Abstract
Replication licensing ensures once per cell cycle replication and is essential for genome stability. Overexpression of two key licensing factors, Cdc6 and Cdt1, leads to overreplication and chromosomal instability (CIN) in lower eukaryotes and recently in human cell lines. In this report, we analyzed hCdt1, hCdc6, and hGeminin, the hCdt1 inhibitor expression, in a series of non-small-cell lung carcinomas, and investigated for putative relations with G1/S phase regulators, tumor kinetics, and ploidy. This is the first study of these fundamental licensing elements in primary human lung carcinomas. We herein demonstrate elevated levels (more than fourfold) of hCdt1 and hCdc6 in 43% and 50% of neoplasms, respectively, whereas aberrant expression of hGeminin was observed in 49% of cases (underexpression, 12%; overexpression, 37%). hCdt1 expression positively correlated with hCdc6 and E2F-1 levels (P = 0.001 and P = 0.048, respectively). Supportive of the observed link between E2F-1 and hCdt1, we provide evidence that E2F-1 up-regulates the hCdt1 promoter in cultured mammalian cells. Interestingly, hGeminin overexpression was statistically related to increased hCdt1 levels (P = 0.025). Regarding the kinetic and ploidy status of hCdt1- and/or hCdc6-overexpressing tumors, p53-mutant cases exhibited significantlyincreased tumor growth values (Growth Index; GI) and aneuploidy/CIN compared to those bearing intact p53 (P = 0.008 for GI, P = 0.001 for CIN). The significance of these results was underscored by the fact that the latter parameters were independent of p53 within the hCdt1-hCdc6 normally expressing cases. Cumulatively, the above suggest a synergistic effect between hCdt1-hCdc6 overexpression and mutant-p53 over tumor growth and CIN in non-small-cell lung carcinomas.
Cell division relies on an ordered sequence of molecular events, which ensure the accurate duplication of the genome (during S phase) and its equal distribution to daughter cells (during M phase). Accurate execution of cellular activities at each cell cycle phase is monitored by complex protein networks at checkpoints, which block the passage into the next phase when cells have failed to complete any particular process.1 Among the vital controls acting over the cell cycle are those ensuring once per cell cycle replication; impairment of these regulatory systems may trigger genomic instability,2,3 a common feature of cancer cells. Although our understanding of the pathways governing G1-to-S phase transition and its misregulation in tumor mammalian cells has rapidly advanced throughout the past few years, the mechanisms controlling once per cell cycle replication in malignant cells versus normal cells are less well defined.4
In normal cells, precise DNA duplication is accomplished via licensing of each replication origin once per cell cycle.5 Licensing is essential for initiation of DNA replication at each origin, and represents a highly conserved mechanism among eukaryotes.6–9 Briefly, during late mitosis and early G1, two key elements, Cdc6 (Cdc18 in fission yeast) and Cdt1, are assembled at origins of replication, through interactions with the origin recognition complex (ORC) and mediate the loading of MCM (minichromosome maintenance) proteins 2 to 7 onto chromatin.10–14 This multiprotein-complex corresponds to the prereplicative complex (pre-RC) and characterizes completion of the licensing process. At the G1 to S transition, the pre-RC is activated, and DNA replication is initiated. Activation of the licensed origins requires the action of two protein kinases, cyclin-dependent kinase (CDK) and Dbf4-dependent kinase (DDK), which are believed to modify the pre-RC leading to the unwinding of replication origins. Simultaneously, the origins are converted to an unlicensed state by disassembling the pre-RC leaving only ORC bound to chromatin, which corresponds to the post-RC state. Thus, rereplication of the origins is impeded during the same S phase and in G2, and is only allowed again once cells have completed the following mitosis.
A crucial feature of the step leading to the unlicensed state is the inactivation of the two replication licensing factors Cdc6 and Cdt1. Ectopic expression of Cdc6 and Cdt1 leads to overreplication of the genome and genomic instability in both lower and higher eukaryotes.12,15–18 The importance of the controls acting over Cdt1 is underscored by the finding that NIH3T3 fibroblasts overexpressing Cdt1 form tumors in mice.19 In human cells, the exact control mechanisms acting over hCdt1 and hCdc6 are not yet fully clarified. There is evidence that CDKs and proteosome-dependent degradation is involved in the control of both regulators throughout the cell cycle.9 hCdc6 expression levels remain stable during S, G2, and M phases, whereas phosphorylation by CDKs after the initiation of DNA replication has been proposed to lead to the export of a fraction of the protein to the cytoplasm.20–24 hCdc6 is degraded through APC (anaphase promoting complex)-dependent proteolysis toward the end of M phase.25 On the other hand, hCdt1 protein accumulates specifically during G1.26,27 After the onset of S phase, hCdt1 is subjected to proteolysis,27 probably mediated by the SCF (Skp1-Cdc53-Fbox)28 and regulated by CDK-dependent phosphorylation. hCdt1 is additionally controlled through a molecular inhibitor, hGeminin,29 which can interact with hCdt1 and prevent the recruitment of MCM 2–7 onto origins.26,30 hGeminin is specifically expressed during S, G2, and early M phases, thereby inhibiting refiring of the origins.26,27,29
Recently, Vaziri and colleagues18 demonstrated that, in human cell lines, forced expression of hCdt1 and/or hCdc6, in a p53 mutant background, results in rereplication and favors genomic instability. Moreover, they suggested that rereplication itself activates p53 via the ATM/ATR-Chk2 pathway. Active p53 was capable of inhibiting further rereplication possibly via induction of p21WAF1/CIP1.18
In a series of previous studies, in non-small-cell lung carcinomas (NSCLCs), we set a number of questions regarding the relationship between alterations in nodal elements of the G1 phase network with the kinetic and ploidy characteristics of the tumors.31–36 One of our main findings was that overexpression of the transcription factor E2F-1, in cases harboring defects of the pRb-p53-MDM2 network, correlated with increased tumor growth and aneuploidy.35 Prompted by the observation that the E2F family of activators orchestrate cell cycle progression through G1 by stimulating a range of molecules, including members of the replication licensing machinery, such as Cdc6, Orc1, and MCM proteins,37 we extended our investigation to components of the licensing apparatus in a series of 75 NSCLCs and examined the following: 1) the expression status of the two major licensing components, hCdt1 and hCdc6, as well as of hGeminin, an established inhibitor of hCdt1 activity; 2) the putative relationship between E2F-1 expression and hCdt1/hCdc6; and 3) the potential associations between these licensing elements, tumor kinetics, and ploidy parameters. Furthermore, the results of the above analysis were stratified according to p53 status. To the authors’ awareness, this is the first comparative study of the replication licensing components hCdt1, hCdc6, and hGeminin in human malignancy.
Materials and Methods
Experiments Using Tissue Samples
Tumor Specimens
Frozen and formalin-fixed paraffin-embedded biopsy material from a total of 75 NSCLCs (32 adenocarcinomas, 34 squamous cell carcinomas, and 9 large undifferentiated carcinomas), resected from patients with a mean age of 63 years (range, 45 to 79 years), as well as adjacent healthy lung tissue, were analyzed. Forty-three cancer cases were included in a series of previous studies by our group.31–36 Tumors were classified according to World Health Organization criteria and TNM staging system, and consisted of 30 stage I, 27 stage II, and 18 stage III tumors. During the patients’ follow-up (mean, 24 months), 28 deaths were recorded. All cases were diagnosed and treated at the Sotiria General Hospital, Athens, Greece. Patients had received no chemotherapy, radiotherapy, or immunotherapy before surgery.
Immunohistochemistry (IHC)-Immunofluorescence
Antibodies: The following antibodies (Abs) were used for immunohistochemical analysis: anti-hCdc6 (180.2) (class: IgG1 mouse monoclonal; epitope: full-length Cdc6, human origin; Santa Cruz, Bioanalytica, Athens, Greece); anti-E2F-1 (KH95) (class: IgG2a mouse monoclonal; epitope: Rb-binding domain of E2F-1 p60, human origin; Santa Cruz); anti-p53 (DO7) (class: IgG2b, mouse monoclonal; epitope: residues 1 to 45 of p53; DAKO, Kalifronas, Athens, Greece); anti-Ki-67 (MIB-1) (class: IgG1 mouse monoclonal; epitope: Ki-67 nuclear antigen; DAKO); anti-phospho-pRb (class: IgG rabbit polyclonal; epitope: phospho-pRb (Ser-795), human origin; Santa Cruz); anti-total-pRb (LM95.1) (class: IgG2b, mouse monoclonal; Calbiochem, Biodynamics, Athens, Greece).
Method: IHC was performed according to the indirect streptavidin-biotin-hyperoxidase protocol, as previously described.35 Indirect immunofluorescence against anti-hCdc6 was applied on asynchronous cultured HeLa cells. HeLa cells were subconfluently cultivated on a coverslip in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum in a 5% CO2 atmosphere. The antigen-primary antibody complexes were detected with a fluorescein isothiocyanate-labeled goat anti-mouse secondary antibody, at a dilution 1:250 (Santa Cruz, Bioanalytica). Counterstain was obtained with 100 ng/ml of 4′,6-diamidino-2-phenylindole (Sigma, Athens, Greece). Microscopic observation was performed with a Zeiss Axioplan 2 fluorescence microscope.
Evaluation: Only nuclear staining for E2F-1, total pRb, phosphorylated (p-) pRb, and p53 was considered positive according to criteria presented elsewhere.31,35 hCdc6 positivity was ascribed when cytoplasmic and/or nuclear staining was observed, as shown by the manufacturer company. E2F-1 expression index (EI) and proliferation index (PI), estimated by E2F-1 and Ki67 immunoexpression, respectively, were assessed as the percentage of stained tumor nuclei.32,35 Slide examination was performed by four independent observers (PZ, PK, LV, and VG) with minimal interobserver variability.
Controls: The HeLa (cervical cancer) cell line served as positive control for hCdc6 expression.27 Controls for E2F-1 and p53 staining have been previously described.35 Characterized cases from our previous study,35 exhibiting normal total pRb status and E2F-1 overexpression, were used as positive controls for total pRb and p-pRb. Antibodies of the corresponding IgG fractions with unrelated specificities were used as negative controls in each set of immunoreactions.
Comparative Reverse Transcriptase (RT)-Polymerase Chain Reaction (PCR)
Primers: Each of hCdt1, hCdc6, and hGeminin was amplified by two sets of oligonucleotides. For hCdt1 (accession no. 19923847 and GeneID 81620): forward set A 5′ gtgctctgaagggggtgtcc 3′ and reverse set A 5′ cggagaggagcagcaggtg 3′, forward set B 5′ aaggatcccgcctaccagcgcttcc 3′, and reverse set B 5′ ccaagcttgaaggtggggacactg 3′ (positions 1292 and 1564, 556 and 831, respectively). Product size for A is 273 bp and for B is 276 bp. For hCdc6 (accession no. 19264107 and GeneID 990): forward set A 5′ gccaagaaggagcacaagat 3′ and reverse set A 5′ gggtggggtgtaagagaagaa 3′, forward set B 5′ tgttctccaccaaagcaaggc 3′ and reverse set B 5′tcctgatgacatccatctccc 3′ (positions 1535 and 1894, 406 and 739, respectively). Product size for A is 360 bp and for B is 334 bp. For hGeminin (accession no. 3249004 and GeneID 51053): forward set A 5′ gacaatgaaattgcccgcctga 3′ and reverse set A 5′ atgcaatagggtcccaggattcaaa 3′, forward set B 5′ cttctgtgcttcaccatctaca 3′ and reverse set B agtggaggtaaacttcggcag 3′ (positions 696 and 1081, 291 and 997, respectively). Product size for A is 386 bp and for B is 707 bp.
RNA Extraction and RT-PCR: RNA extraction and cDNA synthesis from frozen samples were performed as previously described.35 mRNA levels of target genes were assessed with the use of a semiquantitative multiplex RT-PCR protocol.35
Controls: To avoid RT-PCR intra-assay fluctuations that might affect the original target-to-reference transcript ratio, twofold serial dilutions of cDNA solutions were subjected to control amplifications. According to these reactions, the maximum acceptable number of thermal cycles was established for the assessment of transcript ratios.
Evaluation: Target mRNA levels were assessed via comparing the relative ratios between tumor and corresponding normal specimens. mRNA expression was characterized normal when tumor-to-normal ratios were between 0.5 and 2.0.35 Values lower than 0.5 and greater than 2.0 were considered as underexpression and overexpression, respectively.
Total and Subcellular Fractionation Protein Extraction—Western Blotting
Method: Total protein, as well as nuclear and cytoplasmic extracts preparation and Western blot analysis on frozen samples were performed as previously described.36
Antibodies and Controls: The following primary antibodies were used: anti-E2F-1 (mouse monoclonal, clone KH95) and anti-hCdc6 (mouse monoclonal, clone 180.2) (all commercially provided by Santa Cruz, BioAnalytica). For hGeminin, an affinity-purified rabbit antibody was used, generated against the C-terminus of hGeminin and specifically recognizing full-length hGeminin in several human cancer cell lines (H Nishitani, Z Lygerou, and T Nishimoto, manuscript submitted for publication). The anti-hCdt1 rabbit polyclonal antibody was previously described.27 Goat anti-mouse (Sigma) and anti-rabbit (Bio-Rad, Athens, Greece) IgG fragments, conjugated with horseradish peroxidase, served as secondary antibodies. The MCF-7 cell line, known to express E2F-1,35 was used as positive control for E2F-1 analysis; the HeLa cell line for hCdt1 and hCdc6; and the K562 cell line for hGeminin.38 Ponceau-S staining and Western blotting of actin ensured equal protein loading per sample.
Mutation Analysis
Microdissection and DNA Extraction: Five-μm serial sections were obtained from archival and frozen specimens and processed as previously described.31
Nested PCR/Single-Strand Conformation Polymorphism and Sequencing Analysis: Nested-PCR/single-strand conformation polymorphism analysis for p53 exons was performed in matched normal and tumor DNA, as previously described.31 Samples yielding electrophoretic patterns suggestive of mutations were subjected to automated sequencing.
Allelic Imbalance [Loss of Heterozygosity (LOH)] Analysis of the pRb Gene
Allelic alterations analysis of the pRb gene was performed using the internal pRb microsatellite marker D13S153, as previously described.32 Analysis was performed on an ABI-PRISM 377 automatic sequencer and results were evaluated using the Genescan and Genotype software (Applied Biosystems). LOH scoring was performed as follows: when the allele ratio values were ≤0.65 or ≥1.54, samples were scored as LOH; whereas, when their values covered the range 0.77 to 1.23, they were scored as negative. Samples exhibiting values between 1.23 to 1.54 or 0.65 to 0.77 were subjected to a second assay and scored as LOH only when a second positive value was obtained.32
Terminal dUTP Nick-End Labeling Assay (TUNEL)
Method: Double-stranded DNA breaks were detected by TUNEL, according to the protocol described elsewhere.32
Controls: Tissue sections incubated with TdT buffer in the absence of enzyme were used as negative controls. Sections incubated with DNase I for 20 minutes before TdT treatment served as positive controls.
Evaluation: Cells exhibiting nuclear staining without cytoplasmic background were regarded as undergoing apoptosis. Apoptotic index (AI) was estimated as the percentage of apoptotic cells in 7 to 10 high-power fields (∼1000 total cells). Slide examination was performed by four independent observers (VG, PK, LV, and PZ) with minimal interobserver variability.
Ploidy Analysis
Ploidy analysis was performed as previously described,32 with certain modifications in the preparation of tissue material. Particularly, the examined tumor cells were collected from three 50-μm sections after trypsin treatment and prepared as cytological material.
Statistical Analysis
Associations of hCdt1, hCdc6, and hGeminin expression with the patients’ clinicopathological parameters, as well as with ploidy and p53 status were assessed using the chi-square statistical test. EI, PI, and GI (= PI/AI) follow normal distribution and so they are described by mean value and SD, in contrast to AI (non-normal distribution) that is presented by median value and the range of values. Therefore, the putative correlations of hCdt1 and hCdc6 with EI and PI were examined by t-test whereas those regarding AI were examined by Mann-Whitney test. Combined patterns of expression between hCdt1, hCdc6, and p53 were used to assess for possible associations with GI, PI, AI, and ploidy status of the tumors. Therefore, cases were grouped according to their molecular expression profiles (see Table 3). Differences among the pattern groups concerning their kinetic parameters and ploidy status were assessed by analysis of variance (for GI, PI; among all groups) and Bonferroni (for GI, PI; between two groups), Kruskal-Wallis (for AI; among all groups), and Mann-Whitney (for AI; between two groups) or Pearson chi-square (for ploidy; among two or more groups) tests, respectively. Survival analysis of follow-up data were performed with the Kaplan-Meier methodology. All tests were performed with the aid of the SPSS-10 statistical package (SPSS Inc., Chicago, IL). Statistical relations characterized by P values lower than 0.05 were considered significant.
Table 3.
Relationship of Cdt1-Cdc6/p53 Patterns with Growth (GI) Index, Its Components Proliferation (PI) and Apoptotic (AI) Indices, and Ploidy Status of the Tumors
| Pattern | (No. of cases) | GI = PI/AI mean value (%) [informative cases] | P | PI mean value (%) [informative cases] | P | AI median value (%) [informative cases] | P | Ploidy status aneuploidy¶ | P |
|---|---|---|---|---|---|---|---|---|---|
| “normal” hCdt1-hCdc6/p53wt | (9) | 12.4 ± 7.8 [8] | 33.3 ± 9.4 [8] | 2.9 [8] | 6/9 (66%) | ||||
| “deregulated” hCdt1-hCdc6/p53wt | (18) | 11.2 ± 8.4 [17] | 24.4 ± 8.4 [17] | 3.1 [17] | 5/18 (28%) | ||||
| “normal” hCdt1-hCdc6/p53mt | (16) | 33.0 ± 30.7 [15] | 0.006* | 33.8 ± 13.7 [15] | 0.006* | 1.25 [15] | 0.013‡ | 9/16 (56%) | 0.034∥ |
| “deregulated” hCdt1-hCdc6/p53mt | (24) | 57.2 ± 63.2 [24] | 38.1 ± 12.9 [24] | 0.9 [24] | 16/22 (73%) | ||||
| “deregulated” hCdt1-hCdc6/p53wt | (18) | 0.008† | 0.003† | 0.001§ | 0.005∥ | ||||
| “deregulated” hCdt1-hCdc6/p53mt | (24) | ||||||||
| “normal” hCdt1-hCdc6/p53wt | (9) | 1.000† | 0.923† | 0.003§ | 0.470∥ | ||||
| “normal” hCdt1-hCdc6/p53mt | (16) |
ANOVA.
Bonferroni test.
Kruskal-Wallis test.
Mann-Whitney test.
Aneuploid cases/total informative cases.
Pearson chi-square test.
wt, Wild-type; mt, mutant.
Experiments Using Cell Lines
In Silico Analysis of the hCdt1 Promoter
The gene structure of the human Cdt1 gene was predicted by comparing the hCdt1 cDNA sequences and expressed sequence tagged sequences with human genomic sequences using BLAST (NCBI), and it was subsequently confirmed by the annotation of the human genome. The transcription start site was predicted using the Gene2Promoter program (Genomatix) and it was confirmed by the human genome annotation. Sequences upstream of the predicted transcription start site were analyzed for consensus transcription-factor binding sites with TFsearch ver1.3 and Genomatix programs.
Plasmids
A 1064-fragment upstream of the hCdt1 gene was cloned from human genomic DNA by PCR using primers: 5′-GCAAGCTTGGCCTTTCAAAGTGCTGGGATTAC-3′ and 5′-CATGTCGACGGAGTGCGCGGCGAAAGC-3′. This PCR product, which extends 1044 nucleotides upstream of the translation initiation codon (ATG) of hCdt1, was cloned into the HindIII and SalI sites of Bluescript (Stratagene) and subsequently into the SmaI and XhoI sites of pGL3 (Promega), generating plasmid hCdt1Prom. Mammalian vectors expressing the six members of the E2F family were kindly provided by Dr. Patrick O Humbert.
Cell Lines
NIH3T3 cells were grown in Dulbecco’s modified Eagle’s medium-high glucose medium with 10% calf serum. For serum starvation, NIH3T3 cells were incubated in the presence of 0.1% serum for 48 hours. NIH3T3 cells were then induced to re-enter the cell cycle by addition of 10% serum.
Transient Transfection and Luciferase Assay
Plasmids were transfected into NIH3T3 cells using lipofectamine according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). In every transfection experiment, 1 μg of hCdt1prom alone or in combination with 0.2 μg of E2F-expressing plasmids was used and the total amount of the DNA transfected adjusted to 2 μg with pBluescript. In addition 0.5 μg of a plasmid expressing the β-galactosidase gene under the control of the CMV promoter was co-transfected. Total protein, luciferase activity, and β-galactosidase activity were assayed as described.39 Luciferase activity was corrected for protein concentration and β-galactosidase activity. Each independent experiment was performed in triplicates.
Results
hCdt1 and hCdc6 Are Significantly Overexpressed in a Subset of NSCLCs
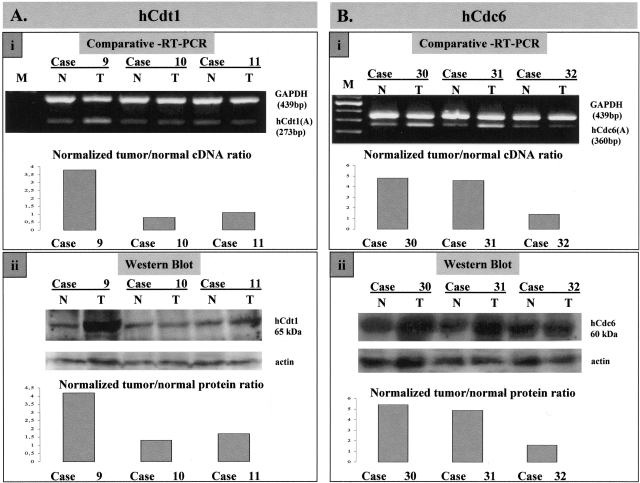
The expression levels of hCdt1 and hCdc6 were examined in all 75 NSCLCs and adjacent healthy lung tissue, at both the mRNA and protein level. For mRNA analysis, multiplex reverse transcription PCR was used to assess the levels of hCdt1 and hCdc6 mRNAs, normalized to the GAPDH mRNA, in total RNA extracted from the tumor and corresponding adjacent healthy tissue. Two sets of primers (A and B; see Materials and Methods), spanning different portions of the cDNAs were used for each of hCdt1 and hCdc6, respectively. Identical results were obtained with both sets of primers. Representative examples of normal tumor pairs are shown in Figure 1, A (i) and B (i), for hCdt1 and hCdc6, respectively. We observed increased (fourfold to fivefold) tumor to normal cDNA ratios in 31 of 72 (43%) informative cases analyzed for hCdt1 and 38 of 75 (50%) informative cases for hCdc6 (Table 1). In the remaining cases the mRNA levels for both molecules were basal.
Figure 1-4261.
mRNA and protein analysis of hCdt1 and hCdc6. A: Representative results of comparative RT-PCR (i) and Western blot analysis (ii) for hCdt1 in paired normal (N)/tumor (T) samples; case 9 with overexpression, and cases 10 and 11 with normal expression of hCdt1. B: Representative results of comparative RT-PCR (i) and Western blot analysis (ii) for hCdc6 in paired normal (N)/tumor (T) samples; cases 30 and 31 with overexpression and case 32 with normal expression of hCdc6. In A and B, the corresponding charts show the compared (as a tumor/normal ratio) and normalized (target to reference) cDNA and protein levels, respectively, of the matched tumor/normal samples. GAPDH mRNA and actin protein were used as reference for RT-PCR and Western blotting analysis, respectively.
Table 1.
Summary of the Clinicopathological Features of the Patients, Status of the Molecules Studied, Tumor Kinetic Parameters, and Ploidy Status of the Tumors
| Histology* |
ADCs: |
32 |
SCCs: |
34 |
UL: |
9 |
| Lymph node invasion* | Yes: | 45 | no: | 30 | ||
| Tumor stage* | I: | 30 | II: | 27 | III: | 18 |
| Survival status† | Deceased (Md): | 26 (14) | Alive (Md): | 39 (18) | ||
| hCdt1 status* | NE: | 41 | OE: | 31 | ||
| hCdc6 status* | NE: | 37 | OE: | 38 | ||
| hGeminin status* | UE: | 8 | NE: | 35 | OE: | 25 |
| E2F-1 index (%) (EI) | M (n): | 39.8% (68) | SD: | 14.2% | Range: | 12–74% |
| p53 status*‡ | P: | 42 | N: | 31 | ||
| Proliferation index (%) (PI) | M (n): | 32.7% (67) | SD: | 12.5% | Range: | 5–70% |
| Apoptotic index (%) (AI) | Md (n): | 1.65% (64) | Range: | 0.1–10.6% | ||
| Growth index (GI = PI/AI) | Md (n): | 22.5% (64) | Range: | 4.0–290.0 | ||
| Ploidy status* | A: | 40 | D: | 30 |
Number of cases.
Follow-up up to 5-years; in months after surgery.
p53 immunopositivity is associated with p53 mutations (P = 0.004, 17 of 23 versus 11 of 35).
P, positive; N, negative; A, D, aneuploid and diploid cases, respectively; ADC, adenocarcinoma; SCC, squamous cell carcinoma; UL, undifferentiated large carcinoma; Md, median value; NE, normal expression; OE, overexpression; UE, underexpression; M, mean value; SD, standard deviation; n, number of informative samples.
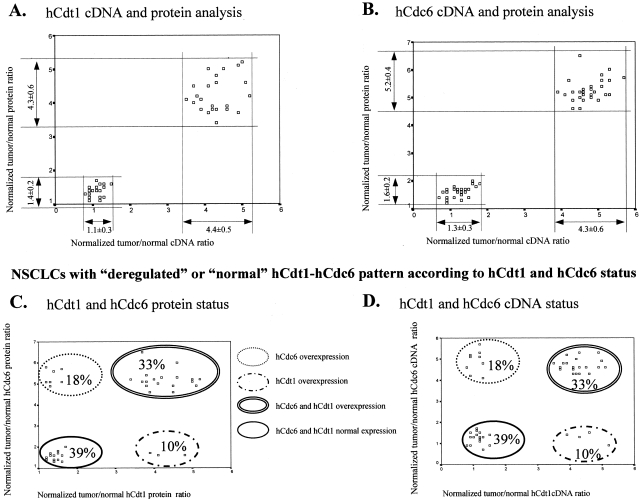
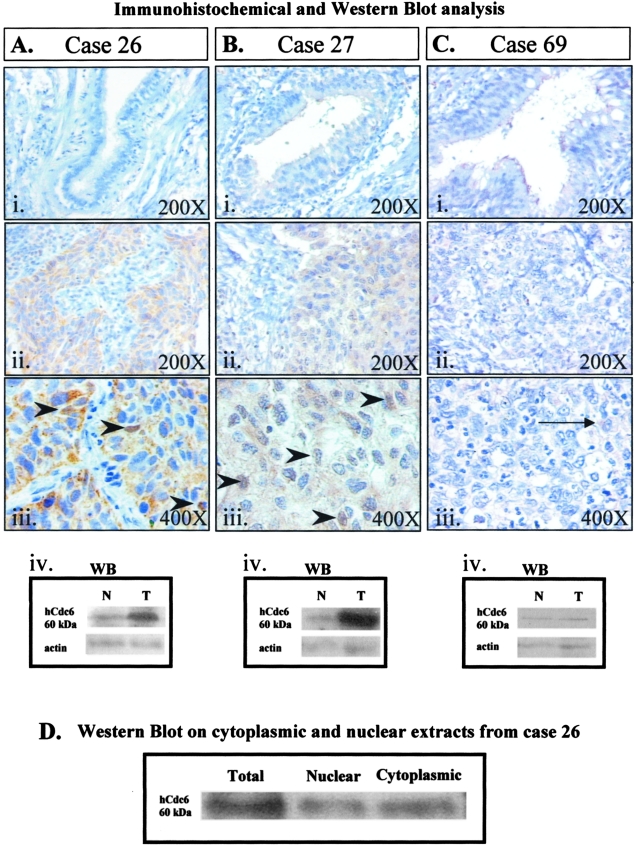
Protein analysis on total extracts from tumor and corresponding normal tissue was performed by Western blotting using antibodies specific for hCdt1, hCdc6, and actin (as loading control). Figure 1, A (ii, hCdt1) and B (ii, hCdc6), show Western blot analysis of the same tumor/normal pairs analyzed by RT-PCR in Figure 1, Ai and Bi, respectively. For both licensing factors examined, cases with high mRNA levels harbor increased protein levels as well (more than or equal to fourfold). Only in a single case was overexpression of hCdt1 protein not accompanied by increased mRNA levels. In Figure 2, scatter plots of tumor/normal ratios of cDNA versus protein are shown for hCdt1 (2A) and hCdc6 (2B). In the rest of the samples, although the expression status of hCdt1 and hCdc6 within the tumor areas was somewhat higher than the corresponding normal tissues in a considerable portion of the cases, the tumor-to-normal ratio never exceeded the value of 2. This marginal increase may be attributed to the higher proliferation rates that neoplastic tissues have in comparison to their corresponding normal ones. It is noteworthy that within the group of tumors harboring overexpressed hCdt1 and/or hCdc6 there is a subgroup of 24 cases (comprising 9 adenocarcinomas, 11 squamous cell carcinomas, and 4 undifferentiated carcinomas) that exhibit clearly elevated expression levels for both factors. Thus, in the next step, we examined whether the expression profiles of hCdt1 and hCdc6 were correlated (Figure 2, C and D). A strong association was established between hCdt1 and hCdc6 expression (24 of 37 hCdt1-overexpressing cases also overexpressed hCdc6 versus 7 of 35 hCdt1 normally expressing ones, P < 0.001 by chi-square test). The expression status of hCdt1 and hCdc6 was independent of histology, lymph node involvement, and disease stage. In sum, our analysis clearly demonstrated the presence of a subset of NSCLCs with markedly increased levels (more than fourfold) of the hCdt1 and hCdc6 licensing factors. Interestingly, when we proceeded to the immunohistochemical study of hCdc6 (hCdt1 IHC was not feasible because the available antibody was inappropriate for paraffin sections), we observed a strong signal in all carcinomas with high levels of hCdc6, previously assessed by Western blotting [Figure 3, A (i to iv) and B (i to iv)]. The obtained signal was cytoplasmic and occasionally nuclear [Figure 3, A (iii) and B (iii)]. Subcellular fractionation of representative cases followed by Western blot analysis revealed hCdc6 in both cytoplasmic and nuclear fractions, confirming the observed immunohistochemical localization (Figure 3D). Using HeLa cells as a control cellular system, known to express high hCdc6 levels,8 we observed in the majority of cells a predominant hCdc6 cytoplasmic signal and in a significantly higher percentage, than the archival tissues examined, nuclear reactivity as well (data not shown). In the remaining, hCdc6 normally expressing tumors faint cytoplasmic signal was observed (Figure 3C, i to iii). Nevertheless, in the latter cases minor intensity differences were seen between normal and tumor areas, as confirmed by immunoblotting.
Figure 2-4261.
Scatter plot of the normalized tumor-to-normal cDNA and protein ratios calculated for the hCdt1 (A) and hCdc6 (B) expression of the examined NSCLCs. According to the hCdt1 and hCdc6 cDNA and protein expression, the carcinomas were subdivided in two groups: one consisted of cases characterized by overexpression at both mRNA and protein levels, whereas the other comprised tumors that exhibited mRNA and protein levels similar to those of their normal counterparts. The mean value and its SD of the corresponding ratios are presented under the arrows that indicate the values’ range for each subgroup. Scatter plots present the NSCLC subgroups according to hCdt1/hCdc6 patterns of the normalized protein (C) or tumor/normal cDNA (D) ratios.
Figure 3-4261.
Representative immunohistochemical results of two cases harboring hCdc6 overexpression and one with normal expression (as scored by RT-PCR and Western blot analysis), alongside their matching normal counterparts (Ai, Bi, and Ci). Case 26 (squamous cell carcinoma) (Aii, Aiii) and case 27 (adenocarcinoma) (Bii, Biii) exhibit strong hCdc6 immunostaining, whereas case 69 (undifferentiated large cell carcinoma) (Cii, Ciii) displays normal hCdc6 immunoreactivity (very weak cytoplasmic staining) (showed by arrow in Ciii). As shown in the accompanying pictures of higher magnification (Aiii and Biii, respectively), the hCdc6 immunostaining was mainly cytoplasmic and occasionally nuclear (positive nuclei are demarcated by arrowheads). hCdc6 immunostaining was in accordance with Western blot analysis (Aiv, Biv, and Civ). D: Western blot analysis for hCdc6 on total, nuclear, and cytoplasmic protein extracts from case 26 verifying the results of the immunocytochemical analysis and implying that the applied hCdc6 antibody detects the cytoplasmic as well as the nuclear form of hCdc6.
hCdt1 Expression Is Associated with E2F-1 Expression in NSCLCs
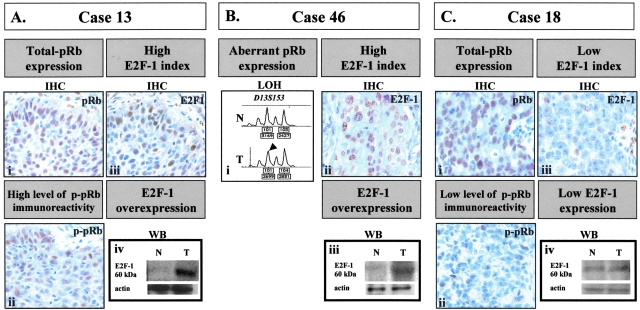
Having in mind that E2F family members play an important role in regulating the transcription of genes required for cell cycle re-entry,37 we searched for any possible association between hCdt1, hCdc6, and E2F-1 expression in our series of NSCLCs. Thus, first, we completed the in situ study of pRb and E2F-1 in our expanded series of NSCLCs. As mentioned in the Materials and Methods section, 32 new cases were included. The overall analysis of previously studied cases35 and of newly added ones revealed that E2F-1 index (EI) ranged from 12 to 47% (mean value, 39.8 ± 14.2%) (Table 1). In Figure 4, three representative cases displaying a high (Figure 4, A and B) and low (Figure 4C) EI are depicted. Expression of E2F-1 was also assessed by Western blot, which confirmed our previous findings of higher E2F-1 levels within the tumor region than the corresponding normal areas35 (Figure 4; Aiv, Biii, and Civ). Furthermore, we observed that phospho-pRb (p-pRb) immunoreactivity followed E2F-1 immunoexpression in cases with intact pRb (total pRb immunoexpression). Bearing in mind that the active and free form of E2F-1 is dependent on the phosphorylation status of pRb,40 we pose that the high degree of p-pRb and E2F-1 co-localization, observed in our series, is indicative that E2F-1 staining, at least in a portion, corresponds to the unbound and active form. In support to the above, it should be mentioned that in all cases with intact pRb, staining for total pRb was found in the majority of tumor cells, whereas the percentage of p-pRb immunoreactivity paralleled E2F-1 labeling index (Figure 4, A and C). In the remaining E2F-1-overexpressing cases, E2F-1 expression was accompanied by aberrant pRb status as assessed by IHC and LOH analysis (Figure 4Bi) [cumulatively 20 of 75 (27%)]. In the next step, analyzing the expression profiles of hCdt1, hCdc6, and E2F-1, we found that hCdt1-overexpressing cases demonstrated increased E2F-1 levels (mean EI, 44.1 ± 15.7%) compared to hCdt1 normally expressing ones (mean EI, 37.0 ± 12.7%; P = 0.048 by t-test).
Figure 4-4261.
Representative results of E2F-1 analysis in two cases (cases 13 and 46) demonstrating high E2F-1 labeling index and a third one (case 18) displaying low E2F-1 expression levels. In case 13, E2F-1 overexpression was associated with inactivation of pRb (as revealed by p-pRb immunoreactivity in serial sections). In case 46, E2F-1 overexpression was accompanied by deregulated total pRb expression (absence of immunostaining that was associated with pRb LOH). For comparison, in case 18 the observed lower EI (as assessed also by Western blot analysis) was associated with a lower degree of p-pRb immunoreactivity. As described in the Materials and Methods section, the following assays are presented in the figure: for pRb status: IHC against total pRb and p-pRb, and LOH analysis using the D13S153 internal pRb microsatellite marker [the arrowhead indicates allelic loss as estimated by the densitometric values (presented in the corresponding boxes below the chart) based on previously described criteria32]; for E2F-1: IHC and Western blot analysis.
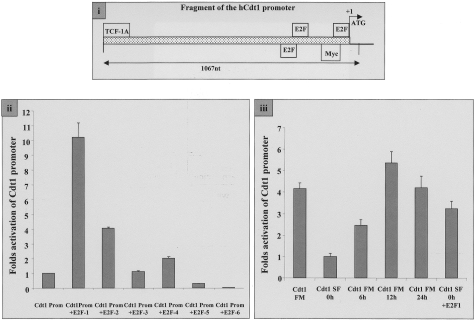
The hCdt1 Promoter Contains Functional E2F-1-Binding Sites
Prompted by the association between hCdt1 and E2F-1 levels in the examined tumors, we sought to clarify whether a direct functional link exists between E2F-1 and hCdt1. In silico analysis of a 1067-nucleotide fragment of the 5′ upstream region of the hCdt1 gene identified three putative E2F-binding sites (Figure 5i), two of which were located in close proximity to the predicted transcriptional initiation site, reminiscent of the promoter organization of the hCdc6 gene.41,42 The 1067-bp fragment of the hCdt1 promoter region was cloned upstream of the luciferase reporter gene, thus creating plasmid Cdt1Prom. When transfected into asynchronously growing NIH3T3 cells, Cdt1Prom exhibited nearly 100-fold higher luciferase activity compared to the vector alone (data not shown). NIH3T3 cells were transfected with Cdt1Prom together with plasmids expressing each of the six known members of the E2F-family (E2F-1 to E2F-6) and luciferase activity was evaluated. Reporter expression downstream of the hCdt1 promoter was 10-fold higher in the presence of E2F-1 than of the promoter construct alone. E2F-2 and E2F-4 activation produced a moderate increase (fourfold and twofold, respectively) whereas E2F-5 and E2F-6 resulted in a reduction of promoter activity (Figure 5ii).
Figure 5-4261.
i: Schematic representation of hCdt1 promoter with putative transcription factor binding sites indicated. The predicted transcription start site is shown by an arrow, whereas the 1067-nucleotide fragment used for further analysis is marked. ii: Differential regulation of the hCdt1 promoter by members of the E2F family. NIH3T3 cells were transfected with 1000 ng of the hCdt1 promoter cloned into pGL3 vector alone (Cdt1Prom) or in combination with 200 ng of vectors expressing the six members of the E2F family (+E2F 1 to 6) together with 500 ng of a β-gal-expressing plasmid as an internal control. Luciferase values, corrected for total protein and transfection efficiency, are expressed as fold increase over the activity exhibited by Cdt1Prom alone. Standard deviations resulted from three independent experiments, each performed in triplicates. iii: hCdt1 promoter activity in synchronized NIH3T3 cells. NIH3T3 cells were transfected with the reporter construct containing the hCdt1 promoter (Cdt1Prom) and subsequently cultured in 0.1% serum for 48 hours (0 hours). Cell proliferation was induced by addition of 10% calf serum and cells were harvested at the indicated time points (6, 12, 24 hours) to assay luciferase and galactosidase activity. Asynchronously growing cells were transfected with hCdt1 promoter containing plasmid and luciferase activity was measured (Cdt1 FM). In addition, an E2F1 expression plasmid enhances transcription from an hCdt1 promoter-driven luciferase construct on serum starvation when co-transfected in NIH3T3 cells (Cdt1 SF 0 hours and Cdt1 SF 0 hours +E2F1). Luciferase values of triplicate experiments are presented as fold increase of the promoter activity observed in serum-starved cells.
Subsequently, NIH3T3 cells transfected with the reporter plasmid containing the hCdt1 promoter region were cultured in the absence of serum for 48 hours and promoter activity was assessed. On exit from the cell cycle, the hCdt1 promoter exhibited a fourfold down-regulation compared to promoter activity in asynchronously growing cells. When serum-starved NIH3T3 cells were induced to re-enter the cell cycle by serum re-addition, hCdt1 promoter activity increased progressively. Co-transfection of the E2F-1 expression plasmid in serum-starved NIH3T3 resulted in an increase in hCdt1 promoter activity, almost reaching the levels observed in asynchronously proliferating cells (Figure 5iii). Cumulatively, our results suggest that E2F-1 is able to trans-activate the hCdt1 promoter.
The Association of hCdt1 and hCdc6 Expression with Tumor Kinetic Parameters and Ploidy Is Dependent on the Status of p53
To investigate whether cases with hCdt1/hCdc6 overexpression attain aggressive biological characteristics, we searched for putative associations with the kinetic parameters and ploidy status of the carcinomas (Table 1). In this context, we regarded hCdt1 and/or hCdc6 overexpression as a deregulated hCdt1-hCdc6 pattern. Based on the fundamental roles of hCdt1 and hCdc6 in S phase progression, we hypothesized that cases with the aberrant hCdt1-hCdc6 pattern might be associated with chromosome instability and increased proliferation. However, this notion was not confirmed, because no association was observed between hCdt1-hCdc6 expression, proliferation (assessed by Ki-67 IHC), apoptosis (assessed by TUNEL), and ploidy (P = 0.428 by t-test, P = 0.652 by Mann-Whitney test and P = 0.613 by chi-square test, respectively).
Prompted by the recent study of Vaziri and colleagues,18 we wished to stratify our cases according to their p53 status. p53 status was assessed by IHC and single-strand conformation polymorphism analysis followed by sequencing (Table 2). Analysis of immunohistochemical and sequencing findings revealed that p53 immunopositivity was significantly associated with the presence of mutations (Tables 1 and 2; P = 0.004 by chi-square test). Having the latter statistical correlation in mind and based on our previous work on NSCLCs, as well as on other relevant studies showing that p53 immunopositivity is strongly associated with p53 mutations,31,32,43 we considered p53 staining as an indicator of p53 mutation.
Table 2.
Results of the p53 Mutation and Immunohistochemical Analysis of the New Added Samples of Our Expanded Series of NSCLCs
| No.* | p53 mutations |
aa sub | p53 IHC | Ploidy status | |
|---|---|---|---|---|---|
| Codon | Point mutation | ||||
| 1 | 135 | tgc to cgc | C to R | P | D |
| 2 | – | N | A | ||
| 3 | 111 | ctg to gtg | L to V | N | D |
| 4 | – | N | D | ||
| 5 | – | N | D | ||
| 6 | – | N | A | ||
| 7 | – | N | A | ||
| 8 | 273 | cgt to cat | R to H | P | D |
| 9 | 157 | gtc to ttc | V to F | P | D |
| 10 | – | P | A | ||
| 11 | – | N | A | ||
| 12 | 280 | aga to ata | R to I | P | A |
| 13 | 314 | tcc to tgc | S to C | P | D |
| 14 | 111 | ctg to gtg | L to V | N | A |
| 15 | 281 | gac to gaa | D to E | P | A |
| 16 | 80 | cct to ctt | P to L | P | A |
| 17 | 90 | tcc to acc | S to T | P | D |
| 18 | – | N | D | ||
| 19 | – | N | A | ||
| 20 | 55 | act to aat | T to N | P | D |
| 21 | 275 | tgt to ttt | C to F | P | A |
| 22 | – | N | A | ||
| 23 | 248 | cgg to tgg | R to W | P | A |
| 24 | 74 | gcc to acc | A to T | P | A |
| 25 | 157 | gtc to ttc | V to F | P | D |
| 26 | – | N | A | ||
Material for mutational analysis was available for 26 of 32 cases.
aa sub; amino acid substitution; P, positive; N, negative; D, diploid; A, aneuploid; –, absence of mutation.
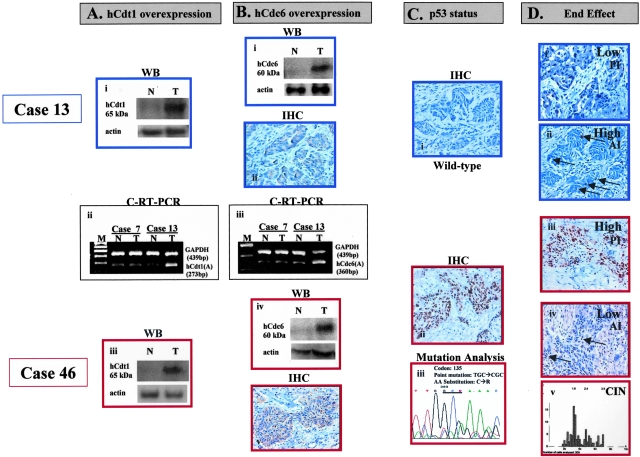
Among the tumors bearing the deregulated hCdt1-hCdc6 pattern (42 cases), p53-mutant (p53mt) cases (24 cases, 57%) were associated with higher tumor growth values and frequent aneuploidy compared with p53-intact (p53wt) ones (P = 0.003 for PI; P = 0.001 for AI; P = 0.008 for GI, and P = 0.005 for ploidy; Table 3). The possibility that the observed associations were merely an artifact of the relations between p53 itself and tumor kinetics or ploidy is highly unlikely, because tumor growth and ploidy were independent of p53 status within the subset of hCdt1-hCdc6 normally expressing cases (Table 3). In Figure 6, hCdt1, hCdc6, p53, PI, AI, and chromosomal instability (CIN) data are shown for two representative cases of hCdt1- and hCdc6-overexpressing tumors. Regarding the aforementioned representative cases, case 13 bears wild-type p53 and exhibits low proliferation and high apoptotic index whereas case 46 bears a mutation in the p53 gene and exhibits a high PI, a low apoptotic index, and CIN. These are the same two cases analyzed for E2F and pRb status in Figure 4.
Figure 6-4261.
Representative results showing downstream events associated with overexpression of hCdt1 and hCdc6 in two cases (cases 13 and 46). In case 13 bearing wild-type p53, low proliferation and high apoptotic indices were observed. On the other hand, in case 46 harboring mutant p53 (C135R), high proliferation and low apoptotic indices, as well as, CIN (aneuploidy) were recorded. As described in the Materials and Methods section the following assays are presented in the figure: for hCdt1: C-RT-PCR and Western blot analysis; for hCdc6: C-RT-PCR, Western blot and IHC analysis; for p53: IHC and sequencing analysis; for proliferation: IHC against Ki-67; for apoptosis: TUNEL assay (apoptotic nuclei are depicted by arrows); for CIN: ploidy analysis (the x axis represents nuclear DNA quantity in c units, whereas y axis corresponds to the number of evaluated nuclei).
The Group of Patients Displaying the Deregulated hCdt1-hCdc6/p53mt Pattern Tends to Carry an Adverse Prognosis as Compared to the Deregulated hCdt1-hCdc6/p53wt One
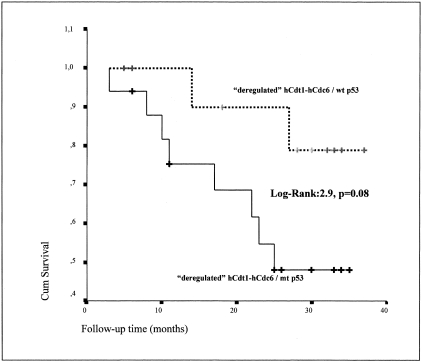
Motivated by the aforementioned observations, we performed survival analysis of the 29 cases exhibiting the aberrant hCdt1-hCdc6 expression pattern according to p53 status. The group exhibiting the deregulated hCdt1-hCdc6/p53-mt profile consisted of 17 patients, of which 8 died in the course of this study (mean survival, 25 months). The group exhibiting deregulated hCdt1-hCdc6 in a p53 wild-type background included 12 patients, of which only 2 died during the present study (mean survival, 34 months). Interestingly, and in support of our previous results, the outcome of the patients harboring p53 mutations tended to be worse compared to those with wild-type p53 (P = 0.08 by Kaplan-Meier) (Figure 7). Importantly, in the subgroup of tumors with normal hCdt1-hCdc6, no correlation between p53 status and patients’ outcome was found. Nevertheless, this difference did not reach statistical significance by Cox regression analysis, among all parameters tested, disease stage was the only one associated with the patients’ outcome (P = 0.013).
Figure 7-4261.
Survival curves of patients with deregulated hCdt1-hCdc6 stratified according to p53 status. In this subgroup of carcinomas with deregulated hCdt1-hCdc6, mutant p53, as compared to wild-type p53, tended to be associated with adverse prognosis (P = 0.08 by Kaplan-Meier methodology).
hGeminin Is Aberrantly Expressed in NSCLCs—Association between Increased hGeminin Levels and hCdt1 Overexpression—Relationship with Ploidy
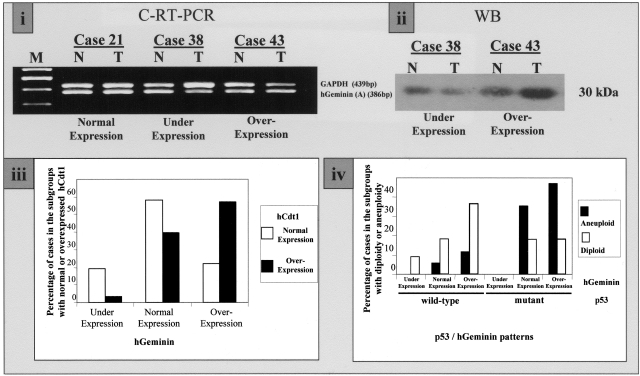
hGeminin is a negative regulator of hCdt1.26,30 We therefore wished to analyze our collection of NSCLCs for expression levels of hGeminin. The mRNA and protein levels of hGeminin were assessed by comparative RT-PCR and Western blot analysis, respectively. The results were consistent by both methods and revealed hGeminin underexpression in 8 (12%) and up-regulation in 25 (37%) specimens from a total of 68 informative ones (Figure 8, i and ii; Table 1). There was no correlation between hGeminin status, histology, lymph node involvement, or disease stage. In addition, no correlation between hGeminin status and tumor growth values or aneuploidy was observed. Interestingly, an association between hGeminin and hCdt1 overexpression was observed (P = 0.025 by chi-square test). More specifically, cases with normal hCdt1 status exhibited under-, normal and overexpression of hGeminin in 20, 58, and 22%, respectively, compared to corresponding percentages of 4, 39, and 57% in hCdt1-overexpressing cases (Figure 8iii). Considering the above findings, suggesting a pivotal effect of p53 protein status in hCdt1/hCdc6 deregulated tumors, as well as that hGeminin suppresses hCdt1,26,30 we sought to examine the relationship between ploidy and p53/hGeminin patterns in hCdt1-overexpressing tumors. We observed that among diploid hCdt1-overexpressing tumors, the p53wt/hGeminin-overexpressing pattern was the most common, in contrast to the aneuploid group of cases, which frequently exhibited the p53mt/hGeminin-overexpressing pattern. Notably, no p53mt/hGeminin-underexpressing cases were recorded (Figure 8iv).
Figure 8-4261.
i: Representative C-RT-PCR analysis of matched normal (N)-tumor (T) tissues from cases 21, 38, and 43, showing normal, underexpression, and overexpression of hGeminin, respectively. ii: Representative Western blot analysis of two paired N-T cases (cases 38 and 43), confirming the previously observed underexpression and overexpression of hGeminin by C-RT-PCR in i. iii: Bar chart depicting the percentage of tumors exhibiting normal (white bars) and overexpressed hCdt1 (black bars) when hGeminin was evaluated as underexpressed, normal, and overexpressed. iv: Bar chart depicting the percentage of aneuploid (black bars) and diploid (white bars) cases according to the p53/hGeminin patterns.
Discussion
Legitimate replication licensing represents a critical prerequisite for secure cellular proliferation. In lower eukaryotes, deregulation of licensing, notably by overexpression of Cdc6 and Cdt1, brings about overreplication of the genome.12,15–17 Similarly, inactivation of the licensing inhibitor Geminin in cultured Drosophila cells leads to overreplication,44 whereas forced expression of hCdc6 drives the human megakaryoblastic cell line HEL into endoreduplication.38 Very recently, it has been demonstrated in cultured human cells that forced expression of the licensing factors hCdt1 and hCdc6, occurring within a background of inactive p53, leads to rereplication, suggesting that this phenomenon may provide seeds for genomic instability.18 However, there has been no study of hCdt1 and hCdc6 overexpression and ploidy status in human tumors. We therefore set to examine whether hCdt1/hCdc6 overexpression is actually present in human malignancy and whether it is coupled with CIN. Toward this aim, we used a series of lung carcinomas, which has been extensively investigated by our group for possible associations between G1 phase network defects, tumor kinetics, and ploidy.32–35
Protein and mRNA analysis of hCdt1 and hCdc6 exposed two subsets of carcinomas regarding their expression levels. One subgroup exhibited a tumor-to-normal protein ratio that was always lower than 2, whereas in the other, expression levels of hCdt1 and hCdc6 were markedly increased, exceeding in the majority of cases a fourfold increase in tumor tissues compared to normal ones (Figure 1, A and B; Figure 2, A and B). More specifically, overexpression of hCdt1 and hCdc6 was seen in 43% and 50% of the tumors, respectively (Table 1). Having in mind that cancerous tissues are generally characterized by higher proliferation rates than their normal counterparts, we anticipated a normal-to-tumor variation. Nevertheless, the presence of a subset of NSCLCs with considerably increased levels of these licensing factors implies that this group of carcinomas may possess a more aggressive biological behavior. Such a behavior has been marked by Williams and colleagues45 and Bonds and colleagues46 in cervical cancer, in which a proportional increase between hCdc6 labeling index and grade of the cervical premalignant and malignant lesions was demonstrated. In contrast, underexpression of hCdc6 has been reported in prostate cancer.47 Our immunohistochemical results were in agreement with Western blotting data, demonstrating strong in situ staining for hCdc6 exclusively in cases with hCdc6 overexpression [Figure 3, A (ii and iii) and B (ii and iii)]. hCdt1 immunostaining was not feasible because of the absence of an available antibody for paraffin sections. The hCdc6 signal was mainly cytoplasmic and occasionally nuclear (Figure 3, Aiii, Biii, and D). This is consistent with reports in human tumor cell lines, showing cytoplasmic localization of hCdc6 in S phase and G2, especially when the protein is overexpressed.21–25 Using HeLa cells as a control cellular system, known to express high hCdc6 levels,20 we stained for hCdc6 and observed in the majority of the cells a predominant cytoplasmic signal and in a significant higher percentage, than the archival tissues we examined, nuclear reactivity as well (data not shown). This result is in line with the reported subcellular hCdc6 pattern by Saha and colleagues,21 and Fujita and colleagues,22 but in contrast with the findings of Williams and co-workers20 who found hCdc6 to be expressed only in the nuclear fraction of HeLa cells. An explanation for the above discrepancies may be the use of different antibodies, which possibly demonstrate divergent staining capabilities depending on the subcellular fraction and tissue processing. The almost absolute concordance noticed between mRNA and protein levels of hCdt1 and hCdc6 is suggestive of a direct deregulation at the transcriptional level for both proteins. However, in one case, overexpression of hCdt1 protein was not accompanied with increased mRNA levels, implying that posttranslational mechanisms may also contribute to this deregulation. Furthermore, our analysis revealed a strong correlation between hCdt1 and hCdc6 expression (P < 0.001), although differential expression was observed in a number of cases (20 of 72 cases). Explanations for this phenomenon include the presence of common mechanisms regulating the levels of both proteins, a possible positive feedback loop between the two or a synergistic participation of the two proteins in lung carcinogenesis.
To shed light on the mechanisms underlying hCdt1/hCdc6 overexpression, we searched for a putative association with E2F-1, bearing in mind the regulatory effect of E2F-family members over several licensing components in cultured cells.37 Before proceeding to the above statistical analysis, we showed that E2F-1 expression was accompanied by p-pRb immunoreactivity in serial sections of cases with normal total pRb status, implying that E2F-1 staining possibly reflects to a significant extent the pRb-unbound, active form of E2F-1 (Figure 4A). Statistical evaluation revealed a link of E2F-1 index (EI) with hCdt1 (P = 0.048), but not hCdc6, expression. Urged by this observation, we hypothesized that hCdt1 may represent a downstream target of E2F-1. This was further supported by the presence of one perfect and two imperfect putative E2F-binding sites on the hCdt1 promoter. To investigate this further, we performed transient transfection assays using all six members of the E2F family,37 which demonstrated that E2F-1 and, to a lesser degree, E2F-2 were capable of triggering the transcription of a reporter gene downstream of the hCdt1 promoter, suggesting that at least one of the above-mentioned sequences was actually E2F-1 responsive (Figure 5, ii and iii). Taken together, the aforementioned data support a functional link between E2F-1 and hCdt1. This notion gains additional support by a recent report showing that transactivation of human Cdt1, Geminin, and MCM7 is induced by E2F-1 to -4 and not by E2F-5 to -7 transcription factors.48 Furthermore, the Drosophila Cdt1 homologue, double-parked, has also been shown to be regulated by E2F.49 Our data indicate that overexpression of hCdt1 in NSCLCs may be due, at least in part, to an E2F-1 mediated effect. The absence of a correlation between E2F-1 and hCdc6 in NSCLCs, despite its well-documented regulation by E2F41,42 implies that other members of the E2F family may play a role in its regulation. Consistent with this interpretation, a link between hCdc6 transcriptional regulation and the E2F-2 and E2F-3 family members, rather than E2F-1, has been reported.50,51 Notably, mouse embryonic fibroblasts bearing an E2F-3 and not an E2F-1 knockout exhibit misregulated Cdc6 expression.50
In the next step of our study, we examined our main hypothesis regarding whether the group of carcinomas exhibiting the deregulated hCdt1-hCdc6 expression pattern (cases with hCdt1 and/or hCdc6 overexpression) is associated with increased proliferation and aneuploidy. This assumption was mainly based on the following: 1) the ability of hCdt1, hCdc6, and the E2F-1 target, cyclin A,37 to induce endoreplication in cultured human cells;18 2) our previously reported link of E2F-1 with increased proliferation and aneuploidy in NSCLCs;35 and 3) the herein presented associations between E2F-1 and hCdt1 (P = 0.048), as well as hCdt1 and hCdc6 (P < 0.001). Altogether, these facts prompted us to speculate that, between E2F-1 overexpression and the end effects of proliferation and aneuploidy observed in our series of lung carcinomas, lies the aberrant hCdt1-hCdc6 expression pattern. Nevertheless, no association was found between this expression profile, kinetic parameters, and ploidy status of the carcinomas, thus declining this oversimplified scenario. Incited by the results of Vaziri and colleagues,18 we re-examined the aforementioned associations stratifying our specimens according to p53 status. It is worth noting that the NSCLCs used in our study represent a similar cellular environment to the lung cancer-derived cell lines used by Vaziri and colleagues.18 This approach uncovered that the deregulated hCdt1-hCdc6 pattern was strongly correlated to higher tumor growth values (P = 0.008) and aneuploidy (P = 0.005) in p53-defective tumors compared to those bearing intact p53 (Table 3). Interestingly, among all groups of carcinomas, the one exhibiting the deregulated hCdt1-hCdc6/p53wt profile included the lowest number of aneuploid cases, even compared to the group bearing normal hCdt1-hCdc6 expression and wtp53 (Table 3). The possibility that the observed associations were merely an artifact of the relations between p53 itself and tumor kinetics or ploidy is highly unlikely, because tumor growth and ploidy were independent of p53 status within the subset of hCdt1-hCdc6 normally expressing cases (Table 3). These findings support the notion that rereplicated DNA, resulting from hCdt1 and hCdc6 overexpression, leads to DNA damage, which, in turns, activates the p53 checkpoint network. In cases with functional p53, cell cycle arrest or apoptosis is induced, whereas as a result of p53 inactivation (through point mutations, deletions, or methylation)52 rereplicated DNA may conduct breakage-fusion-bridge cycles, eventually leading to a state of CIN. In accordance to this hypothesis are the survival analysis data, revealing that patients bearing the aberrant hCdt1-hCdc6/p53mt pattern tend to carry a worse prognosis than those harboring wtp53 (P = 0.08, Figure 7). However, the fact that a significant fraction of tumors demonstrating the normal hCdt1-hCdc6/p53wt pattern were also aneuploid (Table 3) underscores that defects in other pathways, irrelevant to the licensing apparatus or the status of p53, may trigger genomic instability. Candidates for these may include other E2F-1 targets (eg, chromosome condensation factors smc2 and smc4, spindle checkpoint proteins bub3 and mad2, or securin).53
The final stage of our study was devoted to the investigation of hGeminin expression in relation to the licensing components hCdt1 and hCdc6, p53, tumor kinetics, and ploidy. Despite its well-documented function as a suppressor of hCdt1 activity,26,30 hGeminin has exhibited a somewhat unexpected behavior in cell lines27,54,55 and has even been proposed as a marker of proliferation in normal tissues, as well as in breast, cervical, and colon malignancies.55 In our series of NSCLCs, aberrant expression of hGeminin was noted in 49% of the tumors, consisting of 37% overexpressing and 12% underexpressing cases (Figure 8, i and ii; Table 1). Most hGeminin-overexpressing cases exhibited high levels of hCdt1, yielding a positive correlation between the two proteins (Figure 8iii) (P = 0.025). This observation lies in accordance with the study of Vaziri and colleagues,18 who reported an analogous paradoxical increase of hGeminin levels in response to hCdt1-overexpression, and may suggest a positive feedback loop between Cdt1 and Geminin levels in the cell. A similar correlation has been reported in Drosophila cultured cells, in which reducing the levels of Geminin by RNA interference resulted in a concomitant decrease in the levels of Cdt1.44 A mechanism for co-regulating the levels of hCdt1 and its inhibitor hGeminin may be important for ensuring tight control over hCdt1 in normal cells. In NSCLCs, the observed overexpression of hGeminin may not be sufficient to counteract the effects of increased hCdt1 levels, in accordance with the apparent inability of hGeminin up-regulation to prevent rereplication in hCdt1/hCdc6-overexpressing cultured human cells.18 We cannot exclude the possibility that overexpressed Geminin may be functionally inert, because of incorrect localization, protein modification, or mutational inactivation. Despite the proposed role of hGeminin to inhibit rereplication, we observed no correlation between low hGeminin levels and increased ploidy, which, in turn, was only dependent on p53 status. It is worth noting that the p53mt/hGeminin-overexpressing pattern was the most common among aneuploid tumors (Figure 8iv). In conclusion, our findings suggest a synergistic effect between hCdt1-hCdc6 overexpression and mutant p53 over tumor growth and CIN in NSCLCs.
Acknowledgments
We thank Dr. Thanos D. Halazonetis for helpful comments, Dr. Athanassios Kotsinas for excellent technical assistance and support to this work, and Dr. Patrick O. Humbert for providing the E2F-expressing vectors.
Footnotes
Address reprint requests to Vassilis G. Gorgoulis, M.D., Ph.D., Antaiou 53 Str., Lamprini, Ano Patissia 111 46, Athens, Greece. E-mail: histoclub@ath.forthnet.gr.
Supported in part by the Association for International Cancer Research, the Hellenic Anticancer Institute, the Empeirikeion Foundation, the SARG of the University of Athens (grants 70/4/4591 70/4/4281) and the University of Patras Research Committee (K. Karatheodoris Program).
P.K., S.T., and L.V.V. contributed equally to this study.
References
- Paulovich AG, Toczyski DP, Hartwell LH. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Nyberg KA, Michelson RJ, Putnam CW, Weinert TA. Toward maintaining the genome: DNA damage and replication checkpoints. Annu Rev Genet. 2002;36:617–656. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- Shreeram S, Blow JJ. The role of the replication licensing system in cell proliferation and cancer. Prog Cell Cycle Res. 2003;5:287–293. [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Laskey RA. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature. 1988;332:546–548. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- Diffley JF. DNA replication: building the perfect switch. Curr Biol. 2001;11:R367–R370. doi: 10.1016/s0960-9822(01)00196-8. [DOI] [PubMed] [Google Scholar]
- Lei M, Tye BK. Initiating DNA synthesis: from recruiting to activating the MCM complex. J Cell Sci. 2001;114:1447–1454. doi: 10.1242/jcs.114.8.1447. [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Nishitani H, Lygerou Z. Control of DNA replication licensing in a cell cycle. Genes Cells. 2002;7:523–534. doi: 10.1046/j.1365-2443.2002.00544.x. [DOI] [PubMed] [Google Scholar]
- Cocker JH, Piatti S, Santocanale C, Nasmyth K, Diffley JF. An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature. 1996;379:180–182. doi: 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- Coleman TR, Carpenter PB, Dunphy WG. The Xenopus Cdc6 protein is essential for the initiation of a single round of DNA replication in cell-free extracts. Cell. 1996;87:53–63. doi: 10.1016/s0092-8674(00)81322-7. [DOI] [PubMed] [Google Scholar]
- Nishitani H, Lygerou Z, Nishimoto T, Nurse P. The Cdt1 protein is required to license DNA for replication in fission yeast. Nature. 2000;404:625–628. doi: 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- Maiorano D, Moreau J, Mechali M. XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature. 2000;404:622–625. doi: 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Knapp D, Nasmyth K. Loading of an Mcm protein onto DNA replication origins is regulated by Cdc6p and CDKs. Cell. 1997;90:649–660. doi: 10.1016/s0092-8674(00)80526-7. [DOI] [PubMed] [Google Scholar]
- Nishitani H, Nurse P. p65cdc18 plays a major role controlling the initiation of DNA replication in fission yeast. Cell. 1995;83:397–405. doi: 10.1016/0092-8674(95)90117-5. [DOI] [PubMed] [Google Scholar]
- Muzi Falconi M, Brown GW, Kelly TJ. cdc18+ regulates initiation of DNA replication in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1996;93:1566–1570. doi: 10.1073/pnas.93.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanow SK, Lygerou Z, Nurse P. Expression of Cdc18/Cdc6 and Cdt1 during G2 phase induces initiation of DNA replication. EMBO J. 2001;20:4648–4656. doi: 10.1093/emboj/20.17.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri C, Saxena S, Jeon Y, Lee C, Murata K, Machida Y, Wagle N, Hwang DS, Dutta A. A p53-dependent checkpoint pathway prevents rereplication. Mol Cell. 2003;11:997–1008. doi: 10.1016/s1097-2765(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Arentson E, Faloon P, Seo J, Moon E, Studts JM, Fremont DH, Choi K. Oncogenic potential of the DNA replication licensing protein CDT1. Oncogene. 2002;21:1150–1158. doi: 10.1038/sj.onc.1205175. [DOI] [PubMed] [Google Scholar]
- Williams RS, Shohet RV, Stillman B. A human protein related to yeast Cdc6p. Proc Natl Acad Sci USA. 1997;94:142–147. doi: 10.1073/pnas.94.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha P, Chen J, Thome KC, Lawlis SJ, Hou ZH, Hendricks M, Parvin JD, Dutta A. Human CDC6/Cdc18 associates with Orc1 and cyclin-cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol Cell Biol. 1998;18:2758–2767. doi: 10.1128/mcb.18.5.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Yamada C, Goto H, Yokoyama N, Kuzushima K, Inagaki M, Tsurumi T. Cell cycle regulation of human CDC6 protein Intracellular localization, interaction with the human mcm complex, and CDC2 kinase-mediated hyperphosphorylation. J Biol Chem. 1999;274:25927–25932. doi: 10.1074/jbc.274.36.25927. [DOI] [PubMed] [Google Scholar]
- Petersen BO, Lukas J, Sorensen CS, Bartek J, Helin K. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 1999;18:396–410. doi: 10.1093/emboj/18.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coverley D, Pelizon C, Trewick S, Laskey RA. Chromatin-bound Cdc6 persists in S and G2 phases in human cells, while soluble Cdc6 is destroyed in a cyclin A-cdk2 dependent process. J Cell Sci. 2000;113:1929–1938. doi: 10.1242/jcs.113.11.1929. [DOI] [PubMed] [Google Scholar]
- Petersen BO, Wagener C, Marinoni F, Kramer ER, Melixetian M, Denchi EL, Gieffers C, Matteucci C, Peters JM, Helin K. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev. 2000;14:2330–2343. doi: 10.1101/gad.832500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- Nishitani H, Taraviras S, Lygerou Z, Nishimoto T. The human licensing factor for DNA replication Cdt1 accumulates in G1 and is destabilized after initiation of S-phase. J Biol Chem. 2001;276:44905–44911. doi: 10.1074/jbc.M105406200. [DOI] [PubMed] [Google Scholar]
- Li X, Zhao Q, Liao R, Sun P, Wu X. The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J Biol Chem. 2003;278:30854–30858. doi: 10.1074/jbc.C300251200. [DOI] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- Tada S, Li A, Maiorano D, Mechali M, Blow JJ. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat Cell Biol. 2001;3:107–113. doi: 10.1038/35055000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis VG, Zacharatos P, Kotsinas A, Liloglou T, Kyroudi A, Veslemes M, Rassidakis A, Halazonetis TD, Field JK, Kittas C. Alterations of the p16-pRb pathway and the chromosome locus 9p21-22 in non-small-cell lung carcinomas: relationship with p53 and MDM2 protein expression. Am J Pathol. 1998;153:1749–1765. doi: 10.1016/S0002-9440(10)65690-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis VG, Zacharatos P, Kotsinas A, Mariatos G, Liloglou T, Vogiatzi T, Foukas P, Rassidakis G, Garinis G, Ioannides T, Zoumpourlis V, Bramis J, Michail PO, Asimacopoulos PJ, Field JK, Kittas C. Altered expression of the cell cycle regulatory molecules pRb, p53 and MDM2 exert a synergetic effect on tumor growth and chromosomal instability in non-small cell lung carcinomas (NSCLCs) Mol Med. 2000;6:208–237. [PMC free article] [PubMed] [Google Scholar]
- Mariatos G, Gorgoulis VG, Zacharatos P, Kotsinas A, Vogiatzi T, Rassidakis G, Foukas P, Liloglou T, Tiniakos D, Angelou N, Manolis EN, Veslemes M, Field JK, Kittas C. Expression of p16(INK4A) and alterations of the 9p21-23 chromosome region in non-small-cell lung carcinomas: relationship with tumor growth parameters and ploidy status. Int J Cancer. 2000;89:133–141. doi: 10.1002/(sici)1097-0215(20000320)89:2<133::aid-ijc6>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Tsoli E, Gorgoulis VG, Zacharatos P, Kotsinas A, Mariatos G, Kastrinakis NG, Kokotas S, Kanavaros P, Asimacopoulos P, Bramis J, Kletsas D, Papavassiliou AG, Kittas C. Low levels of p27 in association with deregulated p53-pRb protein status enhance tumor proliferation and chromosomal instability in non-small cell lung carcinomas. Mol Med. 2001;7:418–429. [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis VG, Zacharatos P, Mariatos G, Kotsinas A, Bouda M, Kletsas D, Asimacopoulos PJ, Agnantis N, Kittas C, Papavassiliou AG. Transcription factor E2F-1 acts as a growth-promoting factor and is associated with adverse prognosis in non-small cell lung carcinomas. J Pathol. 2002;198:142–156. doi: 10.1002/path.1121. [DOI] [PubMed] [Google Scholar]
- Kotsinas A, Evangelou K, Zacharatos P, Kittas C, Gorgoulis VG. Proliferation, but not apoptosis, is associated with distinct beta-catenin expression patterns in non-small-cell lung carcinomas: relationship with adenomatous polyposis coli and G(1)-to S-phase cell-cycle regulators. Am J Pathol. 2002;161:1619–1634. doi: 10.1016/s0002-9440(10)64440-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevaux O, Dyson NJ. A revised picture of the E2F transcriptional network and RB function. Curr Opin Cell Biol. 2002;14:684–691. doi: 10.1016/s0955-0674(02)00388-5. [DOI] [PubMed] [Google Scholar]
- Bermejo R, Vilaboa N, Cales C. Regulation of CDC6, geminin, and CDT1 in human cells that undergo polyploidization. Mol Biol Cell. 2002;13:3989–4000. doi: 10.1091/mbc.E02-04-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taraviras S, Schutz G, Kelsey G. Generation of inhibitory mutants of hepatocyte nuclear factor 4. Eur J Biochem. 1997;244:883–889. doi: 10.1111/j.1432-1033.1997.00883.x. [DOI] [PubMed] [Google Scholar]
- Mundle SD, Saberwal G. Evolving intricacies and implications of E2F-1 regulation. EMBO J. 2003;17:569–574. doi: 10.1096/fj.02-0431rev. [DOI] [PubMed] [Google Scholar]
- Yan Z, DeGregori J, Shohet R, Leone G, Stillman B, Nevins JR, Williams RS. Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc Natl Acad Sci USA. 1998;95:3603–3608. doi: 10.1073/pnas.95.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hateboer G, Wobst A, Petersen BO, Le Cam L, Vigo E, Sardet C, Helin K. Cell cycle-regulated expression of mammalian CDC6 is dependent on E2F. Mol Cell Biol. 1998;18:6679–6697. doi: 10.1128/mcb.18.11.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- Mihaylov IS, Kondo T, Jones L, Ryzhikov S, Tanaka J, Zheng J, Higa LA, Minamino N, Cooley L, Zhang H. Control of DNA replication and chromosome ploidy by geminin and cyclin A. Mol Cell Biol. 2002;22:1868–1880. doi: 10.1128/MCB.22.6.1868-1880.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GH, Romanowski P, Morris L, Madine M, Mills AD, Stoeber K, Marr J, Laskey RA, Coleman N. Improved cervical smear assessment using antibodies against proteins that regulate DNA replication. Proc Natl Acad Sci USA. 1998;95:14932–14937. doi: 10.1073/pnas.95.25.14932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonds L, Baker P, Gup C, Shroyer KR. Immunohistochemical localization of cdc6 in squamous and glandular neoplasia of the uterine cervix. Arch Pathol Lab Med. 2002;26:1164–1168. doi: 10.5858/2002-126-1164-ILOCIS. [DOI] [PubMed] [Google Scholar]
- Robles LD, Frost AR, Davila M, Hutson AD, Grizzle WE, Chakrabarti R. Down-regulation of Cdc6, a cell cycle regulatory gene, in prostate cancer. J Biol Chem. 2002;277:25431–25438. doi: 10.1074/jbc.M201199200. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Inoue I. Regulation of Geminin and Cdt1 expression by E2F transcription factors. Oncogene. 2004;23:3802–3812. doi: 10.1038/sj.onc.1207488. [DOI] [PubMed] [Google Scholar]
- Whittaker AJ, Royzman I, Orr-Weaver TL. Drosophila double parked: a conserved, essential replication protein that colocalizes with the origin recognition complex and links DNA replication with mitosis and the down-regulation of S phase transcripts. Genes Dev. 2000;14:1765–1776. [PMC free article] [PubMed] [Google Scholar]
- Humbert PO, Verona R, Trimarchi JM, Rogers C, Dandapani S, Lees JA. E2f3 is critical for normal cellular proliferation. Genes Dev. 2000;14:690–703. [PMC free article] [PubMed] [Google Scholar]
- Schlisio S, Halperin T, Vidal M, Nevins JR. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 2002;21:5775–5786. doi: 10.1093/emboj/cdf577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prives C, Hall PA. The p53 pathway. J Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shreeram S, Sparks A, Lane DP, Blow JJ. Cell type-specific responses of human cells to inhibition of replication licensing. Oncogene. 2002;21:6624–6632. doi: 10.1038/sj.onc.1205910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel JA, Kutok JL, Weng AP, Dutta A. Expression of geminin as a marker of cell proliferation in normal tissues and malignancies. Am J Pathol. 2002;161:267–273. doi: 10.1016/S0002-9440(10)64178-8. [DOI] [PMC free article] [PubMed] [Google Scholar]