Abstract
TMS1/ASC is an intracellular signaling molecule with proposed roles in the regulation of apoptosis, nuclear factor-κB activation, and cytokine maturation. Previous studies have shown that TMS1/ASC is silenced by epigenetic means in human breast tumors. In this study, we examined methylation and expression of TMS1/ASC in glioblastoma multiforme (GBM). Whereas normal brain tissue was unmethylated at the TMS1 locus and expressed TMS1 message, 11 of 23 human GBM cell lines exhibited reduced or absent expression of TMS1 that was associated with aberrant methylation of a CpG island in the promoter of the TMS1 gene. Quantitative analysis showed that there was an inverse correlation between the degree of methylation and level of TMS1 expression. Treatment of GBM cell lines lacking TMS1 expression with the methyltransferase inhibitor 5-aza-2′deoxycytidine resulted in partial demethylation and re-expression of TMS1. Analysis of primary tissues indicated that the TMS1 gene is unmethylated and expressed in normal brain, where its expression is restricted to astrocytes. In contrast, TMS1 was aberrantly methylated in 43% (10 of 23) primary GBM specimens. Tumors that exhibited aberrant methylation of TMS1 generally expressed reduced or absent expression of TMS1 as compared to unmethylated cases. Methylation of TMS1 was not associated with patient age, gender, or treatment status. Although the relationship did not reach statistical significance, there was a trend toward increased overall survival for patients with unmethylated tumors. For one patient, disease progression from astrocytic astrocytoma (World Health Organization grade III) to GBM (World Health Organization grade IV) was associated with selective expansion of TMS1-negative cells. The data suggest a role for the epigenetic silencing of TMS1 in the pathogenesis of human GBM. Methylation of TMS1 may prove to be a useful prognostic marker and/or predictor of patient survival and tumor malignancy.
Glioblastoma multiforme (GBM) is the most prevalent and lethal of human gliomas and accounts for 50 to 60% of primary brain tumors. Despite intensive research throughout the past 2 decades, there has been little progress in the treatment of GBM; the median survival after diagnosis remains at 1 year, although patient outcome can vary considerably from a few months to 5% of patients surviving more than 2 years.1 This clinical variability is likely because GBMs exhibit considerable heterogeneity at both the genetic and biological levels. GBMs can be classified into two groups based on their clinical phenotype: primary, which occur de novo (ie, without having progressed from a less malignant precursor) in older patients;2 and secondary, which are much less frequent (∼10% of cases) and tend to develop from a low-grade or anaplastic astrocytomas in younger patients.2,3 Although these two groups are histologically indistinguishable, they appear to have arisen along distinct genetic pathways.4 Typically, primary GBMs show frequent amplification of EGFR, while only rarely harboring TP53 mutations, whereas secondary GBMs show frequent mutations in TP53 in the absence of EGFR alterations. These relatively few genetic and clinical distinctions provide only limited information beyond the traditional histological classification of GBM and thus far have not significantly improved our ability to predict patient prognosis or response to therapy.
Thus, the impetus remains to seek out additional genetic and epigenetic alterations that might provide insight into mechanisms that contribute to the malignancy of GBMs. Aberrant methylation of CpG island-associated genes is a frequent epigenetic alteration associated with the inactivation of tumor suppressor and other genes in human cancers.5–7 Approximately one-half of human genes contain CpG islands; short stretches of CpG-dense DNA typically associated with the 5′ ends of genes.8 Unmethylated in normal tissues, these regions can become methylated de novo in cancer cells. This change is accompanied by alterations in histone modification and chromatin conformation rendering the CpG island and its embedded promoter transcriptionally inert.9 In human gliomas, such epigenetic mechanisms have been implicated in the silencing of several key regulators of the cell cycle (RB, p16INK4A, p73), DNA repair (O6MGMT), apoptosis (DAP kinase), angiogenesis (THBS1), and invasion (TIMP3).10 CpG island methylation is frequent in low-grade gliomas, preceding many of the aforementioned genetic alterations, and the number of events increase with tumor progression.11 Indeed, methylation of the promoter of the DNA repair gene O6MGMT in gliomas has shown promise as an independent predictor of response to treatment with alkylating agents, and of disease-free survival.12–14 Thus aberrant methylation events have become critical to our understanding of the initiation and progression of human brain malignancies and are showing promise as prognostic tools.
Recently, we identified a novel CpG island-associated gene, TMS1 (for target of methylation-associated silencing), that is aberrantly methylated and silenced in a significant proportion of human breast cancers.15 Also known as ASC,16 TMS1 encodes a bipartite adaptor molecule composed of an N-terminal Pyrin domain and a C-terminal CARD domain, and functions as a mediator of intracellular signaling from apoptotic and inflammatory stimuli.17 In previous studies, we showed that overexpression of TMS1 induces apoptosis and inhibits the growth of breast cancer cells, consistent with a putative tumor suppressor role.15 Recent studies showing that TMS1 modulates the activity of caspase-1 and can block the downstream activation of nuclear factor-κB suggest that methylation-mediated silencing of TMS1 could promote tumorigenesis by allowing cells to bypass apoptosis, to evade a local immune response, and by allowing nuclear factor-κB-dependent survival signals to go unchecked.18–21
The goal of the present study was to determine whether methylation-mediated silencing of TMS1 plays a role in the pathogenesis of human gliomas. We find that TMS1 is aberrantly methylated in a significant proportion of GBM cell lines and primary tumors, and that methylation of the TMS1 gene is correlated with the tumor-specific down-regulation of TMS1 in primary GBMs.
Materials and Methods
Cell Lines and Primary Tumors
Primary GBM tumor samples used in this study were obtained from 25 patients who underwent neurosurgical resection at Emory University Hospital between 1996 and 2001. Patients ranged in age from 27 to 81 years. Tumor specimens were frozen at −80°C immediately after resection. All cases were reviewed by a single pathologist (D.B.) for histological confirmation of GBM (World Health Organization grade IV astrocytoma) before being included in this study. For DNA isolations, frozen tumor specimens were macrodissected at the time of histological verification. For comparison, brain tissue from patients without cancer (n = 5) were obtained at the time of autopsy, frozen at −80°C, and dissected into white and gray matter before DNA and RNA isolation.
Human GBM cell lines were maintained as previously described.22 For 5-aza-2′-deoxycytidine experiments, U251 MG and LN 229 cells were seeded at 106 cells per 75-cm2 flask, allowed to attach overnight, and treated with 0.5 μmol/L 5-aza-2′-deoxycytidine. Cells were harvested after 48 hours treatment, and DNA and RNA were extracted. DNA and RNA from all GBM cell lines and primary tumors were extracted using the DNeasy tissue and RNeasy kits (Qiagen, Valencia, CA) according to the manufacturer’s protocol.
Methylation-Specific Polymerase Chain Reaction (PCR)
DNA was subjected to bisulfite modification and methylation-specific PCR as previously described.15,23 Approximately 50 ng of bisulfite-modified DNA was amplified by PCR with the following reaction conditions: 67 mmol/L Tris-HCl (pH 8.8), 16.6 mmol/L NH4SO4, 6.7 μmol/L ethylenediaminetetraacetic acid (EDTA), 10 mmol/L β-mercaptoethanol, 4.7 mmol/L MgCl2, and 1 μmol/L of each primer in a 25-μl reaction. A hot start was performed (95°C, 5 minutes), followed by the addition of 0.5 U of Taq polymerase (Life Technologies, Inc., Grand Island, NY) and 35 cycles of PCR (95°C, 30 seconds; 58°C, 30 seconds; and 72°C, 30 seconds). Reaction products were separated by electrophoresis on a 6% polyacrylamide/Tris-borate-EDTA gel, stained with ethidium bromide, and photographed. Primers were designed such that they overlay three potential methylation (ie, CpG) sites. Primers used were 5′-GGT TGT AGT GGG GTG AGT GGT-3′ and 5′-CAA AAC ATC CAT AAA CAA CAA CAC A-3′ for unmethylated reaction, and 5′-TTG TAG CGG GGT GAG CGG C-3′ and 5′-AAC GTC CAT AAA CAA CAA CGC G-3′ for the methylated reaction.
Combined Bisulfite Restriction Analysis (COBRA) and Bisulfite Sequencing
COBRA was performed essentially as described.24 Briefly, DNA from GBM cell lines was bisulfite modified and amplified by PCR using the same conditions described above for methylation-specific PCR (MSP) except, in this case, primers were designed to avoid potential methylation sites (CpGs) so that unmethylated and methylated sequences would be amplified equally. DNA from the amplification reaction was purified using a Wizard PCR Clean-Up kit (Promega, Madison, WI) and digested with 10 U of ClaI for 4 hours at 37°C. Reaction products were separated by electrophoresis on a 6% polyacrylamide/Tris-borate-EDTA gel, stained with ethidium bromide, and digitally photographed. If TMS1 is unmethylated, the 557-bp PCR product remains intact. If methylated, this band is digested to 453- and 104-bp products. Percent methylation was determined as the ratio of intensities of the intact band to that of the digested bands using Imagequant v5.2 software (Molecular Dynamics, Sunnyvale, CA). DNA from breast cancer cell lines with well-characterized methylation patterns at the TMS1 locus [MCF7 (unmethylated), T47D (∼50% methylated), and MDA MB231 (100% methylated)]15,25 were included to verify the quantitative nature of the technique. For bisulfite sequencing, the bisulfite modification and PCR reaction were performed as described above again using primers that avoid CpG sites. In this case, the resulting amplicon was subcloned using the TOPO-TA cloning kit (Invitrogen, Carlsbad, CA). Eight to twelve individual subclones were isolated per PCR reaction and sequenced by the University of Michigan Sequencing Facility. Primers used for COBRA and bisulfite sequencing were: 5′-TTG GTG TAA GTT TAG AGA TAA GT-3′ and 5′-ACC ATC TCC TAC AAA CCC ATA-3′.
Conventional and Real-Time Reverse Transcriptase-PCR
Total RNA (6 μg) was pretreated with DNase I (Life Technologies, Inc.) and reverse-transcribed using random hexamer primers and MMLV reverse transcriptase (Life Technologies, Inc.). One-thirtieth of the reverse transcription reaction (200 ng of starting RNA) was used in a PCR reaction. The PCR reaction conditions were: 67 mmol/L Tris-HCl (pH 8.8), 16.6 mmol/L NH4SO4, 6.7 μmol/L EDTA, 10 mmol/L β-mercaptoethanol, 4.7 mmol/L MgCl2, 10% dimethyl sulfoxide, and 400 nmol/L of each primer in a 25-μl reaction. A hot start was performed (5 minutes, 95°C), followed by the addition of 0.5 U of Taq polymerase (Life Technologies, Inc.) and 35 cycles of PCR (95°C, 30 seconds; 50 to 55°C, 60 seconds; and 72°C, 60 seconds). TMS1 primers (5′-TGG GCC TGC AGG AGA TG-3′ and 5′-ATT TGG TGG GAT TGC CAG-3′) were used at an annealing temperature of 50°C. β-Actin primers (5′-CCT TCC TGG GCA TGG AGT CCT G-3′ and 5′-GGA GCA ATG ATC TTG ATC TTC-3′) were used at an annealing temperature of 55°C. Reaction products were separated by electrophoresis on a 6% polyacrylamide/Tris-borate-EDTA gel, stained with ethidium bromide, and photographed.
For real-time analysis, 1 μl of a 50-fold dilution of the reverse transcriptase reaction (10 ng of starting RNA) was amplified per reaction using the iQ SYBR Green Supermix kit (Bio-Rad, Hercules, CA) and the MyIQ real-time detection system. Reaction conditions included a hot start (3 minutes, 95°C), followed by 50 cycles of 95°C, 10 seconds and 55°C, 60 seconds). Melt curve analysis was performed to ensure a single product species. Parallel reactions were performed using primers to 18S rRNA as an internal control. Starting quantities were calculated by comparison to a common standard curve of MCF10A cell cDNA that was included in each run. Primers for real-time PCR analysis were for TMS1, 5′-TCC AGC AGC CAC TCA ACG-3′ and 5′-GCA CTT TAT AGA CCA GCA-′; and for 18S, 5′-GAG GGA GCC TGA GAA ACG G-3′ and 5′-GTC GGG AGT GGG TAA TTT GC-3′.
Generation of TMS1 Antibodies
An affinity-purified rabbit polyclonal antibody was developed against the C-terminus of human TMS1/ASC. A peptide corresponding to amino acids 182 to 195 of human TMS1/ASC was synthesized by the Microchemical Core Facility of Emory University, coupled to KLH via an N-terminal cysteine, and used to immunize New Zealand White rabbits (Cocalico Biologicals, Lancaster, PA). Crude antiserum (EU107) was affinity purified by selection over a column consisting of the same peptide coupled to a solid support (Sulfo-link; Pierce, Rockford, IL) and elution with 100 mmol/L glycine, pH 2.0. Fractions were neutralized with Tris-base, dialyzed against PBS, and analyzed for specificity by Western blot analysis using a partially purified recombinant GST-TMS1 fusion protein.
Immunohistochemical Analysis of TMS1 Expression
Immunohistochemistry was performed on archived formalin-fixed and paraffin-embedded human GBM resection specimens. Tissue sections used for routine histological examination and for immunohistochemistry contained both regions that were diagnostic of GBM as well as adjacent nonneoplastic brain. For immunohistochemical studies, sections were deparaffinized and subjected to antigen retrieval by steaming (20 minutes, 80°C). Slides were then incubated at room temperature with a 1:320 dilution of affinity-purified anti-human TMS1 antibody (EU107). Negative controls included normal saline and irrelevant IgG substitution for the primary antibody. Antibodies were detected using the avidin-biotin complex method, using diaminobenzidine as the chromogen (Envision System; DAKO, Carpinteria, CA). Sections were counterstained with hematoxylin. Staining for human CD3 and CD68 was performed using a CD3 polyclonal antibody (1:80 dilution, DAKO) and the KP-1 monoclonal antibody (1:320 dilution, DAKO), respectively.
Results
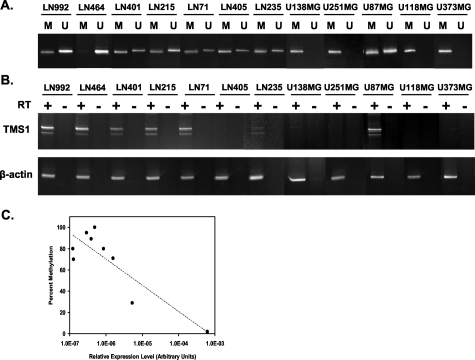
To study the potential role of TMS silencing in pathogenesis of GBM, we first established the expression and the methylation status of the TMS1 gene in normal human brain tissue. Brain tissue derived from cancer-free patients at autopsy was analyzed for the methylation of the TMS1 CpG island by MSP and for TMS1 expression by reverse transcriptase (RT)-PCR (Figure 1). TMS1 was found to be expressed in normal cerebral cortex and white matter (n = 2), brain and the TMS1 CpG island was predominately unmethylated in five of five normal brain specimens. We next examined methylation and expression of TMS1 in 22 GBM cell lines. All but one of the cell lines examined showed some degree of aberrant methylation of the TMS1 CpG island (Figure 2A). One cell line was devoid of methylation at the TMS1 locus; 13 of 22 (59%) GBM cell lines examined were partially methylated and 8 of 22 (36%) cell lines showed only methylated DNA at the TMS1 CpG island. Quantitation of the degree of methylation in a subset of GBM cell lines using a COBRA24 assay confirmed that methylation was indeed extensive in many cases (Table 1). Eleven of twenty-three cell lines examined showed reduced or absent expression of TMS1 by RT-PCR (Figure 2B). Cell lines that were predominantly or completely methylated at the TMS1 promoter expressed little or no TMS1 message, whereas cell lines that were completely unmethylated or exhibited only low levels of methylation expressed TMS1 (Table 1). Quantitative analysis of percent methylation (by COBRA assay) and TMS1 expression by real-time PCR confirmed an inverse correlation between the degree of methylation and the level of gene expression in the GBM cell lines (Table 1 and Figure 2C).
Figure 1-4259.
TMS1 methylation and expression in normal brain. A: Normal brain biopsies were obtained from five cancer-free individuals (00-23, 00-06, 99-08, 01-09, 01-08) at autopsy. DNA was isolated from both gray (G) and white (W) matter from each specimen, and was analyzed for methylation of TMS1 by methylation-specific PCR. Parallel amplification reactions were performed using primers specific to the unmethylated (U) or methylated (M) DNA. B: Total RNA isolated from the normal brain tissue (01-86, 99-98) was reverse transcribed (+RT) and amplified using primers specific for TMS1 (top) or human β-actin (bottom). Control reactions in which reverse transcriptase was omitted (−RT) were amplified under the same conditions.
Figure 2-4259.
Methylation and expression of TMS1 in glioblastoma cell lines. A: DNA from the indicated GBM cell line was bisulfite modified and analyzed for methylation of the TMS1 CpG island by methylation-specific PCR analysis. Parallel amplification reactions were performed using primers specific to the unmethylated (U) or methylated (M) DNA. B: Total RNA isolated from 20 GBM cell lines was reverse transcribed (+RT) and amplified with primers specific for the TMS1 transcript (top) or for the human β-actin transcript (bottom). Control reactions (−RT) were amplified under the same conditions for both TMS1 and human β-actin. C: DNA methylation levels were quantified using a COBRA assay and TMS1 expression by reverse transcription and real-time PCR analysis. The relative expression of TMS1 was determined by comparison to a common human cell cDNA standard included in each run. Shown are the relative levels of TMS1 expression normalized to the levels of 18S rRNA. Each data point represents a different GBM cell line. Dotted line, linear regression; R2 = 0.76.
Table 1.
Methylation and Expression of TMS1 in Human Glioblastoma Cell Lines
| Cell line | CpG island methylation* | % Methylation (COBRA)† | mRNA expression‡ |
|---|---|---|---|
| LN 464 | U | 0 | ++ |
| LN 827 | U/M | nd¶ | ++ |
| LN 71 | U/M | nd | ++ |
| LN 215 | U/M | 71 | ++ |
| LN 401 | U/M | nd | ++ |
| LN 308 | U/M | nd | ++ |
| LN 992 | U/M | 24 | ++ |
| U 87 MG | U/M | 80 | ++ |
| LN 235 | U/M | nd | +/− |
| LN 405 | U/M | nd | − |
| LN 427 | U/M | 100 | − |
| LN 428 | M | nd | +/− |
| LN 18 | M | nd | +/− |
| U 138 MG | M | 89 | − |
| LN 229 | M | nd | − |
| U 343 MG | M | 70 | − |
| U 373 MG | M | 80 | − |
| U 251 MG | M | nd | − |
| U 118 MG | M | 95 | − |
| MCF7§ | U§ | 0 | ++§ |
| MB231§ | M§ | 95 | −§ |
| T47D§ | U/M§ | 51 | +/−§ |
Methylation of the TMS1 gene was analyzed by methylation-specific PCR. U, completely unmethylated; U/M, both unmethylated and methylated copies present; M, completely methylated.
The percent methylation was determined by COBRA analysis.
Expression of TMS1 was determined by RT-PCR. Expression was scored as strong (++); weak (+/−); or undetectable (−−) after 35 cycles of PCR.
Methylation-specific PCR and expression data for the breast cancer cell lines MCF7, MDA MB231, and T47D is derived from references 15 and 25. DNA from these cell lines were used as controls for the COBRA analysis.
nd, not determined.
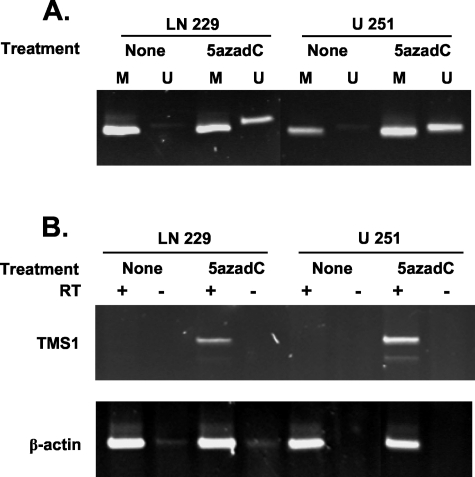
If methylation plays a role in the silencing of TMS1, demethylation of the locus should result in the reactivation of gene expression. Treatment of two GBM cell lines, U251 and LN 229, with the DNA methyltransferase inhibitor 5′-aza-2′-deoxycytidine for 48 hours resulted in the partial demethylation and re-expression of TMS1 (Figure 3). These data indicate that methylation is the only impediment to TMS1 expression and suggest that methylation plays a direct role in the repression of TMS1 in GBM cell lines.
Figure 3-4259.
Demethylation of the TMS1 CpG island restores expression in TMS1-negative glioblastoma cell lines. U 251 MG and LN 229 cells were treated (5azadC) or not treated (none) with 0.5 μmol/L 5′-aza-2′-deoxycytidine for 3 days. The samples were analyzed for methylation of the TMS1 CpG island by methylation-specific PCR (A) and for TMS1 expression by RT-PCR (B) as described in the legend to Figure 2.
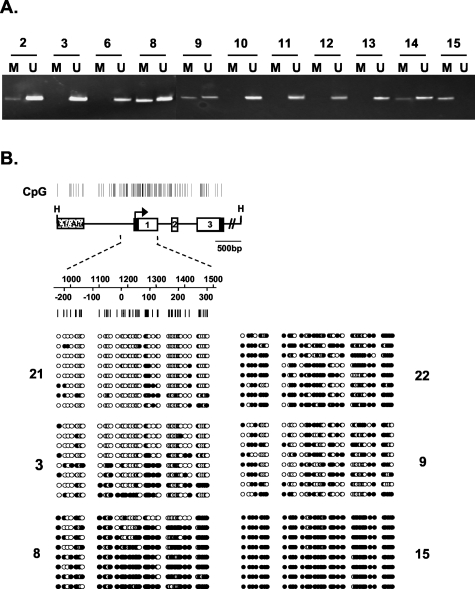
To determine whether aberrant methylation of the TMS1 in GBM cell lines reflects an epigenetic event occurring in primary GBM, we next examined 23 primary GBMs for TMS1 methylation. As mentioned above, brain tissue derived from five unrelated individuals was unmethylated at the TMS1 CpG island. In contrast, 10 of 23 primary GBMs (43%) showed aberrant methylation at TMS1 (Figure 4A). A subset of the primary GBM samples was also analyzed by bisulfite sequencing, which provides a detailed view of the methylation density across individual TMS1 alleles in the tumor cell population (Figure 4B). In general, there was good concordance between the MSP and bisulfite sequencing data in that samples that were predominantly unmethylated by MSP also showed a low density of methylation by bisulfite sequencing (eg, sample 3) and others that were predominantly methylated (eg, sample 15) were densely methylated across all alleles. This analysis also revealed the heterogeneity in methylation patterns, both with respect to individual CpGs across a single allele as well as between independent alleles, in different tumor samples (Figure 4B). It is possible that this heterogeneity reflects the mixed cellularity likely present in grossly dissected primary GBM tumor specimens. Thus, frequent and extensive DNA methylation also affects the TMS1 locus in primary GBM.
Figure 4-4259.
TMS1 CpG island methylation in primary glioblastomas. A: Methylation-specific PCR analysis. DNA was isolated from GBM specimens frozen immediately after surgical resection. Samples were reviewed by a single neuropathologist for histological confirmation of GBM and for tissue integrity. The DNA was bisulfite modified and analyzed for methylation of the TMS1 CpG island by methylation-specific PCR as described in the legend to Figure 2. Shown are 11 representative samples for which methylation data were collected. Of a total of 23 primary GBM specimens examined, 10 of 23 (43%) exhibited aberrant methylation of TMS1. B: Fine methylation mapping of the TMS1 locus in primary GBMs. Top: Genomic map of the TMS1 locus. Open boxes, exons. Top numbers indicate nucleotide position relative to the upstream HindIII site; bottom numbers, the position relative to transcription start (zero). Bottom: Bisulfite sequence analysis was used to analyze 53 CpG sites in the TMS1 promoter. Bisulfite-modified DNA from primary GBM were amplified using primers that avoid CpG sites. The amplification product was then subcloned and sequenced. Each row represents an individual subclone. Open circles, unmethylated CpGs; filled circles, methylated CpGs.
There did not appear to be a significant relationship between TMS1 methylation status and patient age (mean unmethylated, 53; methylated, 59; P = 0.23) or sex (unmethylated, 7 males and 6 females; methylated, 5 males and 5 females). Although only a limited dataset, there was a trend toward increased overall survival time among patients with unmethylated tumors (mean, 12 months; median, 14 months) compared to those with methylated tumors (mean, 9.5 months; median, 8 months); however the relationship did not reach statistical significance (Kaplan-Meier survival analysis comparing unmethylated versus methylated, P = 0.32).
The finding that TMS1 was aberrantly methylated in a significant proportion of primary GBM prompted us to examine the expression of TMS1 protein in these tumors. We developed an affinity-purified antibody directed against human TMS1 for use in immunohistochemical analysis of fixed tissues. Matched paraffin-embedded tissue was available from 18 of the 23 primary GBM tumors previously analyzed for TMS1 methylation status, as well as normal brain tissue collected at autopsy from cancer-free individuals. In normal brain tissue, TMS1 exhibited moderate cytoplasmic staining extending into the processes of individual astrocytes in the cortex (Figure 5) and white matter (not shown). In contrast, neurons and oligodendroglial cells were uniformly negative for TMS1 in all regions of the brain, as was the neuropil. Absence of TMS1 staining in neurons was confirmed by staining of serial sections for the neuron-specific nuclear antigen, Neu-n (not shown). Endothelial cells lining the microvasculature were also negative for TMS1 expression, both in normal and tumor tissue (Figure 6, A and B).
Figure 5-4259.
TMS1 expression in normal brain. Paraffin-embedded tissues were subjected to immunohistochemical analysis using the EU107 anti-human TMS1 antibody (1:360). Immunocomplexes were detected by the avidin-biotin complex method using diaminobenzidine as the chromogen. Sections were counterstained with hematoxylin. Shown is nonneoplastic tissue adjacent to surgically resected glioblastoma from the frontal lobe cortex (A) and temporal lobe cortex (B). Note the cytoplasmic and nuclear staining of astrocytes (arrow), but lack of staining of large cortical neurons (arrowheads), background neuropil, and endothelial cells.
Figure 6-4259.
TMS1 expression in glioblastomas. Paraffin-embedded tissues were subjected to immunohistochemical analysis using the EU107 anti-human TMS1 antibody (1:360). Immunocomplexes were detected by the avidin-biotin complex method using diaminobenzidine as the chromogen. Sections were counterstained with hematoxylin. Shown are three representative primary glioblastomas that were unmethylated at TMS1 (A–C) and three that were methylated at TMS1 (D–F). Note the strong TMS1 staining of all tumor cells (A, B) but absence of staining of microvasculature (A and B, arrows). C: An example of a focally positive tumor. Note absence of TMS1 staining in methylated tumors (D–F) but the retention of TMS1 expression in infiltrating leukocytes (E, arrow) and entrapped normal astrocytes (F, arrow) within TMS1-negative tumor tissue.
In contrast to normal astrocytes, primary GBMs exhibited a range of TMS1 expression levels. Tumors that were unmethylated expressed either high levels of TMS1 throughout the tumor sample (Figure 6, A and B) or were more focally positive wherein a fraction of the cells in the tumor were highly positive and the remaining cells showed little or no staining (Figure 6C). In contrast, tumors that were methylated at TMS1 tended to exhibit reduced or absent expression of TMS1 (Figure 6; D to F). Notably, normal components, such as infiltrating mononuclear cells (Figure 6E) or entrapped normal astrocytes (Figure 5F) retained high TMS1 expression even within the context of a methylated, TMS1-negative tumor.
In general, there was a correlation between the overall degree of TMS1 expression and methylation of the TMS1 promoter in that none of the tumors that were methylated at TMS1 (n = 8) exhibited more than ∼25% of tumor cells that stained positive (Table 2). Conversely, the majority (five of eight) of the unmethylated tumors showed medium to high staining in >20% of cells in the tumors (Table 2). For cases in which both immunohistochemistry and bisulfite-sequencing analyses were available, the overall pattern of expression in the tumor tissue reflected the allelic methylation patterns. For example, tumor sample 21 expressed high levels of TMS1 in most tumor cells (Figure 6A) and showed a low density of methylation (Figure 4B, sample 21). This was also true of sample 3 (compare Figure 6C to Figure 4B, sample 3). Likewise, tumor sample 22 showed no expression of TMS1 and was methylated on nearly every CpG on all alleles (compare Figure 6D with Figure 4B, sample 22). Interestingly, the heterogeneity in expression pattern observed in some of the tumors was also reflected in the methylation pattern (compare Figure 4B, sample 9, with Figure 6, E and F). There were, however, a few examples of tumors that were unmethylated (by MSP) that exhibited low or absent TMS1 expression. Taken together, these data suggest that whereas methylation is associated with the absence of gene expression in many GBM, there may be additional mechanisms that contribute to lack of TMS1 expression in some tumors.
Table 2.
Summary of TMS1 Methylation and Protein Expression in Primary Glioblastomas*
| GBM sample number | TMS1 methylation* | TMS1 expression† |
Cellularity (% of sample composed of tumor cells) | ||
|---|---|---|---|---|---|
| TMS1-positive tumor cells (%) | Nuclear | Cytoplasmic | |||
| 6 | U | 60% | 2+ | 2+ | 80% |
| 21 | U | 50% | 2+ | 2+ | 50% |
| 3 | U | 40% | 1+ | 1+ | 40% |
| 11 | U | 20% | 2+ | 2+ | 75% |
| 19 | U | 20% | 2+ | 2+ | 30% |
| 25 | U | 5% | 1+ | 1+ | 25% |
| 12 | U | 0% | 0 | 0 | 60% |
| 10 | U | 0% | 0 | 0 | 50% |
| 4 | M | 25% | 3+ | 2+ | 90% |
| 8 | M | 20% | 1+ | 1+ | 80% |
| 14 | M | 20% | 2+ | 2+ | 90% |
| 9 | M | 10% | 1+ | 1+ | 90% |
| 15 | M | 10% | 2+ | 1+ | 60% |
| 2 | M | 15% | 1+ | 1+ | 80% |
| 20 | M | 1% | 1+ | 1+ | 80% |
| 22 | M | 0% | 0 | 0 | 60% |
Methylation was determined by MSP on DNA extracted from fresh-frozen resected tumor tissue. U, Unmethylated; M, methylated.
TMS1 expression was determined on matched-fixed tumor specimens by immunohistochemistry, an affinity-purified antipeptide antibody directed against human TMS1 (EU107). Percentages represent the average percent of neoplastic cells staining positive for TMS1. The degree of nuclear and cytoplasmic staining in positive cells was scored on a scale from 0 to 3+. The methylation status of the various tumors was blinded to the pathologist during histochemical analysis.
Considering the mixed cellularity of GBMs (see below), it is not surprising that GBMs exhibit only partial methylation of TMS1 because any normal components (vascular endothelial cells, inflammatory cells, and so forth) would be expected to be unmethylated. Remarkably, there was one tumor that exhibited a densely methylated pattern at TMS1, as determined by both MSP and bisulfite sequencing (sample 15, Figure 3 and Figure 4B). Interestingly, this sample was derived from a GBM that had recurred from an anaplastic astrocytoma (AA) (World Health Organization grade III) diagnosed and resected 1 year earlier and thus represented tumor cells that persist after the patient had been treated with chemotherapy and radiation. Although we were unable to obtain DNA from the AA sample for methylation analysis, fixed tissue was available for immunohistochemical analysis. Side-by-side comparison of TMS1 expression in the AA and GBM from the same patient showed that whereas a significant fraction of the tumor cells in the AA retained TMS1 expression (∼25%) there was a decrease in the percentage of TMS1-positive cells in the GBM (∼10%) (Figure 7). This decrease in the TMS1-positive fraction appeared to result from an increase in the overall cellularity of the tumor without a corresponding increase in the TMS1-positive cells. Although only a single case, these data are consistent with a selective outgrowth of TMS1-negative cells during glioma progression.
Figure 7-4259.
TMS1 expression during glioma progression. Surgically resected anaplastic astrocytoma (World Health Organization grade III) (A) and a recurrent glioblastoma (World Health Organization grade IV) (B) from the same individual. Paraffin-embedded tissues were stained with EU107 anti-human TMS1 antibody (1:360) and detected by the avidin-biotin complex method using diaminobenzidine as the chromogen. Inset: Estimated percent TMS1-positive tumor cells per field. Note the increased density of TMS1-negative tumor cells in B relative to A.
In several GBM cases, we noted prominent TMS1 staining in a perivascular distribution (Figure 8; A to C). This staining appeared to involve neoplastic cells surrounding the vessels rather than the cells lining the vessels because vascular endothelial cells were uniformly negative both in normal brain as well as in the microvasculature of TMS1-positive tumors (Figure 6B). In contrast, pseudopalisading cells, which typically surround areas of necrosis and are thought to be associated with hypoxic regions, were uniformly TMS1-negative (Figure 8C). Perivascular cuffing resulting from infiltrating lymphocytes is common in glioblastomas.26 Serial sections were, therefore, stained with antibodies to TMS1, CD3 (B and T lymphocytes), and CD68 (macrophages) to distinguish inflammatory from neoplastic astrocytes (Figure 8; D to F). TMS-positive perivascular cells were negative for CD3, indicating that these cells were not lymphocytes (Figure 8E). CD68 staining highlighted a population of perivascular macrophages that was more tightly associated with the vessel, and overlapped a subset of the TMS1-positive cells (Figure 8F). However, the distribution of TMS1-positive cells extended further away from vessel than CD68. These more distant cells had a morphology consistent with neoplastic astrocytes. These data indicate that there is a subset of perivascular cells that have up-regulated TMS1 expression relative to the surrounding tumor, possibly in response to a difference in microenvironmental conditions such as local oxygen tension or cytokine concentration.
Figure 8-4259.
Up-regulation of TMS1 in perivascular cells in glioblastomas. A–C: Representative glioblastoma sections stained with anti-human TMS1 antibody (1:360). Note the dense TMS1 staining in a perivascular distribution (arrows) and absence of staining in pseudopalisading cells (arrowhead) surrounding an area of necrosis (asterisk). D–F: Serial sections of a single glioblastoma stained with antibodies to TMS1 (D), CD68 (E), and CD3 (F).
Discussion
TMS1 (also known as ASC) is a bipartite intracellular signaling molecule that has been implicated in the regulation of apoptosis, nuclear factor-κB activation, and the maturation of proinflammatory cytokines. Previous work from our laboratory has shown that TMS1 is a target of epigenetic silencing in human breast cancer cell lines and primary breast tumors.15 Subsequent work has confirmed a role for aberrant methylation of TMS1/ASC in breast cancers, with the frequency of this event ranging from 10 to 40% in different studies.27,28 Here we show that TMS1 is also subject to methylation-associated silencing in human GBMs. Whereas TMS1 was unmethylated and expressed in normal brain tissue, the vast majority of GBM cell lines and 43% of primary GBM tumors exhibited aberrant hypermethylation of a CpG island in the 5′ end of the TMS1 gene. In GBM cell lines, methylation of TMS1 was associated with decreased or absent gene expression, which could be reversed by treatment with the demethylating agent 5-aza-2′deoxycytidine. In primary tumors, TMS1 methylation was associated with reduced TMS1 expression. Taken together, these data implicate methylation-associated silencing in the pathogenesis of human gliomas.
Few studies have addressed the relationship between TMS1 methylation and expression in primary tissues. We found that TMS1 is expressed in normal brain tissue, and is confined to astrocytes, the cells from which gliomas arise. TMS1 was distributed both in the nucleus and cytoplasm, as has been observed in other normal tissues.29 Considering that astrocytes represent a relatively small proportion of the overall cellularity of normal brain tissue, it is difficult to directly compare the levels of expression of TMS1 between normal brain and GBM, where high density of neoplastic astrocytes is characteristic. Nevertheless, among GBM cases, those exhibiting aberrant methylation of TMS1 generally expressed reduced or absent expression of TMS1 relative to unmethylated cases. However, TMS1 expression levels and patterns were more variable among the unmethylated tumors. Whereas several unmethylated tumors expressed high levels of TMS1 throughout the tumor, there were others that expressed high levels of TMS1 in only a subset of tumor cells (focal positive, Figure 6C), or expressed uniformly low levels of TMS1 (Table 2). Thus, although methylation of TMS1 was frequently associated with down-regulation of TMS1, lack of methylation at TMS1 is not always associated with expression. Guan and colleagues30 found a similar relationship between TMS1 methylation and gene expression in primary melanomas. There are several possible explanations for these results. First, although the unmethylated status of the TMS1 promoter may be permissive for transcription, the absolute levels of TMS1 expression in tumors with an unmethylated TMS1 might depend on the degree of activation of particular transcription factors and the local microenvironment. TMS1 is known to be up-regulated in some cells in response to cytokines such as tumor necrosis factor-α and interleukin (IL)-1β.20,31 Indeed, the prominent staining of perivascular cells that we observed in some tumors could be explained by up-regulation of TMS1 in response to high local concentrations of cytokines or oxygen tension.
Alternatively, there may be mechanisms in addition to CpG island methylation that contribute to TMS1 silencing in GBM. At present, the stepwise sequence of events in which genes progress from an unmethylated and active state to a methylated and inactive one in human tumors is not known. One model suggests that the aberrant methylation of DNA leads to binding of methyl CpG-binding proteins and the recruitment of co-repressor complexes containing chromatin-remodeling factors and histone-modifying enzymes, resulting in local changes in chromatin conformation and gene silencing.9 There is also evidence to suggest that in some cases gene silencing and changes at the chromatin level precede DNA methylation.32,33 This latter mechanism might explain cases in which absence of expression is observed in unmethylated tumors. Our detailed analysis of TMS1 methylation in breast tumors has shown that methylation of individual chromosomal copies of TMS1 tends to occur in an all-or-none manner suggesting that at any given time, silencing of TMS1 only affects a subset of cells in the tumor,25 consistent with the observed pattern of interspersed silent and expressing cells (eg, Figure 6). In brain tumors, however, the methylation pattern appeared much more heterogeneous and varied in density across individual alleles in the same sample. This could point to differences in the mechanism(s) that underlie TMS1 silencing between brain and breast tumors or to a difference in the regulation of TMS1 expression in breast epithelium versus astrocytes. Although the precise timing of methylation versus chromatin alterations remains to be determined, once established, methylation appears to play a primary role in the maintenance of gene repression as TMS1 expression is restored by treatment with the methyltransferase inhibitor 5-aza-2′deoxycytidine (this study),34 but is unaffected by treatment with the histone deacetlyase inhibitor, TSA.34
Considering the mixed cellularity of GBMs, it is not surprising that primary GBM samples exhibit only partial methylation of TMS1 because any normal components (vascular endothelial cells, inflammatory cells, and so forth) are expected to be unmethylated. Interestingly, whereas most of the samples in this study were derived from GBMs at first diagnosis, the only fully methylated sample was derived from a GBM that had recurred from an anaplastic astrocytoma diagnosed 1 year earlier, and represented tumor cells that persisted after the patient had failed chemotherapy and radiation treatment. Considering the potential role of TMS1 as a positive mediator of apoptosis, it is tempting to speculate that the fully methylated status of this GBM resulted from the expansion of TMS1-negative (methylated) tumor cells that were selected for because they are resistant to chemotherapy- or radiation-induced cell death. Although the unavailability of DNA from the anaplastic astrocytoma precluded a direct test of its methylation status, analysis of fixed tissue showed that whereas TMS1 was expressed at moderate levels in a subset of cells in the anaplastic astrocytoma, there were far fewer TMS1-positive cells as a percentage of total tumor cells in the recurring glioblastoma (Figure 8). Although only a single case, these data show that the progression from World Health Organization grade III to grade IV glioma can be accompanied by the selective outgrowth of TMS1-negative cells.
In addition to breast tumors and GBMs, recent studies have shown that TMS1/ASC undergoes frequent methylation-associated silencing in lung cancers,28 gastric carcinomas,27 melanomas,30 and colorectal carcinomas.35 One question that remains is how loss of TMS1 contributes to tumorigenesis. TMS1 is composed of a pyrin domain and a CARD domain; two protein interaction surfaces found in proteins that function in the regulation of apoptosis and inflammation. Overexpression of TMS1 promotes apoptosis and inhibits the growth of breast cancer cells,15 whereas enforced decreases in TMS1 protect against drug-induced apoptosis16 and promote the activation of nuclear factor-κB-dependent survival signals.21 Recent studies indicate that TMS1/ASC acts as an adaptor molecule that mediates the clustering and activation of caspase-1 (ICE) and the subsequent maturation of the proinflammatory cytokines IL-1β and IL-18.18–20 These cytokines promote cell-mediated immune responses and are capable of generating anti-tumor responses. For example, IL-18 has been shown to stimulate macrophages to elicit a potent cytotoxic response against glioma cells.36,37 The cytotoxic activity was attributed to increased production of interferon-γ and nitrous oxide.36 Astrocytes/glial cells express caspase-1, IL-18, IL-1β,38,39 and as shown here, TMS1, suggesting that these cells have the potential to secrete the mature cytokines. Epigenetic silencing of TMS1 may thus contribute to the pathogenesis of glioblastomas through decreased production of inflammatory cytokines, allowing tumors to evade the local host immune response. Indeed, elevated IL-1β levels are associated with prolonged survival in GBM patients.40 Although our dataset was limited, the trend toward increased survival time among patients harboring tumors with unmethylated TMS1 is also consistent with this idea.
Footnotes
Address reprint requests to Paula M. Vertino, Ph.D., Winship Cancer Institute, Emory University School of Medicine, 1365-C Clifton Rd., NE, Atlanta, GA 30322. E-mail: pvertin@emory.edu.
Supported by the National Institutes of Health (grants CA077337 to P.M.V., CA86335 to E.V.M., and NS042934 to D.J.B.).
References
- Fine HA. The basis for current treatment recommendations for malignant gliomas. J Neurooncol. 1994;20:111–120. doi: 10.1007/BF01052722. [DOI] [PubMed] [Google Scholar]
- Kleihues P, Burger PC, Scheithauer BW. The new WHO classification of brain tumours. Brain Pathol. 1993;3:255–268. doi: 10.1111/j.1750-3639.1993.tb00752.x. [DOI] [PubMed] [Google Scholar]
- Daumas-Duport C, Scheithauer B, O’Fallon J, Kelly P. Grading of astrocytomas A simple and reproducible method. Cancer. 1988;62:2152–2165. doi: 10.1002/1097-0142(19881115)62:10<2152::aid-cncr2820621015>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Hunter SB, Brat DJ, Olson JJ, Von Deimling A, Zhou W, Van Meir EG. Alterations in molecular pathways of diffusely infiltrating glial neoplasms: application to tumor classification and anti-tumor therapy. Int J Oncol. 2003;23:857–869. [PubMed] [Google Scholar]
- Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16:168–174. doi: 10.1016/s0168-9525(99)01971-x. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Costello JF, Plass C. Methylation matters. J Med Genet. 2001;38:285–303. doi: 10.1136/jmg.38.5.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bird AP, Wolffe AP. Methylation-induced repression—belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- Costello JF. DNA methylation in brain development and gliomagenesis. Front Biosci. 2003;8:s175–s184. doi: 10.2741/1027. [DOI] [PubMed] [Google Scholar]
- Costello JF, Plass C, Cavenee WK. Aberrant methylation of genes in low-grade astrocytomas. Brain Tumor Pathol. 2000;17:49–56. doi: 10.1007/BF02482735. [DOI] [PubMed] [Google Scholar]
- Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- Balana C, Ramirez JL, Taron M, Roussos Y, Ariza A, Ballester R, Sarries C, Mendez P, Sanchez JJ, Rosell R. O6-methyl-guanine-DNA methyltransferase methylation in serum and tumor DNA predicts response to 1,3-bis(2-chloroethyl)-1-nitrosourea but not to temozolamide plus cisplatin in glioblastoma multiforme. Clin Cancer Res. 2003;9:1461–1468. [PubMed] [Google Scholar]
- Komine C, Watanabe T, Katayama Y, Yoshino A, Yokoyama T, Fukushima T. Promoter hypermethylation of the DNA repair gene O6-methylguanine-DNA methyltransferase is an independent predictor of shortened progression free survival in patients with low-grade diffuse astrocytomas. Brain Pathol. 2003;13:176–184. doi: 10.1111/j.1750-3639.2003.tb00017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway KE, McConnell BB, Bowring CE, Donald CD, Warren ST, Vertino PM. TMS1, a novel proapoptotic caspase recruitment domain protein, is a target of methylation-induced gene silencing in human breast cancers. Cancer Res. 2000;60:6236–6242. [PubMed] [Google Scholar]
- Masumoto J, Taniguchi S, Ayukawa K, Sarvotham H, Kishino T, Niikawa N, Hidaka E, Katsuyama T, Higuchi T, Sagara J. ASC, a novel 22-kDa protein, aggregates during apoptosis of human promyelocytic leukemia HL-60 cells. J Biol Chem. 1999;274:33835–33838. doi: 10.1074/jbc.274.48.33835. [DOI] [PubMed] [Google Scholar]
- McConnell BB, Vertino PM. TMS1/ASC: the cancer connection. Apoptosis. 2004;9:5–25. doi: 10.1023/B:APPT.0000012117.32430.0c. [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Poyet JL, Razmara M, Datta P, Zhang Z, Alnemri ES. The PYRIN-CARD protein ASC is an activating adaptor for caspase-1. J Biol Chem. 2002;277:21119–21122. doi: 10.1074/jbc.C200179200. [DOI] [PubMed] [Google Scholar]
- Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- Stehlik C, Lee SH, Dorfleutner A, Stassinopoulos A, Sagara J, Reed JC. Apoptosis-associated speck-like protein containing a caspase recruitment domain is a regulator of procaspase-1 activation. J Immunol. 2003;171:6154–6163. doi: 10.4049/jimmunol.171.11.6154. [DOI] [PubMed] [Google Scholar]
- Stehlik C, Fiorentino L, Dorfleutner A, Bruey JM, Ariza EM, Sagara J, Reed JC. The PAAD/PYRIN-family protein ASC is a dual regulator of a conserved step in nuclear factor kappaB activation pathways. J Exp Med. 2002;196:1605–1615. doi: 10.1084/jem.20021552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N, Maier D, Merlo A, Tada M, Sawamura Y, Diserens AC, Van Meir EG. Frequent co-alterations of TP53, p16/CDKN2A, p14ARF, PTEN tumor suppressor genes in human glioma cell lines. Brain Pathol. 1999;9:469–479. doi: 10.1111/j.1750-3639.1999.tb00536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JJ, Stimson-Crider KM, Vertino PM. Effects of methylation on expression of TMS1/ASC in human breast cancer cells. Oncogene. 2003;22:3475–3488. doi: 10.1038/sj.onc.1206430. [DOI] [PubMed] [Google Scholar]
- Kleihues P, Burger PC, Collins VP, Newcomb EW, Ohgaki H, Cavenee WK. Glioblastoma. Kleihues P, Cavenee WK, editors. International Agency for Cancer Res,; Lyon: Pathology and Genetics: Tumours of the Nervous System. 1997:pp 29–39. [Google Scholar]
- Moriai R, Tsuji N, Kobayashi D, Yagihashi A, Namiki Y, Takahashi H, Watanabe N. A proapoptotic caspase recruitment domain protein gene, TMS1, is hypermethylated in human breast and gastric cancers. Anticancer Res. 2002;22:4163–4168. [PubMed] [Google Scholar]
- Virmani A, Rathi A, Sugio K, Sathyanarayana UG, Toyooka S, Kischel FC, Tonk V, Padar A, Takahashi T, Roth JA, Euhus DM, Minna JD, Gazdar AF. Aberrant methylation of TMS1 in small cell, non small cell lung cancer and breast cancer. Int J Cancer. 2003;106:198–204. doi: 10.1002/ijc.11206. [DOI] [PubMed] [Google Scholar]
- Masumoto J, Taniguchi S, Nakayama J, Shiohara M, Hidaka E, Katsuyama T, Murase S, Sagara J. Expression of apoptosis-associated speck-like protein containing a caspase recruitment domain, a pyrin N-terminal homology domain-containing protein, in normal human tissues. J Histochem Cytochem. 2001;49:1269–1275. doi: 10.1177/002215540104901009. [DOI] [PubMed] [Google Scholar]
- Guan X, Sagara J, Yokoyama T, Koganehira Y, Oguchi M, Saida T, Taniguchi S. ASC/TMS1, a caspase-1 activating adaptor, is downregulated by aberrant methylation in human melanoma. Int J Cancer. 2003;107:202–208. doi: 10.1002/ijc.11376. [DOI] [PubMed] [Google Scholar]
- Shiohara M, Taniguchi S, Masumoto J, Yasui K, Koike K, Komiyama A, Sagara J. ASC, which is composed of a PYD and a CARD, is up-regulated by inflammation and apoptosis in human neutrophils. Biochem Biophys Res Commun. 2002;293:1314–1318. doi: 10.1016/S0006-291X(02)00384-4. [DOI] [PubMed] [Google Scholar]
- Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- Bachman KE, Park BH, Rhee I, Rajagopalan H, Herman JG, Baylin SB, Kinzler KW, Vogelstein B. Histone modifications and silencing prior to DNA methylation of a tumor suppressor gene. Cancer Cell. 2003;3:89–95. doi: 10.1016/s1535-6108(02)00234-9. [DOI] [PubMed] [Google Scholar]
- Stimson KM, Vertino PM. Methylation-mediated silencing of TMS1/ASC is accompanied by histone hypoacetylation and CpG island-localized changes in chromatin architecture. J Biol Chem. 2002;277:4951–4958. doi: 10.1074/jbc.M109809200. [DOI] [PubMed] [Google Scholar]
- Yokoyama T, Sagara J, Guan X, Masumoto J, Takeoka M, Komiyama Y, Miyata K, Higuchi K, Taniguchi S. Methylation of ASC/TMS1, a proapoptotic gene responsible for activating procaspase-1, in human colorectal cancer. Cancer Lett. 2003;202:101–108. doi: 10.1016/j.canlet.2003.08.027. [DOI] [PubMed] [Google Scholar]
- Kito T, Kuroda E, Yokota A, Yamashita U. Cytotoxicity in glioma cells due to interleukin-12 and interleukin-18-stimulated macrophages mediated by interferon-gamma-regulated nitric oxide. J Neurosurg. 2003;98:385–392. doi: 10.3171/jns.2003.98.2.0385. [DOI] [PubMed] [Google Scholar]
- Kikuchi T, Akasaki Y, Joki T, Abe T, Kurimoto M, Ohno T. Antitumor activity of interleukin-18 on mouse glioma cells. J Immunother. 2000;23:184–189. doi: 10.1097/00002371-200003000-00002. [DOI] [PubMed] [Google Scholar]
- Conti B, Park LC, Calingasan NY, Kim Y, Kim H, Bae Y, Gibson GE, Joh TH. Cultures of astrocytes and microglia express interleukin 18. Brain Res Mol Brain Res. 1999;67:46–52. doi: 10.1016/s0169-328x(99)00034-0. [DOI] [PubMed] [Google Scholar]
- Chauvet N, Palin K, Verrier D, Poole S, Dantzer R, Lestage J. Rat microglial cells secrete predominantly the precursor of interleukin-1beta in response to lipopolysaccharide. Eur J Neurosci. 2001;14:609–617. doi: 10.1046/j.0953-816x.2001.01686.x. [DOI] [PubMed] [Google Scholar]
- Cuny E, Loiseau H, Penchet G, Ellie E, Arsaut J, Vital A, Vincendeau P, Demotes-Mainard J. Association of elevated glial expression of interleukin-1beta with improved survival in patients with glioblastomas multiforme. J Neurosurg. 2002;96:294–301. doi: 10.3171/jns.2002.96.2.0294. [DOI] [PubMed] [Google Scholar]