Abstract
Hepatitis C virus (HCV) nonstructural protein 3 (NS3), with its protease, helicase, and NTPase enzymatic activities, plays a crucial role in viral replication, and therefore represents an ideal target for the development of anti-viral agents. We have developed a recombinant human antibody (Fab) that reacts with the helicase domain of HCV NS3. The affinity-purified Fab antibody completely inhibited the helicase activity of HCV NS3 at equimolar concentration. To evaluate the effect of the Fab on HCV replication, the clone encoding the Fab gene was put into an expression vector, which converts Fab into a complete IgG1 antibody. Using a DNA-based transfection model, we demonstrated that intracellular expression of this antibody resulted in significant reduction of HCV-negative strand RNA synthesis. Intracellular expression of this antibody into either a stable cell line replicating subgenomic RNA, or a transient full-length HCV replication model, reduced both HCV RNA and viral protein expression. These results support the use of recombinant antibody fragments to inhibit NS3 enzyme as a novel, feasible, and effective approach for inhibiting HCV replication.
Hepatitis C virus (HCV) infection represents the leading cause of chronic liver disease in the United States and around the world and is considered as a major public health problem.1 The virus persists in the majority of the infected population (85%) whereas only a minority (15%) of patients can mount a successful immune response and clear the virus. Prolonged inflammation in the liver because of HCV infection leads to chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Currently, there is no vaccine available for HCV infection. Interferon in combination with ribavirin is the only approved therapy for HCV. However, this combination therapy benefits only approximately half of HCV-infected patients and produces considerable side effects.1 There is an urgent need to develop a more specific and effective therapy to treat HCV infection.
Several molecular approaches have been designed to inhibit HCV using anti-sense oligonucleotide, ribozymes, or RNA interference.2–8 These strategies have been successful in inhibiting viral gene expression and to some extent viral replication, but have not been efficient for the treatment of some resistant viral strains, including viral quasi-species. Recently, several laboratories have used siRNA to inhibit HCV replication.9,10 The rationale of protein-based therapy to inhibit key viral enzyme function intracellularly may represent an alternative anti-viral therapy for hepatitis C. The development of recombinant antibodies (Fv, scFv, Fab, or IgG) and their expression inside eukaryotic cells (so-called intracellular immunization) can be used to inhibit key viral enzyme activities.9–23 This strategy has certain advantages over the use of anti-sense oligonucleotides or ribozymes or RNA interference because the recombinant antibodies are directed against key enzymes and are thus independent of viral sequence variation. The recombinant antibodies also can be expressed as a single chain, a Fab, or complete antibody. The single chain antibodies are essentially one protein consisting of heavy- and light-chain variable regions of immunoglobulin joined to a synthetic linker. As an alternative to this, antibodies can be expressed as a Fab fragment in which the variable heavy chain along with the first constant domain are associated with the complete light chain. Association of this heterodimer in Fab molecules makes it much more stable than single chain antibodies. In the case of Fab, heavy and light chains are usually expressed from two separate expression cassettes. Both the heavy and light chains can assemble each other intracellularly and bind to antigen with high affinity. The advantage of working with Fab molecules is that they are much more stable as compared to single chain antibody. The rationale of using recombinant antibody fragments to inhibit viral enzyme function may be the best approach toward developing anti-viral therapy for hepatitis C. This approach is supported by the recent development of combinatorial phage libraries for selection of high-affinity antibodies and their applications in anti-viral therapy.15–23 By way of example, recombinant antibody is currently in use against human immunodeficiency virus,17,18 respiratory syncytial virus,19 herpes simplex virus,20 hepatitis B virus,12 and HCV.13
Our study is based on the premise that intracellular expression of recombinant antibody against NS3 should inhibit helicase activity and HCV replication in cultured cells. We developed a human recombinant antibody Fab (HFab-aNS3), which reacts with a conformational epitope of NS3 helicase. The anti-viral properties of this clone were sequentially studied using a cell-free helicase assay, followed by cell culture based on persistent and transient HCV replication models. In this report, we show a successful anti-viral effect of an intracellular expression human antibody clone against HCV.
Materials and Methods
Purification of NS3 Protein
The recombinant clones containing wild-type helicase and DQCH helicase mutant plasmids were provided by Dr. Ding-Shinn Chen, Hepatitis Research Center, National Taiwan University Hospital, Taipei, Taiwan.24 The NS3 plasmid clone contains the RNA helicase domain encoding amino acids 1175 to 1657 (nucleotides 3864 to 5312). Expression of NS3 protein was induced in BL 21(DE3) Escherichia coli by 1 mmol/L isopropyl-1-thio-β-d-galactopyranoside using a standard protocol.24 The purified NS3 protein was run on a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gel and electrotransferred to a nitrocellulose membrane (Hybond ECL, Amersham Biosciences, NJ, USA). The membrane was blocked with 0.1 mol/L phosphate-buffered saline (PBS), pH 7.2, containing 0.05% Tween-20 (PBS-Tween-20) and 5% nonfat dried milk for 1 hour at room temperature. The membrane was then incubated with the rabbit polyclonal anti-NS3 antibody (at 1:1000 dilution) (Virostat Inc., Portland, ME) for 1 hour at room temperature. After washing three times with PBS-Tween-20, the membrane was incubated with peroxidase-conjugated goat anti-rabbit secondary antibody (1:1000 dilution) for 1 hour at room temperature. The membranes were developed using the enhanced chemiluminescence (ECL) detection system (ECL Western blotting system, Amersham Biosciences, NJ, USA) according to the manufacturer’s instructions.
Purification of Fab Antibody
The Fab antibody clone used in this study was obtained by molecular cloning in phage display combinatorial vector of the IgG1/kappa repertoire of an HCV-infected patient (genotype 1b) who suffered from chronic hepatitis.25 This Fab fragment recognizes amino acid residues 1192 to 1457 of the NS3 protein. Gene coding for the anti-NS3 human Fab was inserted into an appropriate expression vector22 and the expression of recombinant Fab was induced in XL1-blue E. coli in the presence of 1 mmol/L isopropyl-1-thio-β-d-galactopyranoside. The Fab fragment was purified using a protein G-anti-human Fab column using a standard published protocol.15 The purity of the fractions was confirmed by Western blot analysis using peroxidase-labeled anti-Fab antibodies (Sigma Chemical Co., St. Louis, MO).
Affinity Measurements
An enzyme-linked immunosorbent assay (ELISA) was used to examine binding affinity of Fab antibody fragment to recombinant NS3 protein expressed in E. coli. Polystyrene plates (Corning Inc., Corning, NY, USA) were coated with 200 μl of purified NS3 protein at a concentration of 10 μg/ml in 50 mmol/L bicarbonate buffer, pH 9.0, by overnight incubation. The wells were then washed with 10 mmol/L PBS, pH 7.2, containing 0.05%Tween-20 (PBS-Tween-20) and blocked with 2% bovine serum albumin solution for 1 hour. Next, the wells were washed three times with PBS-Tween-20 and incubated with 100 μl of serial dilutions of purified Fab antibody for 1 hour at 37°C. The wells were then washed three times with PBS-Tween-20 and incubated with 100 μl of horseradish peroxidase-conjugated goat anti-Fab against human at 1:1000 dilution (Sigma) for 1 hour. After this incubation, wells were washed three times and color was developed after adding 100 μl of orthophenyl diamine (OPD; Sigma) plus hydrogen peroxide for 10 minutes. The reaction was stopped by the addition of 100 μl of 2 mol/L H2SO4. The optical density of each well was taken at 494 nm (ELISA reader). The relative affinity of the purified Fab antibody was determined by competitive ELISA method.26 Briefly, wells of microtiter plates were coated with NS3 protein in bicarbonate buffer at 4°C. The wells were washed and blocked with 2% bovine serum albumin in PBS. The Fab antibody mixed different concentrations of purified NS3 protein and incubated in the ELISA plate for 2 hours. The wells were washed with PBS-Tween-20 (0.05%) and anti-Fab conjugated to horseradish peroxidase was added for 1 hour. The wells were washed and color was developed by the addition of substrate. The optical density of the plate was read as described earlier. The affinity of Fab antibody was determined by measuring the concentrations of NS3 required to inhibit 50% maximum binding (IC50) in the competitive ELISA assay.
Helicase Assay
The helicase activity of recombinant full-length NS3 protein was measured by the extent of unwinding of partial double-stranded RNA as well DNA substrate. Partial double-stranded RNA (dsRNA) substrate was prepared by transcribing in vitro a portion of multiple cloning sequences of a pGem 5Z vector (Promega, Madison, WI) using T7 and Sp6 RNA polymerase. The T7 transcript was labeled with 32P-UTP whereas the other RNA remained unlabeled. The sense and anti-sense RNA were then annealed at a proportion of 1:3. The double-stranded RNA was gel purified and the helicase assay was performed using a previously published protocol with minor modification.27 Partial double-stranded DNA substrate was prepared using two complementary DNA oligonucleotides, 5′-AGAGAGAGAGGTTGAG AGAGAG-AGAGTTTGAGAGAGAGAG-3′ (40-mer template strand) and 5′-CAAA CTCTCTCTCTCTCAACAAAAAA-3′ (26-mer release strand). The release strand was labeled at the 5′-end with [γ-32P]-ATP using T4 polynucleotide kinase (Promega). To prepare the dsDNA hybrids, the two DNA oligonucleotides were combined at a molar ratio of 3:1 (template: release) and annealed by denaturation for 5 minutes at 90°C, followed by slow renaturation at 23°C in buffer containing 20 mmol/L Tris-HCl (pH 8.0), 0.5 mol/L NaCl, and 1 mmol/L ethylenediaminetetraacetic acid (EDTA). The partial duplex DNA substrate was purified by elution from a native 12% polyacrylamide gel using 0.5 mol/L ammonium acetate and 1 mmol/L EDTA. The concentration of labeled DNA was determined (cpm/nmol). The helicase assay was performed in a 20-μl reaction volume containing 25 mmol/L MOPS-NaOH, pH 7.0, 2.5 mmol/L dithiothreitol, 100 μg of bovine serum albumin per ml, 3 mmol/L MgCl2, 1.25 to 40 nmol/L of the NS3 protein, and 1 nmol/L of substrate. After preincubation for 15 minutes at 23°C, 5 mmol/L ATP was added to start the helicase reaction. The reaction was incubated at 37°C for 30 minutes and then stopped by the addition of 5 μl of termination buffer (0.1 mol/L Tris, pH 7.5, 20mmol/L EDTA, 0.5% SDS, 0.1% Nonidet P-40, 0.1% bromophenol blue, and 0.1% xylene cyanol). An aliquot of reaction of mixture (10 μl) was analyzed on a 12% native polyacrylamide gel. Strand separation was visualized by autoradiography using Kodak X-ray film (Eastman-Kodak, Rochester, NY). The specificity of the enzyme reaction was confirmed by using a control reaction, such as the helicase reaction in the absence of ATP, to be sure that strand separation was dependent on hydrolysis of ATP, this being a property of NS3 helicase. To assess anti-viral effect, increasing concentrations of Fab antibody were mixed with 20 nmol/L of NS3 protein for 1 hour at room temperature. The helicase reaction was performed in the presence and in the absence of antibody-NS3 protein complex. The following controls were included side by side to confirm the specificity of enzyme inhibition experiments: the helicase assay was performed using an unrelated purified Fab antibody; and the helicase assay was performed in the presence of known monoclonal antibody against NS3 (Novocastra Laboratories Ltd., Newcastle, UK).
Intracellular Expression of Human Fab
The recombinant Fab antibody clone was expressed in mammalian cells using the expression vector pFab-CMV.28 This vector allows the expression of light and heavy chains of Fab under the control of CMV promoter. This vector has convenient restriction sites for cloning of heavy and light chain genes from vector pComb3, and can also be used for the expression of either Fab or whole IgG1 molecules. First, the SacI-XbaI fragment from the plasmid pComb3-NS3 carrying the light chain gene was subcloned into pFab-CMV, with the resulting plasmid, is called pFab-CMV-NS3 light chain. At the second step, the XhoI-SpeI fragment carrying the heavy chain gene was removed from pComb3-NS3 and cloned into the pFab-CMV-NS3 light chain and the resulting plasmid called pFab-CMV-NS3 (L+H). This vector has heavy chain CH2, CH3, and part of the hinge region from a germline immunoglobulin γ1. This allows conversion of recombinant Fab antibody into a complete IgG1 antibody in the transfected cells. Huh-7 cells (1 × 106) seeded in a 100-mm dish were transfected with 10 μg of control Fab expression plasmid or pFab-CMV-NS3 (L+H) plasmid DNA using the FuGENE 6 transfection reagent (Roche Molecular Biology, Indianapolis, IN). Intracellular expression of anti-NS3 antibodies in the transfected Huh-7 cells was determined by immunostaining using a biotin-conjugated F(ab)2 fragment of human antibody developed in goat as secondary antibody (Sigma). The following protocol was used to demonstrate expression of Fab antibody. Transfected cells were harvested after 72 hours and immobilized on the glass slides by cytospin. The cells were washed with PBS, pH 7.4, twice, air-dried, and fixed with chilled acetone for 10 minutes. The cells were then permeabilized with 0.05% saponin for 10 minutes at room temperature. Blocking was performed with minimum essential medium with 3% normal goat serum for 30 minutes at room temperature. Endogenous biotin-avidin activities were blocked using reagents from the kit (Avidin/Biotin Blocking kit; Vector Laboratories Inc., Burlingame, CA) and blocking for endogenous peroxidase was done with 0.9% H2O2 for 30 minutes at room temperature. The cells were incubated with biotin-conjugated anti-F(ab)2 antibody at 1:500 dilution for overnight at 4°C. The next day slides were washed and incubated for 30 minutes with Elite avidin-biotin peroxidase complex (Vector Laboratories, Inc.). The slides were developed after reaction with diaminobenzidine for 10 minutes. Counterstaining was performed with hematoxylin for a minute. The slides were mounted with permount after dehydration and observed under an Olympus light microscope.
Anti-Viral Effect of Human Fab Clone in HCV Cell Culture Models
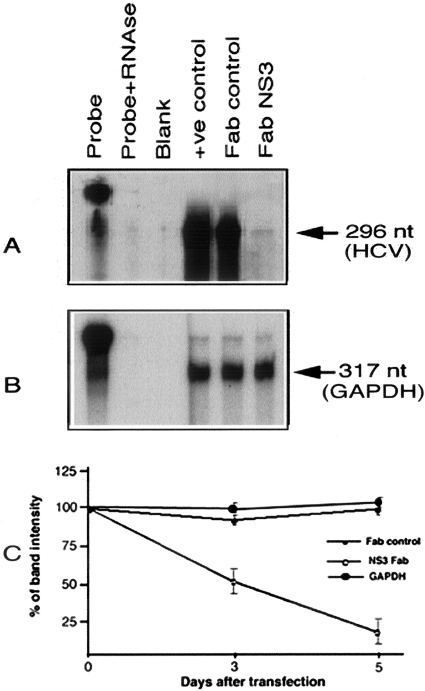
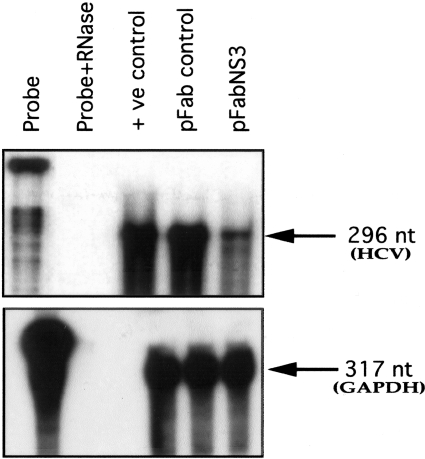
Anti-viral properties of this Fab clone were determined using the highly efficient replicon cell line.29,30 This model has been widely used by different laboratories for studying HCV replication. Huh-7 cells were transfected with HCV subgenomic replicon RNA31 and selected with G-418 (500 μg/ml) for ∼4 weeks. Stable cell lines replicating subgenomic RNA were selected and used for all subsequent experiments. Replicon cell line Con-15 was transfected with 10 μg of pFab-CMV-NS3 using FuGENE 6 transfection reagent. To make the argument that this was not because of a nonspecific effect of Fab expression inside cells, replicon cells were transfected with another plasmid pCMV-Fab expressing unrelated Fab. The cultures were maintained in minimal essential medium without G-418 medium. Anti-viral efficacy of this antibody clone was studied by measuring HCV-positive strand RNA levels at different time points after transfection. Total RNA was prepared from antibody-transfected replicon cells by the guaridineine isothiocyanate (GITC) method and subjected to ribonuclease protection assay (RPA) for the detection of genomic positive strand HCV RNA using an anti-sense probe targeted to the 5′ UTR. Half of the RNA extracts were subjected to RPA for GAPDH mRNA. For RPA experiments, 25 μg of total RNA was mixed with negative strand RNA probe targeted to the 5′UTR of HCV (1 × 106 cpm) in 10 μl of hybridization solution, denatured for 3 minutes at 95°C, and hybridized overnight at 45°C. RNase digestion was performed in 200 μl of RNase digestion buffer (10 mmol/L Tris, pH 7.5, 5 mmol/L EDTA, and 0.3 mol/L NaCl) containing RNaseA/T1 cocktail at 1:100 dilutions (Ambion Inc., Austin, TX) for ∼1 hour at 37°C. Then it was treated with 10 μl of 25% SDS and 10 μl of proteinase K (10 mg/ml) for 15 minutes. Samples were extracted with phenol:chloroform and precipitated with ethanol. The pellet was suspended in 8 μl of gel loading buffer, heat denatured, and separated on an 8% acrylamide/8 mol/L urea gel. The gel was dried and exposed to X-ray film (Eastman Kodak Company). Appearance of 296- and 317-nucleotide protected fragments indicated the presence of HCV-negative strand and GAPDH mRNA, respectively.
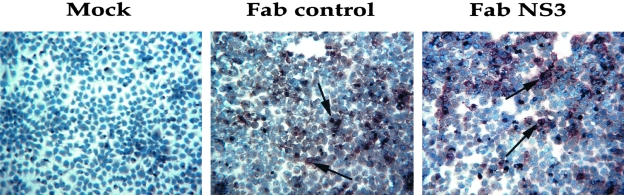
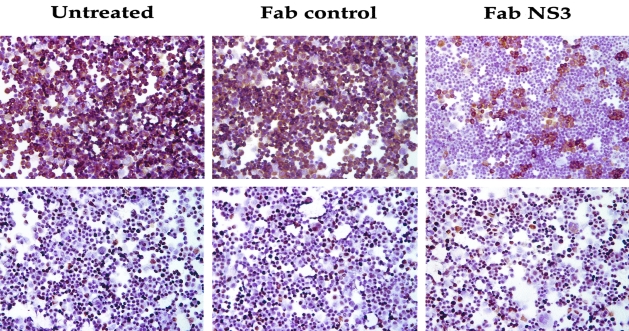
We examined levels of NS3 proteins in the replicon cell line after antibody transfection by the immunocytochemical method. Replicon cell line, Con-15, was transfected either with pFab-CMV-NS3 clone or pCMV-Fab control plasmid using FuGENE 6 transfection reagent. After 72 hours, cells were harvested by trypsin-EDTA. Transfected cells were immobilized on a glass slide by cytospin and immunostaining for NS3 antigen was performed using a monoclonal antibody at 1:50 dilution (Novocastra Laboratories Ltd.) using a standard procedure of our laboratory.
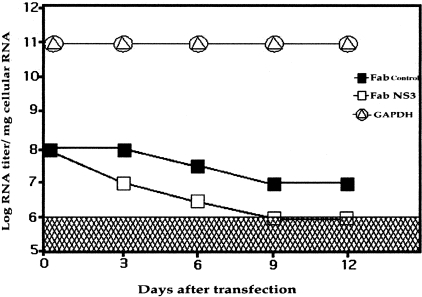
Because this antibody clone efficiently blocks helicase activity, we examined whether it could also inhibit HCV replication in the full-length RNA transfection-based model we developed in HepG2 cells.32,33 HepG2 cells were transfected with 10 μg of full-length in vitro-transcribed HCV RNA by the DEAE-dextran method. After a week, we transfected these cells with antibody expression plasmid. As a negative control, we also transfected plasmid that expresses an unrelated antibody. The levels of HCV in the RNA-transfected cells were measured by quantitative competitive reverse transcriptase-polymerase chain reaction using a primer set targeted to the 5′ NT region.34 To measure the nonspecific inhibition of cellular gene because of the expression of antibody, quantitation for glyceraldehyde phosphate dehydrogenase (GAPDH) mRNA was performed.
Subsequently, the effect of antibody expression on the negative strand RNA synthesis was examined using a DNA-based model established in our laboratory.35,36 For this experiment, Huh-7 cells in a 100-mm dish were infected with AdexCAT7 virus at a multiplicity of infection of 10. After 2 hours, Huh-7 cells were co-transfected with 10 μg of pMO9.6-T7-Rz (carrying full-length HCV cDNA) with or without 10 μg of pFab CMV NS3 (L+H) plasmid or pFab CMV control plasmid using FuGENE 6 transfection reagent (Roche). Three days after transfection, RNA extracts were prepared from transfected HepG2 cells, digested with DNase I (5 U/μg of RNA), and assayed for negative strand RNA using a sense probe targeted to the 5′ UTR. RPA was performed using a standard protocol instructed by the manufacturer (Ambion Inc., Austin, TX, USA).
Results
Recombinant Fab Antibody Reacts with NS3 Protein
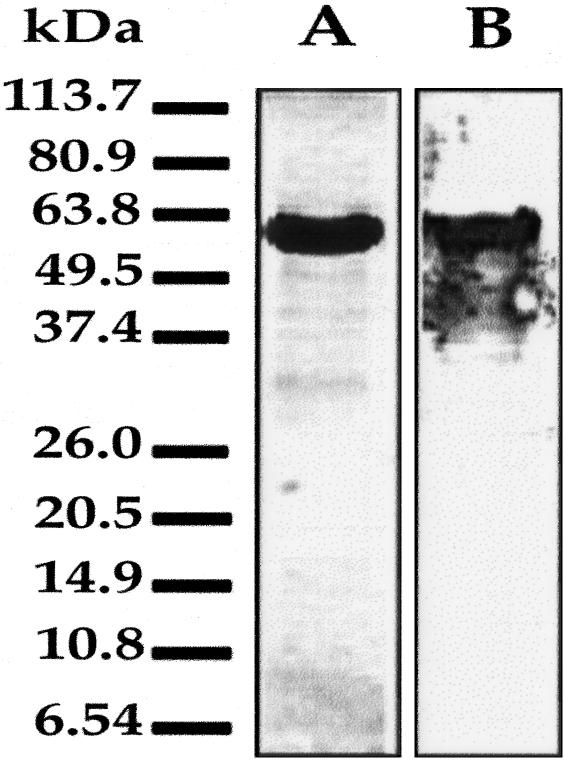
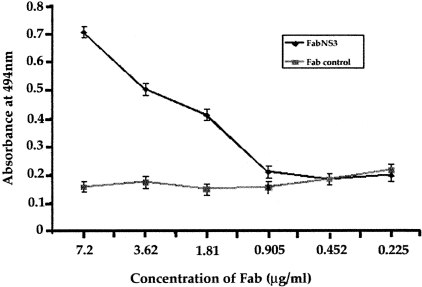
We determined whether the recombinant Fab clone isolated from a human patient could react with NS3 protein and block helicase activity in vitro. For this purpose, NS3 protein with an N-terminal hexahistidine tag was expressed in E. coli and purified using a Ni-NTA column. A single band ∼55 kd in size was seen on SDS-PAGE, which appears to be very pure (Figure 1A). To determine the specificity of this protein, Western blot analysis was performed using a polyclonal antibody to NS3 (Figure 1B). These results suggest that the protein purified from E. coli was HCV NS3. This protein could not have been a carry over from the E. coli because no similar band was present when protein purification was performed using lysates prepared from control DE3 (BL-21) E. coli. Recombinant Fab antibody was also purified from E. coli using a protein G-anti-Fab immunoaffinity column. We demonstrated using ELISA that the purified Fab fragment binds to NS3 protein (Figure 2). The relative affinity of this Fab antibody was determined by competitive ELISA and found to be in the range of 1 nmol/L. Repeated attempts to demonstrate the reactivity of this antibody fragment by Western blot analysis using purified NS3 protein were unsuccessful. This antibody therefore appears to be directed against a conformational epitope.
Figure 1-4241.
Purification of HCV NS3 helicase protein. The HCV NS3 gene was expressed in E. coli and affinity purified using a Ni-NTA affinity column. A: Ten μg of affinity-purified NS3 protein was separated on a SDS-PAGE gel and stained with Coomassie blue. The affinity-purified NS3 helicase is pure because a single band of ∼55 kd is seen after Coomassie staining. B: Western blot analysis showing that the affinity-purified NS3 protein reacts with a polyclonal anti-NS3 antibody.
Figure 2-4241.
ELISA showing that affinity-purified Fab antibody binds to purified HCV NS3 helicase. Polystyrene plates were coated with NS3 helicase in bicarbonate buffer. The wells were washed, blocked with 2% bovine serum albumin, and then incubated with Fab antibody at different concentrations for 1 hour. The wells were washed and incubated with horseradish peroxidase-labeled goat anti-Fab for 1 hour. After this step, wells were washed three times and color was developed after adding 100 μl of OPD solution containing hydrogen peroxide. Finally, the reaction was stopped and optical density at 494 nm was measured. The binding of antibody to NS3 helicase was determined by plotting optical density versus antibody dilution.
Recombinant Fab Antibody Blocks Helicase Activity
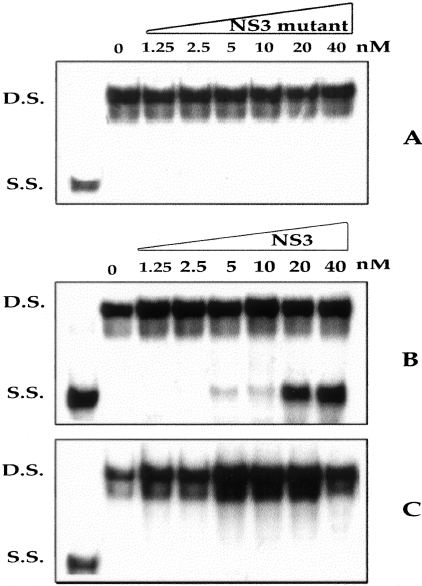
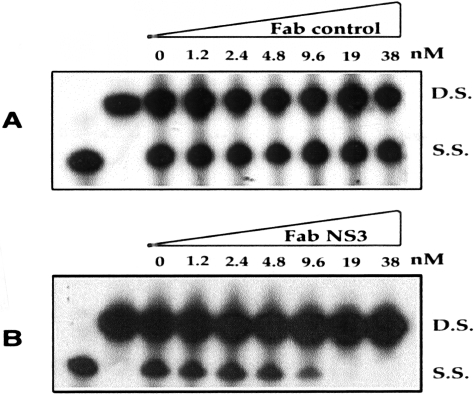
To determine whether NS3 protein purified from E. coli could have activity, a helicase assay was established using double-stranded RNA as well as DNA substrates. In preliminary experiments it was determined that the NS3 protein could unwind either double-stranded RNA or DNA substrates. Subsequent experiments were performed using a partially double-stranded DNA substrate (DS). In the presence of NS3 protein, partial double-stranded DNA substrate was converted to a single strand (SS) as detected by products with altered mobility on acrylamide gel electrophoresis (Figure 3). The specificity of the helicase assay was determined by running a helicase reaction without rATP (Figure 3). The results in Figure 3C, suggest that ATP hydrolysis was essential for the NS3 helicase activity. The helicase assay was performed using mutant NS3 helicase protein (DECH-DQCH). Results of helicase assay using the mutant helicase protein presented in Figure 3A indicate that the helicase activity is not derived from protein contaminants from E. coli. The helicase assay was also performed using different concentrations of NS3 protein (Figure 3B). This experiment indicates that the NS3 helicase activity is saturable at a template DNA to protein ratio of 1:2. These results suggest that the NS3 protein purified from E. coli possesses helicase activity similar to the previously published results of other laboratories. In the next step, specific inhibition of helicase activity of NS3 protein in the presence of different concentrations of purified Fab fragment was studied. Different proportions of purified Fab were incubated with 20 nmol/L of NS3 protein for 1 hour, and then the mixture was added to the helicase assay. The helicase activity was completely blocked by increasing the concentration of Fab, with maximum inhibition at 1:1 molar ratio (Figure 4B). To exclude the possibility of a nonspecific effect of the Fab, the helicase assay was performed in the presence of an unrelated Fab directed against influenza virus (supplied by R. Burioni). This control Fab did not inhibit the helicase activity of HCV NS3 protein (Figure 4A).
Figure 3-4241.
Strand separation helicase activity of purified recombinant NS3 protein using a partial double-stranded DNA substrate. A: Helicase activity of mutant NS3 (DQCH mutant) protein at different concentrations. Mutant helicase protein has no helicase activity. B: Helicase assay performed with purified NS3 helicase protein (wild type) at similar concentration. C: Helicase activity of NS3 protein (wild type) at similar concentration in the absence of rATP. The purified NS3 protein unwinds partially double-stranded DNA substrate; this helicase activity is not because of bacterial contamination and dependent on rATP. Helicase activity is dependent on template to NS3 protein that can be saturable at 1:10 molar ratio.
Figure 4-4241.
Affinity-purified human Fab antibody inhibits helicase of NS3 protein in a concentration-dependent manner. A: Helicase assay was performed in vitro in the presence of different concentrations of unrelated Fab antibody control. Control Fab did not inhibit helicase activity of NS3 protein. B: Helicase assay was performed in vitro in the presence of different concentrations of human Fab to NS3 (HFab-aNS3) antibody. Helicase activity can be inhibited by human Fab at 2:1 molar ratio.
Intracellular Expression of Fab in Hepatic Cells
The ability of the Fab to inhibit helicase activity of HCV NS3 protein prompted us to test its anti-viral effect in HCV cell culture systems. For this purpose, the Fab gene was cloned and expressed in Huh-7 cells as a complete IgG1 antibody molecule using the vector pFab-CMV. The expression of NS3-specific Fab antibody as well as control antibody was examined by immunocytochemical method at 72 hours after transfection. A very strong positive staining was observed in the cytoplasm of most of the Huh-7 cells transfected with either pFab-CMV-NS3 (L+H) plasmid or control Fab plasmid (Figure 5). No staining was present in Huh-7 cells that were mock transfected. These results confirm that the NS3 antibody is expressed in transfected Huh-7 cells.
Figure 5-4241.
Expression of human Fab antibody (HFab-aNS3) in Huh-7 cells. Huh-7 cells were transfected with pFab-CMV-NS3 (H+L) plasmid using FuGENE 6 reagent and after 72 hours the cells were examined for the expression of Fab by immunocytochemistry. Transfected Huh-7 cells were immobilized on glass slides by cytospin, reacted with biotin-labeled anti-human F(ab)2 antibodies, and reacted with ABC reagents. Color was developed with DAB. Cytoplasmic expression of antibody in the transfected cells (brown staining, shown by arrows) was present in the majority of cells transfected with Fab expression plasmid. Mock-transfected Huh-7 cells were negative.
Fab Antibody Inhibits Replication of Subgenomic HCV RNA
Anti-viral efficacy of this recombinant antibody fragment was determined using in vitro cell culture models for HCV. First, the highly efficient replicon Huh-7 cell line replicating chimeric subgenomic HCV RNA was used. This cell line was transfected with chimeric subgenomic HCV RNA that expresses nonstructural genes. Autonomous replication of chimeric subgenomic RNA by the nonstructural proteins in the Huh-7 cells makes the cells resistant to G-418. We used this model to determine whether anti-NS3 antibody could have an effect on full-cycle replication of HCV. The anti-viral effect of this antibody clone was tested using a replicon cell line Con-15. Intracellular expression of NS3 Fab antibody resulted in significant inhibition of HCV RNA levels in the Con-15 replicon cell line (Figure 6). Expression of control Fab did not affect HCV RNA levels, indicating that this might be a specific effect of HFab-aNS3 clone. The GAPDH mRNA levels in the antibody-transfected cells remained to be the same, excluding the possibility of variation in the amount of RNA used in the RPA.
Figure 6-4241.
Intracellular expression of Fab in stable cell line replicating subgenomic RNA inhibits HCV-positive strand RNA. Con-15 cells were transfected with Fab antibody expression plasmid using FuGENE 6 reagent. Total RNA was isolated at 3 and 5 days after antibody transfection. Positive strand HCV-RNA was measured by RPA using a probe targeted to the 5′ UTR region. A: RPA for HCV-positive strand RNA, 5 days after antibody expression in the Con-15 cells. Intracellular expression of HFab-aNS3 in a replicon cell line inhibits HCV-RNA levels. The level of HCV-RNA was not altered after expression of a control Fab antibody. B: RPA for GAPDH mRNA level in the antibody-transfected cells. GAPDH mRNA levels remained unaltered after antibody transfection. C: Potential anti-viral effect of HFab-aNS3 clone against HCV was examined in three separate RPA experiments. The band intensity was measured. Percentage of inhibition was recorded. Data are expressed as mean ± SD.
Fab Antibody Inhibits Viral Protein Expression
We have also examined whether the Fab antibody could alter the level of viral protein expression. Expression of viral NS3 protein in the antibody-transfected cell was examined by an immunocytochemical method. The results of this experiment clearly show that virus protein levels were also reduced after expression of the Fab but not in the control antibody-transfected replicon cells (Figure 7). Taken together, these results suggest that intracellular expression of NS3-specific Fab clone inhibits replication and expression of NS3 protein in a replicon cell line.
Figure 7-4241.
Intracellular expression of Fab antibody in a replicon cell line clears the HCV NS3 protein expression. A replicon cell line Con-15 was transfected with either HFab-NS3 or control Fab expression plasmid. After 72 hours cells were harvested and NS3 antigen expression in replicon cell line was measured by immunocytochemistry using a monoclonal antibody. Top: The expression of NS3 protein. Bottom: The expression of β-actin protein. Expression of HCV NS3 was specifically inhibited in a majority of cells after antibody expression.
Fab Antibody Inhibits Full-Length HCV RNA Replication
Because the replicon cell line does not assemble complete virus particles, we decided to test its anti-viral effect in a full-length RNA transfection-based HCV replication system that was established in our laboratory. We have shown previously that transfection of full-length RNA into HepG2 cells results in the assembly of complete virus particles, which are infectious in chimpanzees.32 This model closely resembles HCV replication in an infected liver. We assessed the anti-viral effect of HFab-aNS3 clone in this model by after inducing antibody expression in HepG2 cells already transfected with full-length HCV genomic RNA. HCV-RNA levels were measured in this system using a competitive reverse transcriptase-polymerase chain reaction method described previously. Results of this analysis suggest that the anti-NS3 Fab clone inhibited full-length RNA in the HepG2 cells, with HCV RNA levels below the detection limit after 6 days (Figure 8). The GAPDH mRNA levels in the HFab-aNS3 or control Fab-transfected cells remained unaltered.
Figure 8-4241.
Intracellular expression of Fab inhibits full-length HCV replication. HepG2 cells were transfected with 10 μg of in vitro transcribed full-length HCV RNA using the DEAE-dextran method. A week later, cells were transfected with antibody expression plasmid (HFab-aNS3). RNA extracts were prepared from transfected cells at 3, 6, 9, and 12 days and HCV RNA levels were measured using competitive reverse transcriptase-polymerase chain reaction assay. Intracellular HCV RNA levels were reduced significantly after antibody transfection without any alteration in the GAPDH mRNA level.
Fab Antibody Inhibits Negative Strand HCV-RNA Synthesis
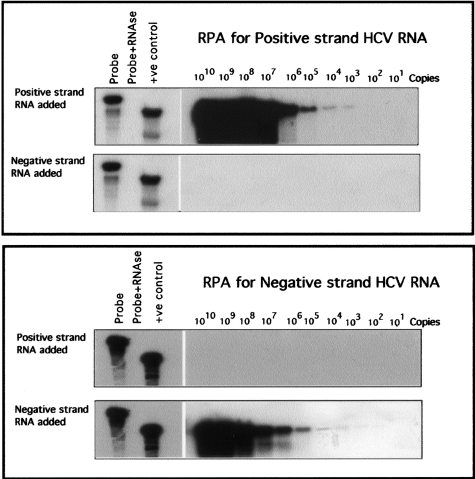
We also examined the effect of HFab-aNS3 expression on negative strand RNA synthesis using a DNA-based model. The antibody expression plasmid was co-transfected with a transcription plasmid containing full-length HCV to Huh-7 cells using a two-step transfection procedure. After 72 hours, total RNA isolated and negative strand RNA was measured by RPA. Intracellular expression of HFab-aNS3 clone blocked synthesis of negative strand RNA formation, with no inhibition seen by control Fab expression (Figure 9). The RPA was performed using a probe targeted to the highly conserved 5′ UTR region (46 to 341). Because this part of the HCV genome has several stem-loop structures, it was important to examine strand specificity of the RNA probes used in the RPA experiment. To evaluate the strand specificity of anti-sense RNA probe used in RPA assay, it was hybridized to in vitro transcribed positive and negative strand genome-length HCV-RNA and RPA was performed. The anti-sense RNA probe hybridized only to positive strand HCV-RNA, and not to negative strand HCV-RNA (Figure 10, top). To evaluate strand specificity of sense RNA probe, it was hybridized to in vitro-transcribed positive and negative strand genome-length HCV-RNA and RPA was performed. The sense RNA probe hybridized only with in vitro-transcribed negative strand HCV-RNA, but not with positive strand HCV-RNA (Figure 10, bottom).
Figure 9-4241.
Intracellular expression of HFab-aNS3 inhibits HCV-negative strand. Huh-7 cells were infected with a replication-defective adenovirus carrying T7 RNA polymerase gene. These cells were then co-transfected with transcription plasmid and antibody expression plasmid. After 72 hours, RNA extracts were prepared, digested with DNase, and tested for negative strand HCV RNA by RPA assay. Top: RPA for HCV-negative strand RNA in the cells transfected with HFab-aNS3. Bottom: RPA for GAPDH mRNA. Lane 1, Probe alone; lane 2, probe digested with RNase; lane 3, cells transfected with pMO9.6T7-Rz alone; lane 4, cells co-transfected with pMO9.6T7-Rz and pFab CMV (control); lane 5, cells co-transfected with pMO9.6T7-Rz and pFab-CMV-NS3 (H+L).
Figure 10-4241.
Strand specificity of RPA. Positive and negative strand full-length HCV RNA transcripts were prepared by in vitro transcription reaction. Tenfold serially diluted HCV RNA were hybridized with either a gel-purified negative strand RNA probe or positive strand RNA probe targeted to the 5′UTR (nucleotides 45 to 341). RPA was performed using a commercially available RPA kit. Top: Negative strand RNA probe only hybridized to positive strand HCV RNA. Bottom: Positive strand RNA probe only hybridized to negative strand HCV RNA. The RPA assay used in our experiment is strand-specific.
Discussion
Studies on the development of experimental therapies on HCV infection have been enhanced by the availability of infectious clones and cell culture models. HCV is a positive strand RNA virus that replicates via synthesis of negative strand RNA in the cytoplasm of infected cells. The genome of HCV is ∼9600 nucleotides in length and encodes a large polyprotein of ∼3010 amino acids in length. This large polyprotein is co- and posttranslationally processed by cellular and viral proteases to produce three structural proteins (with the exception of small P7 protein) and six nonstructural proteins.37 The nonstructural protein 3 (NS3) of HCV is a multifunctional protein, having protease activity at the N-terminus and helicase and NTPase activities at the C-terminus.38,39 The helicase/NTPase activity of NS3 protein are presumed to be involved in replication of viral RNA by unwinding the double-stranded RNA intermediates. The chimpanzee model has shown that the protease, helicase, and NTPase activities of NS3 are absolutely critical for replication processes.40 Therefore it is expected that an agent that could block any of these three activities of viral replication should be an excellent anti-viral HCV agent.
In this study, we attempted to block helicase activity of NS3 protein by the use of a human Fab antibody. To test the anti-viral properties of this clone in vitro, we first expressed NS3 protein in E. coli and purified it by Ni-affinity column. The purified protein possessed helicase activity similar to the full-length NS3 protein from other HCV isolates. To determine whether this Fab antibody could block the enzyme activities in vitro, this Fab was expressed in E. coli and affinity-purified using a protein G-anti-human Fab column. We showed that affinity-purified Fab could block helicase activity of HCV NS3 protein. This inhibition of helicase activity is specific because a control Fab purified using the identical procedure did not inhibit the helicase activity of NS3 protein. The affinity of this recombinant Fab antibody was determined by competitive ELISA method and found to be in the range of 1 nmol/L. The affinity of this antibody was comparable to a mouse monoclonal antibody (Novocastra Laboratories Ltd.) used in the immunostaining found to be 10 nmol/L. In our experience, antibodies generated by phage display repertoire library cloning are usually directed against conformational epitopes and low-affinity antibodies. This seems to be the case for this particular human Fab antibody. It is also possible that NS3 antigen bound to an ELISA plate could have resulted in an additional change to the native conformation of NS3 protein, which could have contributed to the low reactivity. These data led to further study of the anti-viral effects of this Fab antibody clone on virus replication in cell culture models.
In cell culture models, we expressed anti-NS3 antibody clone in cells using vector pFab-CMV (H+L). This vector allows intracellular cytoplasmic expression of IgG1 in transfected cells. We examined anti-viral effect of the Fab clone in a stable cell line that replicates subgenomic RNA. Continuous replication of HCV in the cytoplasm of this stable cell line makes it resistant to G-418 drug. We showed that expression of Fab antibody in this cell line inhibited HCV-RNA levels and protein expression in a majority of cells. This effect seems specific to this Fab clone because the control (unrelated) Fab did not inhibit HCV. We also showed that the difference in the RPA results is not because of amounts of RNA present in the extracts, because GAPDH-mRNA levels of the two samples were the same. Anti-viral effect of this Fab clone was tested in a cell culture model we established in HepG2 cells. Interestingly, we found that HCV-RNA levels in the HepG2 cells were reduced by intracellular expression of this Fab using co-transfection studies. We also demonstrated that intracellular expression of anti-NS3 human antibody clone inhibited HCV-negative strand RNA formation. Our study has used all of the HCV in vitro cell culture models that are currently available. Taken together, results of this study demonstrate that inhibition of HCV replication by the use of intracellular immunization by recombinant antibodies may be an alternative approach for these patients who remain resistant to interferon therapy.
Recombinant antibody fragments have been reported to be therapeutically useful in the treatment of viral as well as nonviral diseases.41–43 Use of recombinant antibodies against core protein of HCV has been reported.44 A series of human Fab targeted to the envelope protein of HCV have been developed.45,46 There are some reports of using recombinant antibody fragment to inhibit enzyme activities of NS3 protein.47 In most of these studies, including ours, single chain antibodies have been used to inhibit either protease or helicase activity of NS3 protein and polymerase activity of NS5B protein.13,47,48 However, the single chain variant of this mouse monoclonal antibody was found to lose the ability to bind helicase as compared to the parent antibody. Previously, we have developed adenovirus constructs expressing single chain antibodies against NS3 and NS4A protein.13 The NS3 scFv inhibits helicase activity and HCV levels in a primary hepatocyte culture model. Later, we found that none of these antibodies could inhibit HCV replication in the highly efficient replicon-based cell culture models. We thought it might be because of low affinity of scFvs, therefore, this human Fab antibody was selected. We found that this antibody appears to have good anti-viral activity against HCV. Finally, the antibody used in this study is fully human, limiting the possibility of an immune response by the host against cells producing this molecule.
In summary, inhibition of helicase activity of HCV NS3 protein by a specific human antibody reported here is an important step toward developing an intracellular therapy for HCV. It is possible that antibodies can be directed against other key enzymes to treat viral infection. Intracellular delivery of antibodies may be problematic. These issues can be resolved by future research to develop methods to deliver efficiently therapeutic antibodies in vivo either by the use of viral vector or by direct delivery of antibodies. Intracellular expression of antibodies targeting HCV proteins and inhibiting important viral functions may represent a new direction to follow for the development of effective therapy for HCV infection.
Acknowledgments
We thank Dr. Su-Chen Li, Department of Biochemistry, Tulane University Health Sciences Center, New Orleans, LA, for her help in experiments with thin layer chromatography; Pietro Paolo Sanna, Department of Neuropharmacology, The Scripps Research Institute, La Jolla, CA, USA) for providing Fab expression plasmid; Ding-Shinn Chen, Hepatitis Research Center, National Taiwan University Hospital, Taipei, Taiwan, for providing pET21a-NS3 plasmids; George Liu, University of Kentucky, Lexington, KY, for providing replicon clone; Donald Olivares, Department of Pathology, Tulone University Health Sciences Center, New Orleans, for helping in the digital photography; and Dr. William H. Robichaux, Thibodaux Regional Medical Center, Thibodaux, LA, USA for critically reading the manuscript.
Footnotes
Address reprint requests to Srikanta Dash, Ph.D., Associate Professor, Department of Pathology and Laboratory Medicine, Tulane University Health Sciences Center, 1430 Tulane Ave., New Orleans LA 70112. E-mail: sdash@tulane.edu.
Supported by the National Cancer Institute (grants CA54576 and CA89121 to S.D.) and the Tulane Cancer Center.
This article is dedicated to the memory of Dr. Michael A. Gerber.
References
- Strader DB, Wright T, Thomas DL, Seeff LB. Diagnosis, management, and treatment of hepatitis C. Hepatology. 2004;39:1147–1171. doi: 10.1002/hep.20119. [DOI] [PubMed] [Google Scholar]
- Alt M, Renz R, Hofschneider PH, Paumgartner G, Caselmann WH. Specific inhibition of hepatitis C viral gene expression by antisense phosphorothioate oligodeoxynucleotides. Hepatology. 1995;22:707–717. [PubMed] [Google Scholar]
- Hanecak R, Brown-Driver V, Fox MC, Azad RF, Furusako S, Nozaki C, Ford C, Sasmor H, Anderson KP. Antisense oligonucleotide inhibition of hepatitis C virus gene expression in transformed hepatocytes. J Virol. 1996;70:5203–5212. doi: 10.1128/jvi.70.8.5203-5212.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakita T, Wands JR. Specific inhibition of hepatitis C virus expression by antisense oligonucleotides. J Biol Chem. 1994;269:14205–14210. [PubMed] [Google Scholar]
- Macejak DG, Jensen KL, Jamison SF, Domenico K, Robert EC, Chaudhary N, von Carlowitz I, Bellon L, Tong MJ, Conrad A, Pavco PA, Blatt LM. Inhibition of hepatitis C virus (HCV)-RNA-dependent translation and replication of a chimeric HCV poliovirus using synthetic stabilized ribozymes. Hepatology. 2000;31:769–776. doi: 10.1002/hep.510310331. [DOI] [PubMed] [Google Scholar]
- Oketani M, Asahina Y, Wu CH, Wu GY. Inhibition of hepatitis C virus-directed gene expression by a DNA ribonuclease. J Hepatol. 1999;31:628–634. doi: 10.1016/s0168-8278(99)80341-9. [DOI] [PubMed] [Google Scholar]
- Leiber A, He CY, Polyack SJ, Gretch DR, Barr D, Kay MA. Elimination of hepatitis c virus RNA in infected hepatocytes by adenovirus-mediated expression of ribozymes. J Virol. 1996;70:8782–8791. doi: 10.1128/jvi.70.12.8782-8791.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch PJ, Tritz R, Yei S, Leavitt M, Yu M, Barber J. A potential therapeutic application of hairpin ribozymes: in vitro and in vivo studies of gene therapy for hepatitis C virus infection. Gene Ther. 1996;3:994–1001. [PubMed] [Google Scholar]
- Kapadia SB, Brideau-Andersen A, Chisari FV. Interference of hepatitis C virus RNA replication by short interfering RNA. Proc Natl Acad Sci USA. 2003;100:2014–2018. doi: 10.1073/pnas.252783999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall G, Grakoui A, Rice CM. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc Natl Acad Sci USA. 2003;100:235–240. doi: 10.1073/pnas.0235524100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biocca S, Neuberger MS, Cattaneo A. Expression and targeting of intracellular antibodies in mammalian cells. EMBO J. 1990;74:101–108. doi: 10.1002/j.1460-2075.1990.tb08085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Hayashi N, Takehara T, Ueda K, Mita E, Tatsumi T, Sasaki Y, Kasahara A, Hori M. Intracellular single chain antibody against hepatitis B virus core protein inhibits replication of hepatitis B virus in cultured cells. Hepatology. 1999;30:300–307. doi: 10.1002/hep.510300105. [DOI] [PubMed] [Google Scholar]
- Sullivan DE, Mondelli M, deHaard H, Curiel D, Krasnykh V, Mikheeva G, Dash S, Gerber MA. Construction and characterisation of intracellular single chain human antibody to hepatitis C virus non-structural 3 protein. J Hepatol. 2002;37:660–668. doi: 10.1016/s0168-8278(02)00270-2. [DOI] [PubMed] [Google Scholar]
- Piche A, Kasono K, Johanning F, Curiel TJ, Curiel DT. Phenotypic knockout of the latent membrane protein 1 of Epstein-Barr virus by an intracellular single-chain antibody. Gene Ther. 1998;5:1171–1179. doi: 10.1038/sj.gt.3300706. [DOI] [PubMed] [Google Scholar]
- Barbas CF, Kang AS, Lerner RA, Benkovic SJ. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc Natl Acad Sci USA. 1991;88:7978–7982. doi: 10.1073/pnas.88.18.7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasco WA, La Vecchio J, Winkler A. Human anti-HIV-1 tat scFv intrabodies for gene therapy of advanced HIV-1 infection and AIDS. J Immunol Methods. 1999;23:223–238. doi: 10.1016/s0022-1759(99)00159-3. [DOI] [PubMed] [Google Scholar]
- Burton DR, Barbas CF, Persson MA, Koenig S, Chanock RM, Lerner RA. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci USA. 1991;88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberger P, Shutton JK, Rader C, Elia M, Barbas CF. Generation and characterization of a recombinant human CCR5-specific antibody. J Biol Chem. 2000;275:36073–36078. doi: 10.1074/jbc.M002765200. [DOI] [PubMed] [Google Scholar]
- Barbas CFD, Crowe JE, Jr, Cababa D, Jones TM, Zebedee SL, Murphy BR, Chanoc RM, Burton DR. Human monoclonal Fab fragments derived from a combinatorial library bind to respiratory syncytial virus F glycoprotein and neutralize infectivity. Proc Natl Acad Sci USA. 1992;89:10164–10168. doi: 10.1073/pnas.89.21.10164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burioni R, Williamson RA, Sanna PP, Bloom FE, Burton DR. Recombinant human Fab to glycoprotein D neutralizes infectivity and prevents cell-to-cell transmission of herpes simplex viruses 1 and 2 in vitro. Proc Natl Acad Sci USA. 1994;91:355–359. doi: 10.1073/pnas.91.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson RA, Burioni R, Sanna PP, Partridge LJ, Barbas CF, Burton DR. Human monoclonal antibodies against a plethora of viral pathogens from single combinatorial libraries [published erratum appears in Proc Natl Acad Sci USA 1994, 91:1193] Proc Natl Acad Sci USA. 1993;90:4141–4145. doi: 10.1073/pnas.90.9.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson RA, Persson MA, Burton DR. Expression of a human monoclonal anti-(rhesus D) Fab fragment in Escherichia coli with the use of bacteriophage lambda vectors. Biochem J. 1991;277:561–563. doi: 10.1042/bj2770561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Co MS, Deschamps M, Whitley RJ, Queen C. Humanized antibodies for antiviral therapy. Proc Natl Acad Sci USA. 1991;88:2869–2873. doi: 10.1073/pnas.88.7.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai C-L, Pan W-C, Liaw S-H, Yang U-C, Hwang L-H, Chen D-S. Structure based mutational analysis of the hepatitis C virus NS3 helicase. J Virol. 2001;75:8289–8297. doi: 10.1128/JVI.75.17.8289-8297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaisant P, Burioni R, Manzin A, Solforosi L, Candela M, Gabrielli A, Fadda G, Clementi M. Human monoclonal recombinant Fabs specific for HCV antigens obtained by repertoire cloning in phage display combinatorial vectors. Res Virol. 1997;148:165–169. doi: 10.1016/s0923-2516(97)89904-9. [DOI] [PubMed] [Google Scholar]
- Rath S, Stanley CM, Steward MW. An inhibition enzyme immunoassay for estimating relative affinity and affinity heterogeneity. J Immunol Methods. 1988;106:245–249. doi: 10.1016/0022-1759(88)90204-9. [DOI] [PubMed] [Google Scholar]
- Gallinari P, Brennan D, Nardi C, Brunetti M, Tomei L, Steinkuhler C, De Francesco R. Multiple enzymatic activities associated with recombinant NS3 protein of hepatitis C virus. J Virol. 1998;72:6758–6769. doi: 10.1128/jvi.72.8.6758-6769.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanna PP, Samson ME, Moon JS, Rozenshteyn R, De Logu A, Williamson RA, Burton DR. pFab-CMV, a single vector system for the rapid conversion of recombinant Fabs into whole IgG1 antibodies. Immunotechnology. 1999;4:185–188. doi: 10.1016/s1380-2933(98)00022-0. [DOI] [PubMed] [Google Scholar]
- Lohmann V, Korner F, Koch J-O, Herian U, Theilmann L, Bartenschalager R. Replication of subgenomic hepatitis C RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- Luo G, Xin S, Cai Z. Role of the 5′-proximal stem-loop structure of the 5′untranslated region in replication and translation of hepatitis C virus RNA. J Virol. 2003;77:3312–3318. doi: 10.1128/JVI.77.5.3312-3318.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash S, Kalkaeri G, McClure H, Garry RF, Clejan S, Thung SN, Murthy KK. Transmission of HCV to a chimpanzee using viral particles produced in an RNA-transfected HepG2 cell culture. J Med Virol. 2001;65:276–281. doi: 10.1002/jmv.2030. [DOI] [PubMed] [Google Scholar]
- Dash S, Halim AB, Tsuji H, Hiramatsu N, Gerber MA. Transfection of HepG2 cells with infectious hepatitis C virus genome. Am J Pathol. 1997;151:363–373. [PMC free article] [PubMed] [Google Scholar]
- Dash S, Rege TA, Tsuji H, Gaglio P, Garry RF, Saxena R, Thung SN. HCV RNA levels in hepatocellular carcinomas and adjacent non-tumorous livers. J Virol Methods. 2000;90:15–23. doi: 10.1016/s0166-0934(00)00199-3. [DOI] [PubMed] [Google Scholar]
- Myung J, Khalap N, Kalkeri G, Garry RF, Dash S. Inducible model to study negative strand RNA synthesis and assembly of hepatitis C virus from a full-length cDNA clone. J Virol Methods. 2001;94:55–67. doi: 10.1016/s0166-0934(01)00278-6. [DOI] [PubMed] [Google Scholar]
- Prabhu R, Joshi V, Garry RF, Bastian F, Haque S, Regenstein F, Thung SN, Dash S. Interferon alpha-2b inhibits negative strand RNA and protein expression from full-length HCV1a infectious clone. Exp Mol Pathol. 2004;76:242–252. doi: 10.1016/j.yexmp.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Reed KE, Rice CM. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr Top Microbiol Immunol. 2000;242:55–84. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]
- Bartenschlager R. The NS3/4A proteinase of the hepatitis C virus: unravelling structure and function of an unusual enzyme and a prime target for antiviral therapy. J Viral Hepatol. 1999;6:165–181. doi: 10.1046/j.1365-2893.1999.00152.x. [DOI] [PubMed] [Google Scholar]
- Grakoui A, McCourt DW, Wychowski C, Feinstone SM, Rice CM. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolykhalov AA, Mihalik K, Feinstone SM, Rice CM. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J Virol. 2000;74:2046–2051. doi: 10.1128/jvi.74.4.2046-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddleston M, Rajapakse S, Rajakanthan JS, Sjostrom L, Santharaj W, Thenabadu PN, Sheriff MH, Warrell DA. Anti-digoxin Fab fragments in cardiotoxicity induced by ingestion of yellow oleander: a randomised controlled trial. Lancet. 2000;355:967–972. doi: 10.1016/s0140-6736(00)90014-x. [DOI] [PubMed] [Google Scholar]
- Krawczynski K, Alter MJ, Tankersley DL, Beach M, Robertson BH, Lambert S, Kuo G, Spelbring JE, Meeks E, Sinha S, Carson DA. Effect of immune globulin on the prevention of experimental hepatitis C virus infection. J Infect Dis. 1996;173:822–828. doi: 10.1093/infdis/173.4.822. [DOI] [PubMed] [Google Scholar]
- Mhashilkar AM, Bagley J, Chen SY, Szilvay AM, Helland DG, Marasco WA. Inhibition of HIV-1 Tat-mediated LTR transactivation and HIV-1 infection by anti-Tat single chain intrabodies. EMBO J. 1995;14:1542–1551. doi: 10.1002/j.1460-2075.1995.tb07140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintges T, zu Putlitz J, Wands JR. Characterization and binding of intracellular antibody fragments to the hepatitis C virus core protein. Biochem Biophys Res Comm. 1999;263:410–418. doi: 10.1006/bbrc.1999.1350. [DOI] [PubMed] [Google Scholar]
- Burioni R, Bugli F, Mancini N, Rosa D, Di Campli C, Moroncini G, Manzin A, Abrignani S, Varaldo PE, Clementi M, Fadda G. Nonneutralizing human antibody fragments against hepatitis C virus E2 glycoprotein modulate neutralization of binding activity of human Fabs. Virology. 2001;288:29–35. doi: 10.1006/viro.2001.1014. [DOI] [PubMed] [Google Scholar]
- Burioni R, Plaisant P, Manzin A, Roza D, Deli Carri V, Bugli F, Solforosi L, Abrignani S, Varaldo PE, Fadda G, Clementi M. Dissection of human humoral immune response against hepatitis C virus E2 glycoprotein by repertoire cloning and generation of recombinant Fab fragments. Hepatology. 1998;28:810–814. doi: 10.1002/hep.510280331. [DOI] [PubMed] [Google Scholar]
- Ueno T, Misawa S, Ohba Y, Matsumoto M, Mizunuma M, Kasai N, Tsumoto K, Kumagai I, Hayashi H. Isolation and characterization of monoclonal antibodies that inhibit hepatitis C virus NS3 protease. J Virol. 2000;74:6300–6308. doi: 10.1128/jvi.74.14.6300-6308.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradpour D, Bieck E, Hugle T, Wels W, Wu JZ, Hong Z, Blum HE, Bartenschlager R. Functional properties of a monoclonal antibody inhibiting the hepatitis C virus RNA-dependent RNA polymerase. J Biol Chem. 2002;277:593–601. doi: 10.1074/jbc.M108748200. [DOI] [PubMed] [Google Scholar]