM-assisted multiplex real-time RT-PCR. Shown are the steps involved in generating gene expression profiles of tissue subcompartments and small clusters of cells from frozen sections isolated by laser-dissection microscopy. For details, see Materials and Methods.
Multiplex Pre-Amplification Using Gene-Specific Primers
Isolation of small amounts of RNA such as the amounts obtained after LDM requires pre-amplification to yield sufficient amplifying material. However, pre-amplification potentially biases any down-stream attempts of quantification through unequal amplification. The following experiments were conducted to ensure that the pre-amplification step used by this approach does not bias the final real-time RT-PCR measurements and to determine the optimal number of multiplex pre-amplification cycles for LDM-assisted gene expression profiling (Figure 2). RNA obtained from fresh-frozen tonsils after cryo-sectioning, staining, and LDM-assisted tissue isolation was reverse-transcribed as outlined in Materials and Methods. Eighty-eight genes were amplified from four identical aliquots of this sample that underwent 15, 20, 26, and 30 cycles, respectively, of pre-amplification. The results showed a strong linear correlation between the numbers of pre-amplification cycles performed and the Ct values obtained after the second (real-time RT-PCR) amplification step (Figure 2). Non-linear correlations for some genes were observed when <15 or 30 or more pre-amplification cycles were run, consistent with a previous study.21 Amplification done with gene non-specific primers never resulted in good linear amplification over these numbers of cycles (data not shown). Importantly, optimal numbers of pre-amplification cycles were determined similarly with each different primer mix used to ensure linear amplification for every amplification condition used. Because of the small amounts of RNA isolated by laser-dissection microscopy, 26 pre-amplification cycles (well within the linear range of the assay, Figure 2) were chosen for all experiments presented here to maximize assay sensitivity.
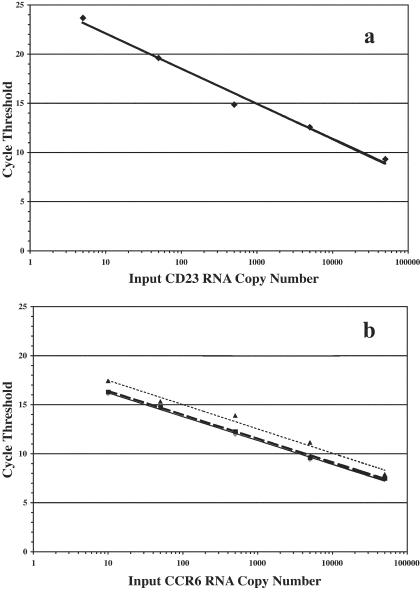
We amplified 10 to 100,000 copies of a randomly chosen control RNA (CCR6) in the presence or absence of 100,000 RNA copies each of 10 other randomly cloned control RNA (Figure 3, a and b), to determine the effects of the simultaneous pre-amplification of large numbers of genes. There was no difference in the threshold of detection, nor the linearity of the assay, when amplification of CCR6 occurred in the presence of amplification of 10 other genes added at 100,000 copies each to the reaction (Figure 3b). There appeared to be a slight decrease in Ct values when primers for 90 genes were used. The presence of large numbers of primers increased Ct values by between 0.4 and 1.7 Ct (1.3- to 3.2-fold), without significantly affecting the linearity of the assay. Thus, multiplexing at the pre-amplification step has only a minimal effect on the threshold levels of detection and allows the analysis of hundreds of genes from extremely small sample sizes.
Figure 3.
Strong correlation of gene copy number and cycle threshold in multiplex RT-PCR. Indicated numbers of RNA copies obtained from cloned PCR products (CD23 for top, CCR6 for bottom) were amplified by real-time RT-PCR in the absence (a, r2 = 0.9862) or presence (b) of an excess of 10 different control RNA (100,000) copies) (diamonds, solid line; r2 = 0.9957) and 10 (squares, dashed line; r2 = 0.9987), or 90 (triangles, speckled line; r2 = 0.9845) distinct primer sets. None of those conditions significantly affected the linearity or sensitivity of the assay.
Relationship between Gene Copy Number and Ct-Value
Real-time RT-PCR has a large linear range of measurement and a low threshold of detection, criteria that are crucial for accurately determining the gene expression profiles of small amounts of biological material. To analyze the relationship between Ct values and RNA gene copy number, we generated RNA from the cloned PCR products of 11 randomly chosen genes. A known amount of RNA was then amplified identically to the steps used for amplification of cellular RNA (Figure 1). RNA input amounts (5 to 50,000 copies) inversely correlated with the obtained Ct values in a strong linear fashion (r2 = 0.99) (Figure 3a). Furthermore, the assay is exquisitely sensitive, detecting as little as 5 to 10 copies RNA.
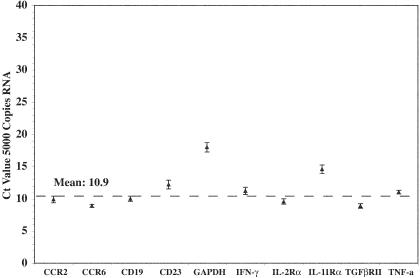
Repeated amplification of the same control RNA (Figure 4) or of RNA extracted from biological samples (not shown) demonstrated the high reproducibility of the assay. Using control RNA we estimated assay-to-assay variation of 0.17 to 1.3 Ct (ie, 1.13- to 2.5-fold differences in gene expression), with most variations being in a range well below 1.0 Ct (median 0.67 Ct or 1.6-fold, Figure 4). Next we determined whether amplification of the same amount of RNA from different genes would result in similar Ct values, thus whether Ct values could be converted into biologically more meaningful gene copy numbers. For most of the amplified genes, 5000 copies of RNA resulted in a Ct value of 10.9 (±1.8) (Figure 4). However, of the 11 genes that were analyzed repeatedly, two (GAPDH and IL11Rα) gave consistently higher Ct values with two different preparations of RNA and despite the fact that their sequence was a perfect match for the primers. Further analysis showed that one of the outer GAPDH primers could form a hairpin structure, providing a possible explanation for the reduced amplification of GAPDH compared to the other genes. Thus, although for most genes a conversion of Ct values in absolute gene copy number can be done, this is not the case for all genes and should be avoided unless a standard RNA is available for all amplified genes.
Figure 4.
Relationship between RNA input copy number and Ct value. Shown are the mean ± SD Ct values obtained by repeated amplification of 5000 copies of cloned PCR product (RNA) from the indicated genes. Although Ct values are similar for each gene, differences in Ct values exist between amplification of same amounts of RNA from different genes. Titration of control RNA for each gene showed a near perfect correlation between input RNA and cycle threshold value (Figure 3 and data not shown).
Gene Expression Differences within Similar Tissue Structures of the Same Patient
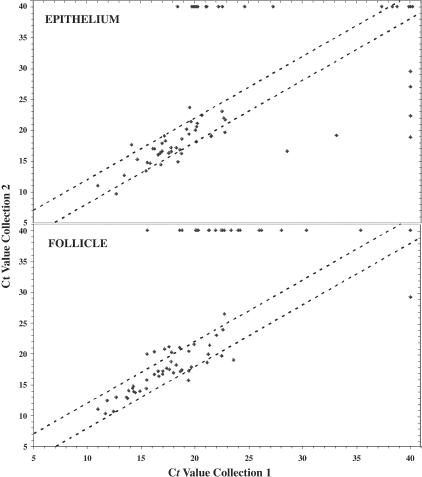
We determined the gene expression profiles for two different follicular and epithelial areas taken from one human tonsil. For each tissue compartment 88 genes were studied encoding cell surface markers, cytokines, growth factors, chemokines and their respective receptors, and other effector molecules. Differences between Ct values obtained from repeated sampling were very small (Figure 5) and not significant when genes were studied that yielded mean normalized Ct values of <27 (P = 0.05 and 0.68 for follicle and epithelial data, respectively). In contrast, genes that gave Ct values above 27 showed larger variation. Overall, the data were not statistically significantly different for the epithelium (P = 0.68) but did reach apparent statistical significance for the follicular data (P = 0.019). Similarly normalized Ct values of 25 and above represent very small input RNA copy numbers for most of the genes under study (Figure 3). These experiments also showed an extremely high degree of reproducibility for gene copy numbers down to <5 copies of RNA, thus larger differences likely represent true sample-to-sample variation. These differences are unlikely to be of biological significance, since they represent overall very small differences in RNA input amount.
Figure 5.
Strong correlation between gene expression levels and tissue of origin. The degree of variation for expression of individual genes within similar tissue structures from the same patient was studied. For this amplification of 88 genes was conducted for two different epithelial and follicular areas from the same human tonsil isolated by LDM. Shown are the Ct values for each of the two collections, plotted against each other. The tightest correlations were found for genes that resulted in Ct values below 25 (area between dotted lines marks Ct value differences of <2). Higher Ct values often resulted in one analysis not showing any gene expression (Ct, 40).
Relationship between Numbers of Cells Captured by LDM and Cycle Threshold Ct-Value of Housekeeping Genes Measured by Real-Time RT-PCR
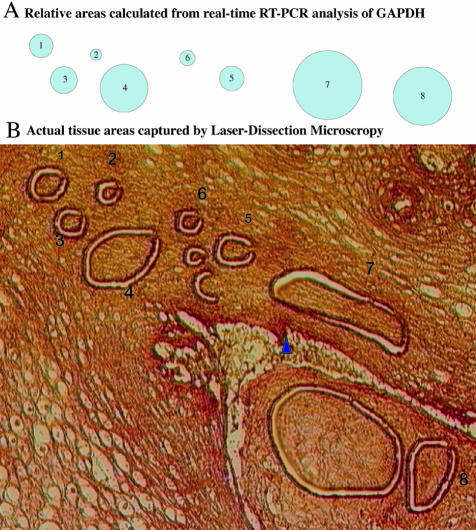
Data must be normalized when comparing gene expression in small tissue samples isolated by LDM to account for possible differences in the amounts of mRNA present in the starting material. Genes that are expressed constitutively at roughly equal levels in all cell types (“housekeeping genes”) are the preferred method for data normalization when RNA concentrations cannot be measured directly. When properly applied, the use of housekeeping genes is fast, highly reproducible, and requires little input RNA.17 Of eight different housekeeping genes tested, GAPDH was one of the most stably expressed and was therefore used for normalization purposes (see Materials and Methods, data not shown). Because the primer sets for GAPDH amplified less efficiently compared to those for other genes (Figure 3), we could use them to amplify this abundant gene under the same conditions as used for all other genes. Importantly, there was a direct relationship between the size of the tissue area (oral mucosa from HIV+ patients) from which RNA was isolated and the Ct values for GAPDH that were obtained (Figure 6 and Table 1). Figure 6A visualizes the relative tissue areas as calculated by the raw GAPDH values obtained for the indicated capture (Table 1). Figure 6B shows how closely related the areas of the actual tissue capture are with those mathematically generated from the GAPDH values. Similar analysis on small cell numbers sorted by flow cytometry (1 to 100 cells) confirmed the precise relationship between input cell numbers and GAPDH Ct values (data not shown and21). Furthermore, amplification of known amounts of cloned GAPDH PCR products consistently gave the same Ct values (Figure 4). For comparisons of tissue-specific gene expression in various patient groups (see below) we therefore attempted to isolate RNA from similar size tissue areas. The raw GAPDH Ct value was used to judge whether similar amounts of tissue were taken and patient materials were compared that gave similar raw GAPDH values (within the range of 2 Ct).
Figure 6.
Amplification of GAPDH accurately reflects the size of captured tissue areas. B: Indicated areas marked 1 to 8 of normal oral mucosa from HIV+ patients were captured by laser-dissection microscopy, RNA was extracted and expression levels for GAPDH were determined as described in Materials and Methods and shown in Table 1, column 2. From this relative areas of tissue were calculated as “relative diameters” shown in Table 1, column 4 and are displayed graphically in (A) (see also Table 1).
Table 1.
Correlation of Measured Ct Values and Size of Captured Tissue Area
| Sample* | Ct value GAPDH | Relative RNA amounts† | Relative diameter of corresponding circle‡ |
|---|---|---|---|
| 1 | 20.06 | 1.00 | 1.00 |
| 2 | 22.15 | 0.23 | 0.48 |
| 3 | 19.69 | 1.29 | 1.14 |
| 4 | 18.01 | 4.13 | 2.03 |
| 5 | 19.93 | 1.09 | 1.05 |
| 6 | 21.30 | 0.42 | 0.65 |
| 7 | 16.96 | 8.55 | 2.92 |
| 8 | 17.45 | 6.12 | 2.47 |
, Numbers refer to tissue areas shown in Figure 6B.
, Calculated by setting sample 1 captured area arbitrarily as 1.00. Relative RNA amounts of other captures were then calculated as fold-differences: 2 raised to the power of (Ct sample 1—Ct sample x).
, Graphic representation is shown in Figure 6A. Relative diameter calculated as diameter for sample x divided by diameter for sample 1. Diameter was calculated as two times the radius (radius = square root of (relative area divided by pi).
Highly Tissue Substructure-Specific Effects of Human Papillomavirus Infection
To study the extent and quality of the local immune response to HPV infection of HIV+ patients on HAART, we performed a detailed gene expression analysis on the stratum spinosum and two adjacent tissue layers (basal layer/superbasal epithelial layer and superficial connective tissue) of HPV32-induced oral warts. Results from the gene expression analysis were compared to similarly LDM-dissected tissue layers from non-diseased oral mucosa of HIV-infected individuals on HAART (Figure 7). Analysis focused on cell surface receptors, cytokines, chemokines, and other effector molecules as well as their receptors (Table 2).
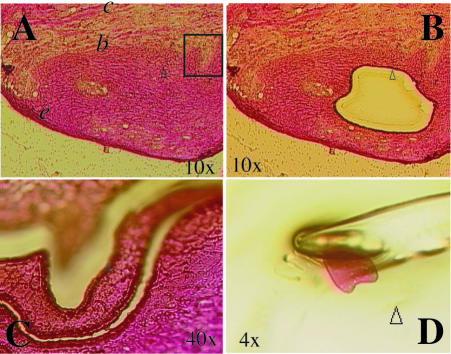
Figure 7.
Isolation of mucosal tissue substructures by LDM. Fresh-frozen normal buccal mucosa is shown before sectioning by LDM (A) and following removal of (e) stratum spinosum (B) and (b) basal layer, and suprabasal stratum spinosum (C). Box in (A) indicates area shown in (C). Isolation of superficial connective tissue was done from area marked (c). Isolated tissue samples were verified by microscopy as shown in (D) for stratum spinosum and processed for RNA isolation as outlined in Materials and Methods.
Table 2.
Differential Gene Expression in Stratum Spinosum of Oral Warts and Normal Mucosa
| Gene group* | Sig. diff.† | Genes analyzed‡ | Fold increased (decreased) gene expression in warts§ |
|---|---|---|---|
| Chemokine receptors (P = 0.46) | No | CCR1, CCR2, CCR5, CCR7 CCR8, CXCR3, CXCR4, CXCR5, IL8Rβ | |
| Yes | CCRL2 | 2,200x | |
| CCR3 | n160 | ||
| CCR6* | (28) | ||
| IL8Rα | 390 | ||
| Chemokines(P = 0.0041) | No | CCL1, CCL16, CCL17, CCL18, CCL23, CXCL13, Eotaxin, IL8, IP10, MCP1, MCP4, MIF, MIP1β, MIP3α, MIP3β, S100A8, SDF, SLC | |
| Yes | MIG | 110x | |
| MIP1α | 100 | ||
| RANTES* | (39) | ||
| S100A9 | 5.4 | ||
| Cytokine and growth factor receptors (P = 0.0034) | No | CSF1R, EDG2, EDG4, EGFR, FGFR4, IFNR, IFNγR2, IL1R type II, IL2Rα, IL2Rβ, IL2Rγ, IL3Rα, IL5Rα, IL6Rα, IL7R, IL9Rα, IL11Rα, IL12Rβ1, IL13Rα1, IL13Rα2, IL15Rα, IL17R, IL18R, MDFA, MGFA, MGFB, TGFβR1, TGFβR2, TGFβR3, TNFR | |
| Yes | EDG1* | (25x) | |
| IFNγR1 | 8.9 | ||
| IL10Rα | 24 | ||
| IL4Rα | 32 | ||
| Cytokines and growth factors (P < 0.0001) | No | CSF1, ENA78, Endothelin 1, Endothelin 3, FGF6, G-CSF, IFNγ, IK, IL1β, IL2, IL4, IL5, IL6, IL9β, IL10, IL11, IL12α, IL12β, IL13, IL14, IL15, IL17, IL18, IL19, IL21, IL22, SCF, LEF1, LIF, Lmphotoxin B, Osteopontin, TGFβ1, TGFβ2, TGFβ3, TNFα, VEGF | |
| Yes | EGF | 960 | |
| EGR1 | 58 | ||
| FGF1 | 230 | ||
| GM-CSF | 2,200 | ||
| HBEGF | 8 | ||
| IL1α | 67 | ||
| IL16 | 24 | ||
| TNFβ* | (59) | ||
| Effector molecules(P = 0.0013) | No | Anexin, Defensin α1, Defensin α3, Defensin α4, Defensin α5, Defensin β1, Granulysin, Granzyme A, Granzyme B, IL1 receptor antagonist, Interferon regulatory factor 1, Myeloperoxidase, NOS2A, Perforin, SLP1, TRAIL | |
| Yes | Defensin α6 | 450x | |
| Defensin β2 | 190 | ||
| Gelatinase B | 120 | ||
| PMN elastase | 140 | ||
| Surface markers(P = 0.17) | No | CD1α, CD4, CD8, CD14, CD16, CD19, CD23, CD27L, CD34, CD40L, CD44, CD56, CD62E, CD62L, CD69, CD80, CD86, FcɛR1, Hevin, ICAM1, ICAM2, ICAM3, KIT, PECAM1, TCRα, TCRδ, Thy1, VCAM1, VLA4 | |
| Yes | CD3γ | (430x) | |
| CD62P | 270 | ||
| CD79α | 3,000 | ||
| TCRβ* | (30) |
, P-value indicates significance level for overall expression differences between oral warts and normal mucosa for all genes within the indicated group.
, No, P > 0.05; Yes, P < 0.05 (bold gene names: P < 0.0003, highly significant after Bonferoni correction) for individual genes.
, Increased gene expression in stratum spinosum of oral warts compared to normal buccal mucosa, except for those marked with an asterisk (*).
, Calculated as 2 raised to the power of the absolute value of delta Ct (delta Ct of 1 equals a 2-fold change in gene expression, see Supplemental Table 1 at http://ajp.amjpathol.org), and shown in parentheses if expression is lower in the oral warts compared to normal buccal mucosa.
Within the patient groups, overall gene expression profiles of each tissue layer differed significantly from each other (P < 0.0001), with significant differences (P < 0.0001) for all but one gene family (effector molecules). Thus, this method allows us to specifically monitor distinct patterns of tissue substructure-specific gene expression with high sensitivity.
The value of tissue sublocation specific measurements became apparent, albeit only on a statistical basis, in the comparison of oral warts and normal mucosa. Using all of the >7000 comparisons (166 genes in 15 patients and 3 tissues), no significant differences (P > 0.77) in gene expression profile of HPV affected and non-affected mucosa were measurable. More detailed comparisons between individual tissue subcompartments, however, identified significant differences in gene expression in the stratum spinosum from oral warts and normal mucosa (P = 0.0056), but not in the basal layer/superbasal stratum spinosum and superficial connective tissue (P = 0.59 and 0.73, respectively). Thus, HPV replication in the epithelium of the oral mucosa affects host gene expression at that particular anatomical location, but does not seem to induce significant alterations even in the immediately adjacent underlying tissues.
This was in strong contrast to the underlying HIV infection itself. A similar analysis performed on normal oral mucosa from HIV-infected and HIV-non-infected patients showed no significant differences (P = 0.998) in expression of the 166 genes when each of the three individual tissue layers were compared separately. However, combined analysis of gene expression for the nearly 10,000 comparisons (166 genes in 20 patients and 3 tissues) resulted in significant (P < 0.0001) differences. For this comparison, the two gene groups associated with identification of cell subsets, namely surface markers and cytokine receptors, showed no significant differences (P = 0.9515 and P = 0.2074). All other gene groups (see Table 2 for listing of gene groups and genes) showed statistical significant differences (data not shown). Thus, HIV infection and/or HAART affect the overall steady-state gene expression in the oral mucosa. The changes induced in these patients appear subtle and not tissue sublocation-specific because large number of measurements (combined measurements of all layers) were required for those differences to reach significance. Whether these overall subtle differences could explain the increased susceptibility of this patient group to HPV infection remains to be studied.
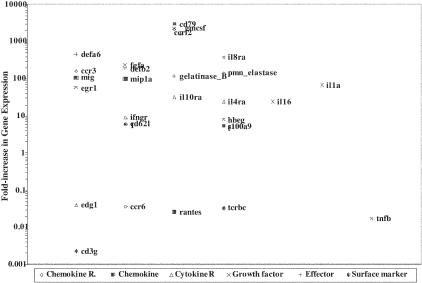
Since significant alterations in gene expression between normal mucosa and HPV32-induced oral warts were found only in the stratum spinosum further analysis concentrated on the changes at this tissue site. Significant differences in the gene expression profile between non-HPV-affected and HPV-affected stratum spinosum existed for all but two of the gene groups tested, chemokine receptors and surface markers (Table 2). Of the 166 individual genes tested, 28 showed significant differences in gene expression in oral warts compared to normal mucosa (Figure 8, Table 2, and Supplemental Table 1 at http://ajp.amjpathol.org). Three major observations were made: genes involved in HPV-induced epithelial growth and differentiation were increased (particularly EGF, FGF1, and IL1α); genes associated with local innate immune responses were increased suggesting polymorphonuclear cell accumulation/activation (CCR3, IL8α receptor, P-selectin, PMN-elastase, gelatinase-B (matrix metalloproteinase 9), and macrophage activation (MIG, MIP1α and IL10Rα, HBEGF, and GM-CSF); and no increase in expression of genes involved in antiviral effector T cell and NK cell responses were found (Table 2).
Figure 8.
Significant differences in gene expression of stratum spinosum from oral warts and normal mucosa from HIV-infected patients. Shown is a graphic representation of the fold-differences in gene expression (listed in Table 2) between the stratum spinosum of normal oral mucosa and of oral warts both from HIV-infected patients on HAART.
Immunohistochemical analysis confirmed a lack of strong inflammatory infiltrate in and around HPV-induced warts in these HIV-infected patients (data not shown). However, IL1α, a strong pro-inflammatory cytokine known to also promote HPV-induced cell growth in vitro22,23 and a gene induced by IL1α, defensin β2,24 showed highly increased expression in oral wart epithelium compared to that normal control mucosa (Table 2, Figure 8). Importantly, increased expression of the IL1 receptor antagonist was found in the underlying connective tissue (770-fold increase in gene expression compared to normal mucosa, P = 0.0002) but not in the stratum spinosum (Table 2) of oral warts or in the normal mucosa from HIV-non-infected patients (data not shown), suggesting that this differences is HPV-induced.
Taken together, this analysis identifies remarkable tissue sublocation-specific effects of HPV32 infection on local gene expression in oral mucosa. The underlying HIV infection in these patients appeared to have much more subtle effects on steady-state expression for the analyzed genes and seems to affect all tissue layers equally.
Discussion
We describe the development and application of a novel approach for studying alterations in gene expression for hundreds of genes in a highly tissue-specific manner from small amounts of biopsy material. The technique enables comprehensive and highly sensitive measurements on local biological processes in precise anatomical subcompartments. This method provides an additional tool for performing gene expression studies on precious clinical materials. It can be used as a cost-effective stand-alone technology if known groups of genes are to be analyzed. Alternatively, it could become a method of choice for confirming and extending microarray studies. Applying this approach to HPV32 infection in those patients with underlying HIV disease, we demonstrate the highly tissue sublocation-specific effects of HPV 32 on the oral epithelium. Our results are consistent with the hypothesis that HPV actively down-modulates local immune responses to evade elimination by the host.
Because real-time RT-PCR has a larger dynamic range and is more sensitive than microarray-based assays,17 it is a superior assay for defining changes in gene expression. However, setting-up large numbers of individual real-time RT-PCR reactions is labor intensive and requires large amounts of starting material, making it unsuitable for large-scale gene expression studies. The approach detailed here provides a technique that uses the advantages of the real-time RT-PCR approach, while enabling analysis of hundreds of genes simultaneously. In theory there is no limit to the number of genes that can be analyzed simultaneously by this assay and we have successfully analyzed more than the 166 genes studied here (Szubin R, Baumgarth N, unpublished observations). Careful evaluations of all reagent cocktails are necessary however to ensure that none of the reagents are amplification-rate limiting. Particularly, the quality and amount of dNTP’s and enzyme added at the pre-amplification step need careful adjustment to ensure optimal performance. Furthermore, the use of gene-specific primers is required21 (Szubin R, Baumgarth N, unpublished observations) to achieve linearity between input RNA amount and Ct values. This also points to a potential problem associated with microarray analyses on samples isolated by LDM. In that case, pre-amplification has to be done with non-specific primers. Confirmation of results obtained by microarray analyses by other means is therefore of great importance. The assay described here avoids this problem and thus provides a powerful alternative to evaluating multi-gene alterations in small samples. The assay might even be used to determine absolute RNA copy numbers within groups of individual cells. However, as shown in Figure 4, it is necessary to include standard RNA for each of the genes analyzed as amplification efficiencies between genes might vary.
Although this technology can be used to analyze gene expression on individual FACS-purified cells21 (Szubin R, Baumgarth N, unpublished observations), we have not been able to reliably do this on LDM-isolated single cells (Szubin R, Baumgarth N, unpublished observations). There might be a number of reasons for this, including the fact that in many cases sectioning of the tissue might cut through cells causing loss of RNA, or that slight RNA degradation through the cell isolation process does occur (see Materials and Methods).11 Nonetheless, we have obtained reliable results from as few as five to six cells captured by LDM. Efforts are now focusing on combining this approach with immunofluorescence to more accurately identify the phenotype of cells before capture.
Using the LDM-real-time RT-PCR approach we demonstrate that HPV infection induces highly localized alterations in gene expression solely in the stratum spinosum and not in the immediately adjacent tissue substructures. These changes are related to cell biological processes affecting epithelial cell growth and differentiation and to innate immune responses, including stimuli that can promote DC activation and maturation. Transfection of HPV16-derived E6 and E7 oncoproteins into keratinocytes resulted in changes in gene expression similar to those induced by in vivo infection with HPV32 revealed by this study.25 In contrast, comprehensive expression analysis by microarray on in vitro HPV11- and 31-infected keratinocytes and a HPV33-expressing cell line26,27,28 did not reveal significant induction of EGF, FGF, or EGR1 in vitro.26,27 Indeed, a moderate reduction in EGR1 and EGR2 gene expression and an up-regulation of TGFβ was reported. Thus HPV11 and HPV33 might differentially affect cell growth and differentiation compared to HPV32. Virus subtype specific differences have been reported for a number of different HPV subtypes.26
We believe the highly localized effects in gene expression seen in the oral warts point to HPV-initiated effects and are not due to the underlying HIV infection, since HIV infection plus HAART itself appears to induce only minor changes. However, because we did not have access to paired oral wart and adjacent normal mucosa samples from the same patient we cannot exclude the possibility that, in the HPV-lesion-carrying patients, HIV has effects different from those in the HPV-non-infected patients, or from HPV-infected but non-lesion developing patients. Future work is aimed at addressing these issues. A comparison of HPV-lesions and normal mucosa that studies gene expression in whole biopsies (non-dissected) versus the analysis performed here, namely combining measurements obtained from each individual tissue layer, might further demonstrate the sensitivity of the approach taken here.
Similar to the present study with tissues from HIV-infected patients, previous reports noted an absence of inflammatory infiltrates around HPV lesions in HIV-negative fully immunocompetent patients,9 suggesting that HPV infection might selectively inhibit local immune responses. Evidence for direct inhibition of immunity by HPV is indeed mounting,9,26 although in the present study we cannot exclude the possibility that the underlying HIV infection might have been responsible for the inability to mount normal immune effector function. Our clinic has not seen HIV-negative patients with oral warts, unless otherwise immunocompromised, hence we were unable to address this question experimentally.
LDM-assisted gene expression analysis did show significant differences for a number of genes related to innate immune responses, but immunohistochemistry performed on those sections did not show significant increases in inflammatory infiltrates (data not shown). Increased expression of genes was noted that suggests polymorphonuclear cell (PMN) accumulation/activation, namely CCR3, IL8α receptor, P-selectin, PMN-elastase, gelatinase-B (matrix metalloproteinase 9); and macrophage activation namely MIG, MIP1α and IL10Rα, HBEGF, and GM-CSF. The strongly increased GM-CSF expression in oral warts is noteworthy, since it was suggested that HPV infection reduces expression of GM-CSF by keratinocytes, thereby preventing the induction of DC activation/maturation.29 In contrast, our data show strong induction of GM-CSF gene expression in HPV-infected epithelial cells or cells that infiltrate this layer. Post-transcriptional modification of protein expression, however, cannot be excluded. We found no evidence for reduced DC presence in the oral warts by gene expression analysis or immunohistochemistry (not shown). Expression of two defensins, defensin-α6 and defensin-β2 was increased in oral warts. Defensin-β2 is expressed by epithelial cells on various mucosal surfaces.30 Its increased expression during inflammation is thought to provide direct antimicrobial activity and to attract immature DC to the site,31,32 thus providing another indication that signals for the migration of immature DC to the oral warts are present. Thus far, defensin-α6 production has been reported only for Paneth cells in the small intestine.33 Identification of the exact cell type that expresses this gene in the oral mucosa will be of importance.
If innate immune responses are induced, including the activation of DC at the site of HPV infection, why then is there no evidence for the presence of antiviral T cell effector responses? This study indicates that these responses might be actively down-modulated. Remarkable is the down-modulation of TNFβ, a potent pro-inflammatory cytokine and the induction of the IL1R antagonist in the connective tissue underlying the stratum spinosum in the presence of large increases of Il-1α expression at that latter site. This presumably inactivates the pro-inflammatory capacity of this cytokine, while the growth-promoting capacity of IL-1 for HPV in the epithelium is maintained. There is an increasing list of viruses that employ mechanisms to achieve exactly this objective. Given the importance of HPV-induced lesions and malignancies, identifying possible targets of the virus to inhibit immune responses will be significant. The technology developed in this study could help to achieve this goal.
In summary, the new technological strategy outlined in this study provides a means to broadly evaluate highly localized alterations in gene expression within defined tissue substructures. While it cannot replace existing microarray technologies, it provides significant advantages in ensuring linear pre-amplification of very small amounts of input RNA/cDNA, exquisite specificity, higher levels of sensitivity, and broader dynamic measurement ranges than microarray analyses. Thus, for studies requiring the analysis of hundreds of known genes or multiple candidate genes identified by microarray, this technology provides an important additional tool for gene expression studies.
Supplementary Material
Acknowledgments
We thank Piri Veluppillai for help obtaining the biopsy material, the Tissue Core, UCSF Comprehensive Cancer Center, for providing the human tonsil material, Drs. Joan Hilton and Ru-Fang Yeh for their input and discussions with regard to the statistical analysis, Dr. Andrew Fell for comments, and Miguel Relloso for cloning the PCR products.
Footnotes
Address reprint requests to Nicole Baumgarth, D.V.M., Ph.D., Center for Comparative Medicine, University of California, Davis, 1 Shields Avenue, Davis, CA 95616. E-mail: nbaumgarth@ucdavis.edu.
Supported by grant DE-P01 07496 from the National Institutes of Health/Institute for Dental and Craniofacial Research. G.M.D was supported by grants NIH-NHLBI/5 P50 HL66564 and NIH-NIAID/1 P01 AI-00–012.
Supplemental information available at http://ajp.amjpathol.org.
References
- Syrjanen KJ. HPV infections in benign and malignant sinonasal lesions. J Clin Pathol. 2003;56:174–181. doi: 10.1136/jcp.56.3.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffman MH, Castle P. Epidemiologic studies of a necessary causal risk factor: human papillomavirus infection and cervical neoplasia. J Natl Cancer Inst. 2003;95:E2. doi: 10.1093/jnci/95.6.e2. [DOI] [PubMed] [Google Scholar]
- King GN, Healy CM, Glover MT, Kwan JT, Williams DM, Leigh IM, Thornhill MH. Prevalence and risk factors associated with leukoplakia, hairy leukoplakia, erythematous candidiasis, and gingival hyperplasia in renal transplant recipients. Oral Surg Oral Med Oral Pathol. 1994;78:718–726. doi: 10.1016/0030-4220(94)90086-8. [DOI] [PubMed] [Google Scholar]
- Coutlee F, Trottier AM, Ghattas G, Leduc R, Toma E, Sanche G, Rodrigues I, Turmel B, Allaire G, Ghadirian P. Risk factors for oral human papillomavirus in adults infected and not infected with human immunodeficiency virus. Sex Transm Dis. 1997;24:23–31. doi: 10.1097/00007435-199701000-00006. [DOI] [PubMed] [Google Scholar]
- Greenspan D, Canchola AJ, MacPhail LA, Cheikh B, Greenspan JS. Effect of highly active antiretroviral therapy on frequency of oral warts. Lancet. 2001;357:1411–1412. doi: 10.1016/S0140-6736(00)04578-5. [DOI] [PubMed] [Google Scholar]
- King MD, Reznik DA, O’Daniels CM, Larsen NM, Osterholt D, Blumberg HM. Human papillomavirus-associated oral warts among human immunodeficieny virus-seropositive patients in the era of highly antiretroviral therapy: an emerging infection. Clin Infect Dis. 2002;34:641–648. doi: 10.1086/338637. [DOI] [PubMed] [Google Scholar]
- Pakker NG, Notermans DW, Boer RJd, Roos MT, de Wolf F, Hill A, Leonard JM, Danner SA, Miedema F, Schellekens PT. Biphasic kinetics of peripheral blood T cells after triple combination therapy in HIV-1 infection: a composite of redistribution and proliferation. Nat Med. 1998;4:208–214. doi: 10.1038/nm0298-208. [DOI] [PubMed] [Google Scholar]
- Dyer WB, Kuipers H, Coolen MW, Geczy AF, Forrester J, Workman C, Sullivan JS. Correlates of antiviral immune restoration in acute and chronic HIV type 1 infection: sustained viral suppression and normalization of T cell subsets. AIDS Res Hum Retroviruses. 2002;18:999–1010. doi: 10.1089/08892220260235362. [DOI] [PubMed] [Google Scholar]
- O’Brien PM, Saveria Campo M. Evasion of host immunity directed by papillomavirus-encoded proteins. Virus Res. 2002;88:103–117. doi: 10.1016/s0168-1702(02)00123-5. [DOI] [PubMed] [Google Scholar]
- Luxton J, Shepherd P. Human papillomavirus antigens and T-cell recognition. Curr Opin Infect Dis. 2001;14:139–143. doi: 10.1097/00001432-200104000-00005. [DOI] [PubMed] [Google Scholar]
- Luzzi V, Mahadevappa M, Raja R, Warrington JA, Watson MA. Accurate and reproducible gene expression profiles from laser-capture microdissection, transcript amplification, and high density oligonucleotide microarray analysis. J Mol Diagn. 2003;5:9–14. doi: 10.1016/S1525-1578(10)60445-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink L, Kohlhoff S, Stein MM, Hanze J, Weissmann N, Rose F, Akkayagil E, Manz D, Grimminger F, Seeger W, Bohle RM. cDNA array hybridization after laser-assisted microdissection from nonneoplastic tissue. Am J Pathol. 2002;160:81–90. doi: 10.1016/S0002-9440(10)64352-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyagi K, Tatsuta T, Nishigaki M, Akimoto S, Tanabe C, Omoto Y, Hayashi S, Sakamoto H, Sakamoto M, Yoshida T, Terada M, Sasaki H. A faithful method for PCR-mediated global mRNA amplification and its integration into microarray analysis on laser-captured cells. Biochem Biophys Res Commun. 2003;300:915–920. doi: 10.1016/s0006-291x(02)02967-4. [DOI] [PubMed] [Google Scholar]
- Michel C, Desdouets C, Sacre-Salem B, Gautier JC, Roberts R, Boitier E. Liver gene expression profiles of rats treated with clofibric acid: comparison of whole liver and laser-capture microdissected liver. Am J Pathol. 2003;163:2191–2199. doi: 10.1016/S0002-9440(10)63577-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Salunga RC, Guo H, Bittner A, Joy KC, Galindo JE, Xiao H, Rogers KE, Wan JS, Jackson MR, Erlander MG. Gene expression profiles of laser-captured adjacent neuronal subtypes. Nat Med. 1999;5:117–122. doi: 10.1038/4806. [DOI] [PubMed] [Google Scholar]
- Fink L, Seeger W, Ermert L, Hanze J, Stahl U, Grimminger F, Kummer W, Bohle RM. Real-time quantitative RT-PCR after laser-assisted cell picking. Nat Med. 1998;4:1329–1333. doi: 10.1038/3327. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, Preter KD, Pattyn F, Poppe B, Roy NV, Paepe AD, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:34.31–34.11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison EA, Goldberg GL, Kadish AS, Burk RD. Polymerase chain reaction detection of human papillomavirus: quantitation may improve clinical utility. J Clin Microbiol. 1992;30:2539–2543. doi: 10.1128/jcm.30.10.2539-2543.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palefsky JM, Holly EA, Ralston ML, Jay N. Prevalence and risk factors for human papillomavirus infection in (HIV)-positive and HIV-negative homosexual men. J Infect Dis. 1998;177:361–367. doi: 10.1086/514194. [DOI] [PubMed] [Google Scholar]
- Beaudenon S, Praetorius F, Kremsdorf D, Lutzner M, Worsaae N, Pehau-Arnaudet G, Orth G. A new type of human papillomavirus asssociated with oral focal epithelial hyperplasia. J Invest Dermatol. 1987;88:130–135. doi: 10.1111/1523-1747.ep12525278. [DOI] [PubMed] [Google Scholar]
- Dolganov GM, Woodruff PG, Novikov AA, Zhang Y, Ferrando RE, Szubin R, Fahy JV. A novel method of gene transcript profiling in airway biospy homogenates reveals increased expression of a Na+-K+-CL-cotransporter (NKCCI) in asthmatic subjects. Genome Res. 2001;11:1473–1483. doi: 10.1101/gr.191301. [DOI] [PubMed] [Google Scholar]
- Woodworth CD, McMullin E, Iglesias M, Plowman GD. Interleukin 1 α and tumor necrosis factor α stimulate autocrine amphiregulin expression and proliferation of human papillomavirus-immortalized and carcinoma-derived cervical epithelial cells. Proc Natl Acad Sci USA. 1995;92:2840–2844. doi: 10.1073/pnas.92.7.2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrilli G, Tatone D, Diodoro MG, Rosini S, Piantelli M, Musiani P. Interleukin 1α and interleukin 6 promote the in vitro growth of both normal and neoplastic human cervical epithelial cells. Br J Cancer. 1997;75:855–859. doi: 10.1038/bjc.1997.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AY, Destoumieux D, Wong AV, Park CH, Valore EV, Liu L, Ganz T. Human β-defensin-2 production in keratinocytes is regulated by interleukin-1, bacteria, and the state of differentiation. J Invest Dermatol. 2002;118:275–281. doi: 10.1046/j.0022-202x.2001.01651.x. [DOI] [PubMed] [Google Scholar]
- Nees M, Geoghegan JM, Hyman T, Frank S, Miller L, Woodworth CD. Papillomavirus type 16 oncogenes down-regulate expression of interferon-responsive genes and upregulate proliferation-associated and NF-κB-responsive genes in cervical keratinocytes. J Virol. 2001;75:4283–4296. doi: 10.1128/JVI.75.9.4283-4296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang ME, Laimins LA. Microarray analysis identifies interferon-inducible genes and Stat-1 as major transcriptional targets of human papillomavirus type 31. J Virol. 2000;74:4174–4182. doi: 10.1128/jvi.74.9.4174-4182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JT, Oh ST, Terhune SS, Laimins LA. Cellular changes induced by low-risk human papillomavirus type 11 in keratinocytes that stably maintain viral episomes. J Virol. 2001;75:7564–7571. doi: 10.1128/JVI.75.16.7564-7571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruutu M, Peitsaro P, Johansson B, Syrjanen S. Transcriptional profiling of a human papillomavirus 33-positive sqamous epithelial cell line which acquired a selective growth advantage after viral integration. Int J Cancer. 2002;100:318–326. doi: 10.1002/ijc.10455. [DOI] [PubMed] [Google Scholar]
- Havard L, Delvenne P, Frare P, Boniver J, Giannini SL. Differential production of cytokines and activation of NF-κB in HPV-transformed keratinocytes. Virology. 2002;298:271–285. doi: 10.1006/viro.2002.1468. [DOI] [PubMed] [Google Scholar]
- Liu L, Wang L, Jia HP, Zhao C, Heng HH, Schutte BC, McCray PB, Jr, Ganz T. Structure and mapping of the human β-defensin HBD-2 gene and its expression at sites of inflammation. Gene. 1998;222:237–244. doi: 10.1016/s0378-1119(98)00480-6. [DOI] [PubMed] [Google Scholar]
- Yang D, Chertov O, Bykovskaia SN, Chen Q, Buffo MJ, Shogan J, Anderson M, Schroder JM, Wang JM, Howard OM, Oppenheim JJ. β-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science. 1999;286:525–528. doi: 10.1126/science.286.5439.525. [DOI] [PubMed] [Google Scholar]
- Biragyn A, Ruffini PA, Leifer CA, Klyushnenkova E, Shakhov A, Chertov O, Shirakawa AK, Farber JM, Segal DM, Oppenheim JJ, Kwak LW. Toll-like receptor 4-dependent activation of dendritic cells by β-defensin 2. Science. 2002;298:1025–1029. doi: 10.1126/science.1075565. [DOI] [PubMed] [Google Scholar]
- Mallow EB, Harris A, Salzman N, Russell JP, DeBerardinis RJ, Ruchelli E, Bevins CL. Human enteric defensins: gene structure and developmental expression. J Biol Chem. 1996;271:4038–4045. doi: 10.1074/jbc.271.8.4038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.