Abstract
The pathogenesis of hepatitis C virus (HCV)-associated insulin resistance remains unclear. Therefore, we investigated mechanisms for HCV-associated insulin resistance. Homeostasis model assessment for insulin resistance was increased in patients with HCV infection. An increase in fasting insulin levels was associated with the presence of serum HCV core, the severity of hepatic fibrosis and a decrease in expression of insulin receptor substrate (IRS) 1 and IRS2, central molecules of the insulin-signaling cascade, in patients with HCV infection. Down-regulation of IRS1 and IRS2 was also seen in HCV core-transgenic mice livers and HCV core-transfected human hepatoma cells. Carbobenzoxy-l-leucyl-l-leucyl-l-leucinal, a potent proteosomal proteolysis inhibitor, blocked down-regulation of IRS1 and IRS2 in HCV core-transfected hepatoma cells. In human hepatoma cells, HCV core up-regulated suppressor of cytokine signaling (SOCS) 3 and caused ubiquitination of IRS1 and IRS2. HCV core-induced down-regulation of IRS1 and IRS2 was not seen in SOCS3−/− mouse embryonic fibroblast cells. Furthermore, HCV core suppressed insulin-induced phosphorylation of p85 subunit of phosphatidylinositol 3-kinase and Akt, activation of 6-phosphofructo-2-kinase, and glucose uptake. In conclusion, HCV infection changes a subset of hepatic molecules regulating glucose metabolism. A possible mechanism is that HCV core-induced SOCS3 promotes proteosomal degradation of IRS1 and IRS2 through ubiquitination.
Chronic liver diseases are associated with glucose intolerance called hepatogenous diabetes.1 Glucose intolerance impairs sustained response rate to anti-viral therapy in patients with chronic hepatitis C virus (HCV) infection2 and is a risk factor for development of hepatocellular carcinoma3 as well as long-term survival in patients with cirrhosis.4 Several epidemiological studies have revealed an association between HCV infection and type 2 diabetes mellitus (DM) in cirrhotic patients.5–13 Case-cohort analysis confirms an increased risk for type 2 DM in cirrhotic patients with HCV infection.14 Cirrhotic patients with HCV infection are twice as likely to have type 2 DM than patients with hepatitis B virus (HBV) infection.6,7,12 Thus, epidemiological data show that HCV infection antedates type 2 DM. It is, however, difficult to prove that HCV itself triggers glucose intolerance in patients with liver cirrhosis. Various factors such as reduced glucose uptake,15 porto-systemic shunting,16 and impaired glucagon metabolism17 are also involved in glucose metabolism in patients with liver cirrhosis. Although glucose intolerance may occur even in the early stage of HCV infection, changes in glucose metabolism in noncirrhotic patients are not evident. To ascertain if HCV infection directly causes glucose intolerance, changes in glucose metabolism in noncirrhotic patients with various hepatobiliary disorders were investigated.
The liver plays a major role in regulation of glucose metabolism because it is the main source of endogenous glucose and the major site involved in insulin metabolism.18,19 Thus, hepatic factors may be involved in HCV-associated glucose intolerance. However, the pathogenic mechanisms for HCV-associated glucose intolerance remain unclear. Insulin exerts many biological effects through insulin receptor substrate (IRS) 1 and IRS2. Disruption of IRS1 results in insulin resistance, but not DM, because of compensatory hyperinsulinemia.20,21 Disruption of IRS2 results in severe DM because of insulin resistance and disturbance of insulin secretion.22 Thus, IRS1 and IRS2 are the molecules that augment the specificity of the insulin-signaling cascade and play a central role in insulin-mediated glucose metabolism.
HCV chronically infects hepatocytes. HCV may escape from the host immune response by suppressing cytokine signaling. We recently showed that HCV core up-regulates suppressor of cytokine signaling (SOCS) 3 expression.23 Although SOCS3 is known to be a negative regulator for cytokine signaling such as interleukin-6, growth hormone, and interferon-α, the role of SOCS3 on HCV-associated glucose intolerance has never been investigated. The aims of this study were to investigate changes in glucose metabolism in noncirrhotic patients with various hepatobiliary disorders and the molecular mechanisms for HCV-associated glucose intolerance.
Materials and Methods
Materials
All reagents were purchased from Wako Pure Chemical Industries (Osaka, Japan) unless otherwise indicated. Affinity-purified polyclonal rabbit anti-SOCS3 antibody was generated against synthetic peptide SKFPAAGMSRPLDTSLRL (Immuno-Biological Laboratories, Gunma, Japan).
Patients
A total of 357 patients with chronic hepatitis C (n = 158), chronic hepatitis B (n = 54), autoimmune hepatitis (AIH) (n = 36), fatty liver (n = 40), primary biliary cirrhosis (PBC) (n = 49), or histologically normal livers (CON; n = 20) were studied retrospectively during the period from January 1997 to August 2003 at Kurume University Hospital. All of the patients were untreated and hospitalized for diagnostic liver biopsy. All of the diagnoses were based on clinical, serological, and histological evidence. Domestic data were collected at the time of liver biopsy including age, sex, and alcohol use. Body mass index (BMI) was calculated as body weight in kg divided by the square of height in meters (kg/m2). Some liver diseases such as AIH and PBC show gender differences, and it is also possible that HCV infection affects BMI. Therefore, age, sex, BMI, and biochemical parameters were not matched among the groups to reduce selection bias. Patients with other causes of liver disease, in particular those known to be involved in the pathogenesis of diabetes such as hemochromatosis or alcoholic liver disease (on the basis of histology or a history of excessive alcohol consumption) were excluded, as were those who had been taking corticosteroids or with a history of, or evidence of, pancreatitis or a pancreatic tumor. The study protocol was approved by the institutional review board, and informed consent for participation in the study was obtained from each subject. None of the patients was institutionalized.
Laboratory Determinations
Venous blood samples were taken in the morning after a 12-hour overnight fast. Plasma glucose levels were measured by a glucose oxidase method. Serum insulin levels were measured by using a sandwich enzyme immunoassay kit (Eiken Chemical, Tokyo, Japan). β-Cell function and insulin resistance were calculated on the basis of fasting levels of plasma glucose and insulin, according to the homeostasis model assessment (HOMA) method.24 The formulas for the HOMA model are as follows: β-cell function (HOMA-β) = fasting insulin (μU/ml) × 360/(fasting glucose (mg/dl) − 63); insulin resistance (HOMA-IR) = fasting glucose (mg/dl) × fasting insulin (μU/ml)/405.
Determination of HCV Genotype and Measurement of HCV Core
HCV genotype was determined by polymerase chain reaction with type-specific primers and HCV genotypes were classified according to classification system of Simmonds and colleagues.25 Unselected serum samples (n = 58) were assayed for HCV core by using a newly developed HCV core antigen enzyme-linked immunosorbent assay test system (Ortho-Clinical Diagnostics K.K., Tokyo, Japan) as previously described.26 This assay has high stability and reproducibility under all conditions and the detection limit is 44 fmol/L.
Histological Data
For each patient, a liver biopsy specimen was fixed in 10% formalin buffer and stained with hematoxylin and eosin. Liver biopsy specimens were evaluated by a single experienced pathologist who was unaware of the patients’ clinical and laboratory data. The specimens were scored according to the METAVIR scoring system, which is suited for evaluation of chronic hepatitis C.27 Activity was graded according to the intensity of necroinflammatory lesions: 0, no activity; 1, mild activity; 2, moderate activity; and 3, severe activity. The stage of fibrosis was scored as follows: 0, no fibrosis; 1, portal fibrosis without septa; 2, portal fibrosis with few septa; 3, portal fibrosis with many septa; and 4, cirrhosis.
Immunohistochemistry
Paraffin-embedded liver sections from patients with HCV infection were deparaffinized and subjected to immunohistochemical staining using a Vectastain ABC kit (Vector Laboratories, Burlingame, CA) with an anti-human IRS1 polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) or an anti-human IRS2 polyclonal antibody (Santa Cruz Biotechnology), and developed with 3,3′-diaminobenzidine. The primary antibodies for IRS1 and IRS2 were used at a 1:100 dilution. The specificity of IRS1 and IRS2 staining was confirmed by immunization using an excess amount of the N-terminal peptide of IRS1 and IRS2.
Immunoblotting
Immunoblotting was performed as previously described28,29 using antibodies against the following: IRS1, IRS2, insulin receptor (Chemicon, Temecula, CA), SOCS-3, Myc (Santa Cruz Biotechnology), phospho-(Tyr) p85 subunit of phosphatidylinositol 3-kinase (PI3K; Cell Signaling Technologies, Beverly, MA), phospho-(Ser 473)-Akt (Cell Signaling Technologies), signal transducer and activation of transcription (STAT) 5 (Santa Cruz Biotechnology), or ubiquitin (Santa Cruz Biotechnology). Equal amounts of protein (40 μg) from liver homogenates or cell extracts were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis on a 7.5% acrylamide gel. The resolved proteins were transferred electrophoretically onto polyvinylidene difluoride membranes (Amersham Int., Buckinghamshire, UK). The membranes were incubated with the primary antibodies indicated in each figure, and were subsequently incubated by the secondary antibodies: a horseradish peroxidase-conjugated goat anti-mouse IgG (Amersham Int.) and a horseradish peroxidase-conjugated goat anti-rabbit IgG (Amersham Int.). The membranes were then incubated with chemiluminescence reagents (ECL kit; Amersham Int.) and immediately exposed on radiograph film. In the experiment for ubiquitination of IRS1 and IRS2 (Figure 4, b and c), cell extracts were immunoprecipitated with anti-IRS1 or anti-IRS2 antibodies and then immunoblotted with anti-ubiquitin antibody as previously described.23
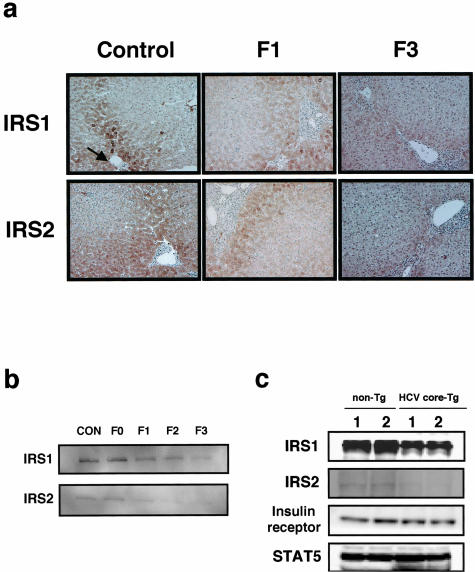
Figure 4.
Protein expression levels of IRS1 and IRS2 in livers from patients with HCV infection and HCV core-transgenic (Tg) mice. a: Immunostaining for IRS1 and IRS2. IRS1 (top column) and IRS2 (bottom column) staining of liver sections from control (left column), F1 (middle column), and F3 (right column). Expression of IRS1 and IRS2 were visualized by 3,3′-diaminobenzidine (brown). Arrow indicates portal vein. b: Immunoblotting for IRS1 and IRS2. Proteins (40 μg) in liver extracts from control and patients with HCV infection were immunoblotted with anti-IRS1 antibodies (top column) or anti-IRS2 antibodies (bottom column). c: Hepatic IRS1 and IRS2 expression in HCV core-Tg mice. Non-Tg littermates and HCV core Tg-mice livers (8 weeks old) were subjected to immunoblotting for IRS1, IRS2, insulin receptor, and STAT5 (as a reference protein). These experiments were repeated three times and representative immunoblotting and immunostaining images are shown. Original magnifications, ×400.
Core and HCV cDNA
The HCV core region (573 nucleotides) was amplified by reverse transcriptase- polymerase chain reaction, using HCV RNA as a template extracted from the serum of a patient with HCV (genotype Ib) infection. The nucleotide sequence is 98% identical and the amino acid sequence is 100% identical to those of HCV strain MD7-1.30 The HCV core region was subcloned into expression vector pcDNA3 with the NH2-terminal Myc tag. Expression of all HCV proteins, including the core, envelope proteins (E1and E2), and nonstructural proteins (NS1, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) were confirmed by immunoblotting in our previous study.23
HCV Core-Transgenic Mice
The transgenic mouse was generated using a construct carrying the HCV core cDNA (genotype 1b) fused to the promoter of the HBV X gene.23 Transgenic mice were developed using conventional methods (C57BL/6 × DBA/2). F1 mice were used to obtain fertilized eggs and those founder mice were mated with C57BL/6 mice for more than five generations. The expression of HCV core was expressed in various tissues including brain, heart, lung, kidney, thymus, and liver. All animal experiments were conducted in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals and were approved by the University of Kurume Institutional Animal Care and Use Committee.
Cells and Transfection
HepG2 cells derived from a human hepatoblastoma and retaining many of the differentiated features of mature hepatocytes,31 Huh 7 and HLF cells derived from hepatocellular carcinomas,32 and primary mouse embryonic fibroblast (MEF) cells from heterozygous (SOCS3−/−) SOCS3 knockout mice and from wild-type (SOCS3+/+) mice were cultured in Dulbecco’s modified Eagle’s medium supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin, and 10% dialyzed fetal bovine serum (Life Technologies, Gaithersburg, MD). The expression vector carrying the HCV core region was transfected using synthetic liposomes (Lipofectamine 2000; Life Technologies) in Opti-MEM I (Life Technologies) as previously described.33,34 Cell extracts for each experiment were prepared 24 hours after transfection. In some experiments, 10 μmol/L of carbobenzoxy-l-leucyl-l-leucyl-l-leucinal (MG132; Peptide Institute, Osaka, Japan), a proteosomal proteolysis inhibitor, was mixed with cDNA of Myc-tagged HCV core and incubated for 1 hour. To examine the effects of HCV core on insulin signaling, stable transfectants of HCV core in the HLF were used.
Assay for 6-Phosphofructo-2-Kinase (EC 2.7.1.1; Fru 6-P,2-Kinase)
The activity of Fru 6-P,2-kinase was assayed by measuring formation of fructose 2,6-bisphosphate as described previously.35
Statistical Analysis
All data are expressed as mean ± SD. Differences between two groups were analyzed using the Mann-Whitney U-test. Statistical comparisons among multiple groups were performed by analysis of variance followed by Scheffé’s post hoc test using StatView Power PC version for Macintosh (version 5.1; SAS Institute, Cary, NC). P values <0.05 were considered significant.
Results
Characteristics of All Patients
We enrolled 337 patients with noncirrhotic chronic hepatobiliary diseases and 20 controls (histologically normal liver). Clinical and laboratory data for these patients are summarized in Table 1. All patients were Japanese. Patients with HBV infection and fatty liver were younger than the other groups and females constituted more than 80% of the group of patients with AIH and PBC. Serum aspartate aminotransferase and alanine aminotransferase levels were increased in all of the groups except for controls. There was no significant difference in BMI among all of the groups. Serum albumin levels and bilirubin levels were normal in all of the groups.
Table 1.
Characteristics of All Patients
| Control | CH-C | CH-B | AIH | Fatty liver | PBC | |
|---|---|---|---|---|---|---|
| Number | 20 | 158 | 54 | 36 | 40 | 49 |
| Age (yr) | 52 ± 10 | 53 ± 8 | 42 ± 9 | 50 ± 11 | 44 ± 7 | 51 ± 8 |
| Sex | ||||||
| Female | 12 (60.0%) | 63 (39.9%) | 29 (53.7%) | 31 (86.1%) | 23 (57.5%) | 41 (83.7%) |
| Male | 8 (40.0%) | 95 (60.1%) | 25 (46.3%) | 5 (13.9%) | 17 (42.5%) | 8 (16.3%) |
| Body mass index (kg/m2) | 22.3 ± 1.9 | 22.8 ± 2.0 | 22.0 ± 1.5 | 22.2 ± 1.8 | 23.2 ± 3.0 | 22.5 ± 1.8 |
| Aspartate aminotransferase (U/l) | 25 ± 16 | 70 ± 29 | 86 ± 57 | 71 ± 39 | 35 ± 37 | 23 ± 12 |
| Alanine aminotransaminase (U/l) | 28 ± 18 | 77 ± 36 | 94 ± 56 | 85 ± 50 | 43 ± 33 | 33 ± 13 |
| Albumin (g/dl) | 3.8 ± 0.3 | 3.8 ± 0.3 | 3.7 ± 0.3 | 3.7 ± 0.4 | 4.0 ± 0.4 | 3.7 ± 0.3 |
| Total bilirubin (mg/dl) | 0.6 ± 0.2 | 0.7 ± 0.2 | 0.6 ± 0.3 | 1.0 ± 0.8 | 0.6 ± 0.2 | 0.7 ± 0.3 |
Note. Data are expressed as mean ± SD or number of patients. All patients were Japanese. All the diagnoses are based on clinical, serological, and histological evidences.
CH-C, chronic hepatitis C; CH-B, chronic hepatitis B; AIH, autoimmune hepatitis; PBC, primary biliary cirrhosis.
Changes in Glucose Metabolism in Patients with Various Chronic Liver Diseases
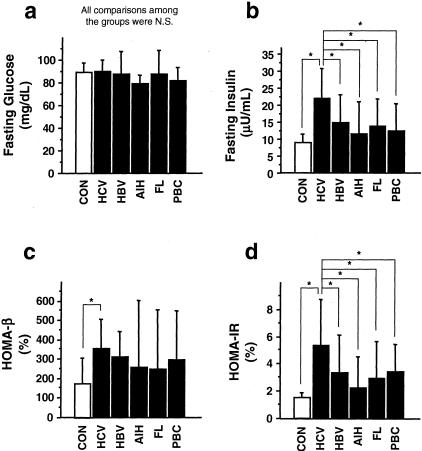
Fasting glucose levels were within normal range in all of the groups and showed no significant differences among the groups. However, fasting insulin levels were ∼1.5 times higher in patients with HCV infection compared to the other groups (Figure 1, a and b). β-Cell function and insulin resistance were evaluated by HOMA-β and HOMA-IR, respectively. HOMA-β levels were increased in patients with HCV infection compared to controls (Figure 1c). HOMA-IR levels were significantly higher in patients with HCV infection compared to the other groups (Figure 1d).
Figure 1.
Fasting glucose and insulin levels, β-cell function, and insulin resistance in patients with various chronic liver diseases. Fasting plasma glucose (a) and fasting serum insulin (b) were measured. HOMA-β (c) and HOMA-IR (d) were calculated (see Materials and Methods). Values were expressed as mean ± SD. The comparisons between the groups were made using analysis of variance with Scheffé’s post hoc test. N.S., not significant. *, P < 0.05. CON, histologically normal livers as controls (n = 20); HCV, chronic hepatitis C virus infection (n = 158); HBV, chronic hepatitis B virus infection (n = 54); AIH, autoimmune hepatitis (n = 36); FL, fatty liver (n = 40); PBC, primary biliary cirrhosis (n = 49).
The Involvement of Virological Factors in HCV-Associated Hyperinsulinemia
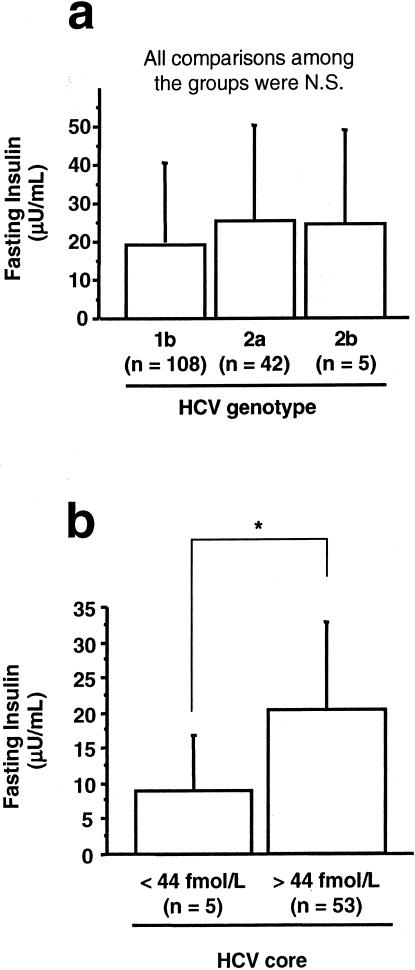
All serum samples from patients with HCV infection were HCV-RNA-positive. HCV genotypes were classified according to the classification system of Simmonds and colleagues.25 Genotype 1a and 3 were not found in any sample. Three samples were excluded in this analysis because of mixed HCV genotypes or undetermined HCV genotype. There was no significant difference in fasting insulin levels among different HCV genotypes (Figure 2a). On the other hand, in patients with >44 fmol/L of HCV core, fasting insulin levels were significantly elevated compared to patients with undetectable levels (<44 fmol/L) of HCV core (Figure 2b).
Figure 2.
The involvement of virological factors in HCV-associated hyperinsulinemia. a: HCV genotypes and fasting insulin levels. Three cases that showed mixed or undetermined HCV genotypes were excluded. Values were expressed as mean ± SD. The comparisons between the groups were made using analysis of variance with Scheffé’s post hoc test. N.S., not significant. b: HCV core and fasting insulin levels. Values were expressed as mean ± SD. The comparison between the two groups was made using the Mann-Whitney U-test. *, P < 0.05.
Histological Parameters and Fasting Insulin Levels
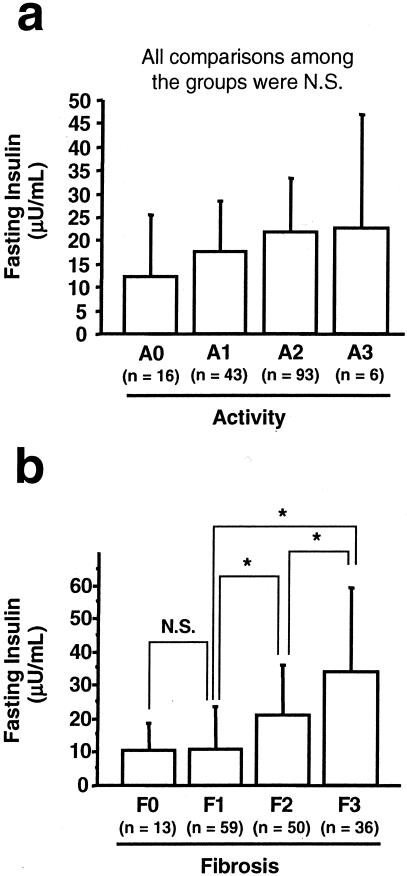
Liver specimens were evaluated according to the METAVIR system.27 There was no significant difference in fasting insulin levels among different activities (Figure 3a). Although no significant difference was seen in fasting insulin levels between F0 and F1, fasting insulin levels were significantly increased in F2 compared to those of F0 and F1. Fasting insulin levels in F3 were elevated more than those of F0, F1, and F2 (Figure 3b). A similar relationship was also found between HOMA-IR levels and the degree of hepatic fibrosis (data not shown).
Figure 3.
Histological parameters and fasting insulin levels in patients with HCV infection. The liver specimens were evaluated according to the METAVIR system (see Materials and Methods). a: Activity and fasting insulin levels. b: Fibrosis and fasting insulin levels. Values were expressed as mean ± SD. The comparisons between groups were made using analysis of variance with Scheffé’s post hoc test. N.S., not significant. *, P < 0.05.
Protein Expression Levels of IRS1 and IRS2 in the Liver from Patients with HCV Infection and HCV Core-Tg Mice
The protein expression levels of IRS1 and IRS2 in liver samples from controls and patients with HCV infection were examined by immunostaining and immunoblotting. In control livers, immunostaining demonstrated that periportal hepatocytes, rather than perivenular hepatocytes, highly expressed both IRS1 and IRS2, showing lobular heterogeneity of IRS1 and IRS2 expression (Figure 4a). Decreased IRS1 and IRS2 expression levels along with progression of hepatic fibrosis were seen in periportal hepatocytes. Immunoblotting showed that IRS1 and IRS2 expression levels decreased with the progression of hepatic fibrosis in livers from patients with HCV infection (Figure 4b), confirming results of immunostaining. We developed HCV core-Tg mice in which the HCV core protein is expressed ubiquitously. The HCV core-Tg mouse is an informative animal model for studying the pathogenesis of HCV infection.36,37 In two independent HCV core-Tg mice, decreased expression of hepatic IRS1 and IRS2, but not of insulin receptor, were also demonstrated compared to two independent wild-type littermates (Figure 4c).
The Effect of HCV Core on IRS1, IRS2, SOCS3, and Insulin Receptor Expression
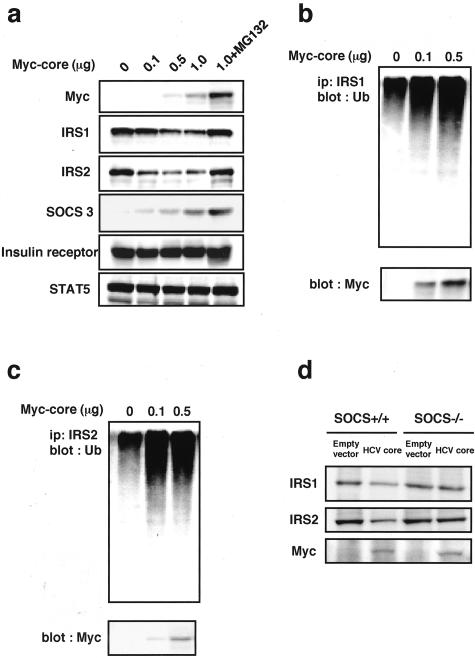
The effects of HCV core on IRS1, IRS2, SOCS3, and insulin receptor expression were examined in HepG2 and Huh7 cells prepared by transient transfection with Myc-tagged HCV core and HLF cells with stable transfection of Myc-tagged HCV core. Expression of Myc-tagged HCV core was confirmed by immunoblotting for Myc in both cells. HCV core dose dependently decreased IRS1 and IRS2 expression in HepG2 cells (Figure 5a). In contrast, SOCS3 expression was dose dependently increased by transient transfection with Myc-tagged HCV core in HepG2 cells. No changes in insulin receptor expression and STAT5 (used as a reference protein) were seen in HepG2 cells transfected with Myc-tagged HCV-core (Figure 5a). We also analyzed the effects of a proteosomal proteolysis inhibitor, MG132. The treatment with MG132 caused an increase in expression levels of IRS1 and IRS2 (Figure 5a). Similar results were obtained in Huh7 cells with transient transfection of HCV core and HLF cells with stable transfection of HCV core (data not shown).
Figure 5.
The effect of HCV core on insulin signaling molecules. a: The effect of HCV core on IRS1, IRS2, SOCS3, and insulin receptor expression. Myc-tagged HCV core was transiently expressed in HepG2 cells. Twenty-four hours after transfection the cell extracts were immunoblotted with the indicated antibodies. MG132 (10 μmol/L) was added with cDNA of Myc-tagged HCV core and incubated for 1 hour. STAT5 was used as a reference protein. b and c: The identification of ubiquitinated IRS1 and IRS2. Whole-cell extracts (40 μg of crude extract) were subjected to immunoprecipitation with IRS1 or IRS2 and followed by immunoblotting using anti-ubiquitin monoclonal antibody. d: The effect of HCV core on IRS1 and IRS2 expression in SOCS3−/− MEF cells. Twenty-four hours after transfection of HCV core, MEF cell extracts were immunoblotted with anti-IRS1 antibodies or anti-IRS2 antibodies. Representative immunoblotting images from three separate experiments are shown.
Role of Ubiquitination in the Regulation of IRS1 and IRS2 Expression
To investigate the involvement of ubiquitination in down-regulation of IRS1 and IRS2 in HepG2 cells transfected with HCV core, whole-cell extracts were immunoprecipitated with anti-IRS1 or anti-IRS2 antibodies and immunoblotted with anti-ubiquitin monoclonal antibodies. HCV core caused an accumulation of ubiquitin-conjugated IRS1 (Figure 5b) and IRS2 (Figure 5c). Expression of Myc-tagged HCV core was confirmed by immunoblotting for Myc (Figure 5, b and c).
Role of SOCS3 in the Regulation of IRS1 and IRS2 Expression
Because SOCS3−/− mice display embryonic lethality, we examined the association between SOCS3 and regulation of IRS1 and IRS2 by using SOCS3−/− MEF cells. HCV core down-regulated IRS1 and IRS2 in SOCS3+/+ MEF cells. On the other hand, HCV core did not cause down-regulation of IRS1 and IRS2 in SOCS3−/− MEF cells (Figure 5d).
The Effects of HCV Core on Insulin Signaling
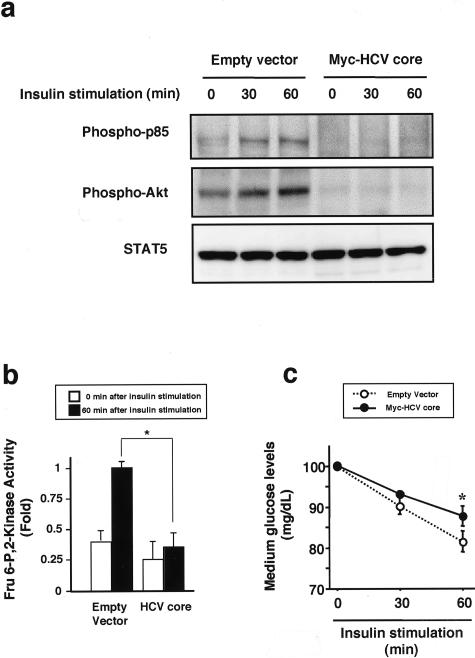
HLF cells with stable transfection of HCV core were treated with insulin (100 ng/ml) from 0 minutes to 60 minutes and phosphorylation of p85 subunit of PI3K and Akt were determined. Insulin-induced phosphorylation of p85 subunit of PI3K and Akt was observed in HLF cells transfected with empty vector. On the other hand, HCV core decreased phosphorylation of p85 subunit of PI3K and Akt at the base line (0 minutes) and inhibited insulin-induced phosphorylation of p85 subunit of PI3K and Akt (Figure 6a). STAT5 protein (reference protein) levels were unchanged by insulin stimulation. Insulin activated Fru 6-P, 2-kinase, a downstream of Akt signal and one of the potent regulators of glycolysis, and decreased medium glucose levels (Figure 6, b and c). HCV core suppresses insulin-induced activation of Fru 6-P, 2-kinase and decreases in medium glucose levels (Figure 6, b and c).
Figure 6.
The effects of HCV-core on insulin signaling. HLF cells with transfection of empty vector or Myc-HCV core were incubated in the presence of insulin (100 ng/ml) for 60 minutes. a: The effect of HCV-core on insulin-induced phosphorylation of p85 subunit of phosphatidylinositol 3-kinase (PI3K) and Akt. Whole-cell lysates were subjected to immunoblotting for phospho-p85 subunit of PI3K and phospho-Akt. STAT5 was used as reference protein. Representative immunoblotting images from three separate experiments are shown. b: The effect of HCV-core on insulin-induced activation of Fru 6-P, 2-kinase. Sixty minutes after insulin stimulation, Fru 6-P, 2-kinase activity in whole-cell lysates was assayed. Values were expressed as mean ± SD. The comparison between the two groups was made using the Mann-Whitney U-test. *, P < 0.05. c: The effect of HCV-core on insulin-induced activation of glucose uptake. Glucose levels in culture medium were measured at 0, 30, and 60 minutes after insulin stimulation. Values were expressed as mean ± SD. The comparisons between groups were made using analysis of variance with Scheffé’s post hoc test. N.S., not significant. *, P < 0.05.
Discussion
In this study, we showed that more severe insulin resistance was present in noncirrhotic patients with HCV infection than in patients with other hepatobiliary diseases. Insulin resistance was associated with the presence of serum HCV core, the severity of hepatic fibrosis and decreased expression of hepatic IRS1 and IRS2 in patients with HCV infection. HCV core down-regulated the expression of IRS1 and IRS2 in human hepatoma cell lines as well as in whole animals. These findings suggest that HCV causes changes in specific hepatic molecules regulating glucose metabolism and results in severe insulin resistance. A possible mechanism is that HCV core-induced SOCS3 promotes proteosomal degradation of IRS1 and IRS2 through ubiquitination.
Although fasting glucose levels were similar among all of the groups, fasting insulin and HOMA-IR levels, a indicator of insulin resistance, were significantly increased in patients with HCV infection compared to the other hepatobiliary disorders. β-Cell function evaluated by HOMA-β was also increased in patients with HCV infection compared to controls. These findings indicate that HCV infection induced insulin resistance and fasting glucose levels were compensated by hyperinsulinemia. Mangia and colleagues38 reported no association between HCV infection and DM in noncirrhotic patients and that the prevalence of DM in noncirrhotic patients is comparable to the expected prevalence in the general population. These results are not in accord with our results and this discrepancy may be explained by different evaluation methods for glucose intolerance. They evaluated glucose intolerance by fasting glucose levels and only patients who had >126 mg/dl of glucose levels were considered as abnormal glucose metabolizers. Because fasting glucose levels are compensated by hyperinsulinemia (Figure 1, a and b), cryptic changes in glucose metabolism can be evaluated by measuring fasting insulin or HOMA-IR levels.24 In our own study, we provide convincing evidence that more severe insulin resistance is present in noncirrhotic patients with HCV infection than in patients with other hepatobiliary disorders. Unique mechanisms may underlie HCV-associated severe insulin resistance.
HCV genotype 2a is specifically linked with extrahepatic manifestations such as cryoglobulinemia and benign monoclonal gammopathy.39,40 Mason and colleagues6 also reported an association between genotype 2a and DM, whereas no association was found between fasting insulin levels and HCV genotypes in our study. The limited number of patients may prevent drawing definite conclusions in both studies. HCV core can modulate cell signaling.23 Therefore, we investigated the relationship between HCV core and fasting insulin levels. In patients with undetectable levels of HCV core, fasting insulin levels were within the normal range. In contrast, in patients with detectable levels of HCV core, fasting insulin levels were increased. Thus, HCV core seems to play a crucial role in HCV-associated insulin resistance.
Then, we examined an association between histological parameters and insulin resistance. Although no significant association was found between activity and insulin resistance, serum insulin levels and HOMA-IR levels were significantly increased with the severity of hepatic fibrosis. These data are in good agreement with recent report by Hui and colleagues.13 HOMA-IR is independently associated with an increased rate of fibrosis progression.13,41 Insulin stimulates hepatic stellate cells to proliferate and secrete extracellular matrix.42 Thus, it appears that insulin resistance contributes to fibrotic progression in patients with HCV infection. To innovate therapies for prevention of fibrotic progression, it is important to investigate the molecular mechanisms for HCV-induced insulin resistance.
IRS1 and IRS2 act as important mediators of insulin action and down-regulation of hepatic IRS1 and IRS2 results in an increase in hepatic insulin resistance. Knockout of the IRS1 gene induces insulin resistance and subsequent compensatory hyperinsulinemia.20 Knockout of the IRS2 gene causes severe diabetes as a consequence of insulin resistance and disturbance of insulin secretion.22 We showed that decrease in expression of IRS1 and IRS2 was associated with the progression of hepatic fibrosis. Down-regulation of IRS1 and IRS2 was also seen in livers from HCV core-Tg mice. These data suggest that down-regulation of IRS1 and IRS2 is responsible for compensatory hyperinsulinemia and progression of hepatic fibrosis.
The effects of HCV core on the expression of IRS1 and IRS2 were investigated by simplified in vitro experiments. HCV is a positive-strand RNA virus consisting of a putative structure (core, E1, E2/p7) and at least six nonstructural proteins (NS2, NS3, NS4A, NS5A, NS5B). HCV core is implicated in cellular transformation.37 In this study, HCV core decreased expression of IRS1 and IRS2 in human hepatoma cell lines. These in vitro findings add weight to the results of our human studies.
One of the negative modulation mechanisms of IRS1 and IRS2 is proteosomal degradation.43 On the other hand, we previously reported that HCV core induced SOCS3 in mouse fibroblast NIH 3T3 cells.23 In the current study, HCV core-induced SOCS3 was also validated in HepG2 cells as well as Huh7 cells and HLF cells. The SOCS family of proteins has similar structural characteristics referred to collectively as a “SOCS box,” a unique NH2-terminal domain of variable length, a central Src-homology 2 domain, and a COOH-terminal, with this structural resemblance reflecting functional similarities among SOCS proteins. The SOCS box acts as an adaptor to facilitate the ubiquitination of signaling proteins and their subsequent targeting to the proteosome by complexing with Elongins B and C.43,44 These facts and our findings led to the assumption that HCV core-induced SOCS3 promoted proteosomal degradation of IRS1 and IRS2 through ubiquitination. To test this hypothesis, HCV core-transfected cells were incubated with MG132, a potent proteosomal proteolysis inhibitor. MG132 blocked HCV core-induced decrease of IRS1 and IRS2 in HepG2. Ubiquitination of IRS1 and IRS2 was increased by transfection of HCV core. Moreover, HCV core did not cause down-regulation of IRS1 and IRS2 in SOCS3−/− MEF cells. All of these data support our hypothesis. Transient overexpression of SOCS3 in mouse liver induces fasting hyperglycemia, and fasting hyperinsulinemia. These changes are returned to normal as SOCS3 expression subsided.43 Thus, studies in whole animals lend added credence to our hypothesis that HCV core-induced SOCS3 promotes proteosomal degradation of IRS1 and IRS2 through ubiquitination.
To verify the biological significance of these studies on IRS1 and IRS2, the effects of HCV core on insulin signaling were examined. Insulin phosphorylates the p85 subunit of PI3K and Akt, which are downstream components of IRS in liver.45 Akt activates Fru 6-P,2-kinase, one of the key enzymes of glycolysis, and glucose uptake.46 HCV core inhibited insulin-induced phosphorylation of p85 subunit of PI3K and Akt, activation of Fru 6-P,2-kinase, and glucose uptake. Thus, HCV core-transfected HLF cells were resistant to insulin stimulation compared with empty vector-transfected HLF cells, suggesting biological significance of HCV core-induced down-regulation of IRS1 and IRS2.
Recently, Aytug and colleagues47 studied changes in the upstream insulin-signaling molecules in the liver specimens obtained from patients with chronic HCV infection and showed impairments of insulin-stimulated PI3K activity and Akt phosphorylation, which were in good agreements with our findings. However, our data for IRS1 differed from those reported by Aytug and colleagues.47 They showed increased IRS1 expression with reduction in tyrosine phosphorylation. Although the reason for this discrepancy remains unclear, one possibility is that their HCV-infected patients show higher BMI values than those of ours. Obesity is a well-recognized risk factor for the development of insulin resistance. Adipocytes secrete a large number of factors with diverse functions. Tumor necrosis factor-α and free fatty acids are secreted by adipocytes and are known to impair insulin signaling by reducing IRS1 tyrosine phosphorylation.48,49 Increased IRS1 expression may be for an adaptation to reduction of IRS1 tyrosine phosphorylation. Moreover, leptin, which is also secreted by adipocytes, is involved in the development of HCV-associated insulin resistance.50 Thus, there may exist several molecular mechanisms for HCV-associated insulin resistance.
In conclusion, more severe insulin resistance was present in noncirrhotic patients with HCV infection than in patients with other hepatobiliary diseases. We uncovered a unique mechanism of HCV-associated insulin resistance. HCV core-induced SOCS3 may promote proteosomal degradation of IRS1 and IRS2 through ubiquitination.
Acknowledgments
We thank Kyuichi Tanikawa, M.D., Ph.D., and Takahisa Eguchi, M.D., Ph.D., for helpful discussions; and Yumi Ogo, Takako Ohotsubo, Syunichi Hattori, and Hidetoshi Itoya (Ortho-Clinical Diagnostics, K.K.) for technical assistance.
Footnotes
Address reprint requests to Takumi Kawaguchi, M.D., Ph.D., Second Department of Medicine, Kurume University School of Medicine. 67 Asahi-machi, Kurume 830-0011, Japan. E-mail: takumi@med.kurume-u.ac.jp.
Supported, in part, by Grants-in-Aid for Scientific Research (C) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (no. 15590963 to T.K., no. 16590648 to M.S.).
T.K. and T.Y. contributed equally to this work.
A part of this study was presented at the 54th Annual Meeting of the American Association for the Study of Liver Diseases (Boston, MA) in October 2003 and published in abstract form (Hepatology 2003: 34:456A).
References
- Megyesi C, Samols E, Marks V. Glucose tolerance and diabetes in chronic liver disease. Lancet. 1967;2:1051–1056. doi: 10.1016/s0140-6736(67)90334-0. [DOI] [PubMed] [Google Scholar]
- Romero-Gomez M, Corpas R, Grande L, Castellano-Megias VM, Sanchez-Munoz D, del Rocio V, Vazquez-Albertino R. Insulin resistance impairs sustained response rate to antiviral therapy in patients with chronic hepatitis C. Hepatology. 2003;38:747A. [Google Scholar]
- Hassan MM, Hwang LY, Hatten CJ, Swaim M, Li D, Abbruzzese JL, Beasley P, Patt YZ. Risk factors for hepatocellular carcinoma: synergism of alcohol with viral hepatitis and diabetes mellitus. Hepatology. 2002;36:1206–1213. doi: 10.1053/jhep.2002.36780. [DOI] [PubMed] [Google Scholar]
- Bianchi G, Marchesini G, Zoli M, Bugianesi E, Fabbri A, Pisi E. Prognostic significance of diabetes in patients with cirrhosis. Hepatology. 1994;20:119–125. doi: 10.1016/0270-9139(94)90143-0. [DOI] [PubMed] [Google Scholar]
- Mehta SH, Brancati FL, Sulkowski MS, Strathdee SA, Szklo M, Thomas DL. Prevalence of type 2 diabetes mellitus among persons with hepatitis C virus infection in the United States. Ann Intern Med. 2000;133:592–599. doi: 10.7326/0003-4819-133-8-200010170-00009. [DOI] [PubMed] [Google Scholar]
- Mason AL, Lau JY, Hoang N, Qian K, Alexander GJ, Xu L, Guo L, Jacob S, Regenstein FG, Zimmerman R, Everhart JE, Wasserfall C, Maclaren NK, Perrillo RP. Association of diabetes mellitus and chronic hepatitis C virus infection. Hepatology. 1999;29:328–333. doi: 10.1002/hep.510290235. [DOI] [PubMed] [Google Scholar]
- Fraser GM, Harman I, Meller N, Niv Y, Porath A. Diabetes mellitus is associated with chronic hepatitis C but not chronic hepatitis B infection. Isr J Med Sci. 1996;32:526–530. [PubMed] [Google Scholar]
- Grimbert S, Valensi P, Levy-Marchal C, Perret G, Richardet JP, Raffoux C, Trinchet JC, Beaugrand M. High prevalence of diabetes mellitus in patients with chronic hepatitis C. A case-control study. Gastroenterol Clin Biol. 1996;20:544–548. [PubMed] [Google Scholar]
- Ozyilkan E, Erbas T, Simsek H, Telatar F, Kayhan B, Telatar H. Increased prevalence of hepatitis C virus antibodies in patients with diabetes mellitus. J Intern Med. 1994;235:283–284. doi: 10.1111/j.1365-2796.1994.tb01075.x. [DOI] [PubMed] [Google Scholar]
- Simo R, Hernandez C, Genesca J, Jardi R, Mesa J. High prevalence of hepatitis C virus infection in diabetic patients. Diabetes Care. 1996;19:998–1000. doi: 10.2337/diacare.19.9.998. [DOI] [PubMed] [Google Scholar]
- Zein NN, Abdulkarim AS, Wiesner RH, Egan KS, Persing DH. Prevalence of diabetes mellitus in patients with end-stage liver cirrhosis due to hepatitis C, alcohol, or cholestatic disease. J Hepatol. 2000;32:209–217. doi: 10.1016/s0168-8278(00)80065-3. [DOI] [PubMed] [Google Scholar]
- Caronia S, Taylor K, Pagliaro L, Carr C, Palazzo U, Petrik J, O’Rahilly S, Shore S, Tom BD, Alexander GJ. Further evidence for an association between non-insulin-dependent diabetes mellitus and chronic hepatitis C virus infection. Hepatology. 1999;30:1059–1063. doi: 10.1002/hep.510300416. [DOI] [PubMed] [Google Scholar]
- Hui JM, Sud A, Farrell GC, Bandara P, Byth K, Kench JG, McCaughan GW, George J. Insulin resistance is associated with chronic hepatitis C and virus infection fibrosis progression. Gastroenterology. 2003;125:1695–1704. doi: 10.1053/j.gastro.2003.08.032. [DOI] [PubMed] [Google Scholar]
- Mehta SH, Brancati FL, Strathdee SA, Pankow JS, Netski D, Coresh J, Szklo M, Thomas DL. Hepatitis C virus infection and incident type 2 diabetes. Hepatology. 2003;38:50–56. doi: 10.1053/jhep.2003.50291. [DOI] [PubMed] [Google Scholar]
- Petrides AS, De Fronzo RA. Failure of glucagon to stimulate hepatic glycogenolysis in well-nourished patients with mild cirrhosis. Metabolism. 1994;43:85–89. doi: 10.1016/0026-0495(94)90161-9. [DOI] [PubMed] [Google Scholar]
- Smith-Laing G, Sherlock S, Faber OK. Effects of spontaneous portal-systemic shunting on insulin metabolism. Gastroenterology. 1979;76:685–690. [PubMed] [Google Scholar]
- Sherwin R, Joshi P, Hendler R, Felig P, Conn HO. Hyperglucagonemia in Laennec’s cirrhosis. The role of portal-systemic shunting. N Engl J Med. 1974;290:239–242. doi: 10.1056/NEJM197401312900502. [DOI] [PubMed] [Google Scholar]
- Kadowaki T. Insights into insulin resistance and type 2 diabetes from knockout mouse models. J Clin Invest. 2000;106:459–465. doi: 10.1172/JCI10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki K, Yamauchi T, Tamemoto H, Tobe K, Yamamoto-Honda R, Kaburagi Y, Akanuma Y, Yazaki Y, Aizawa S, Nagai R, Kadowaki T. Restored insulin-sensitivity in IRS-1-deficient mice treated by adenovirus-mediated gene therapy. J Clin Invest. 2000;105:1437–1445. doi: 10.1172/JCI7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki E, Lipes MA, Patti ME, Bruning JC, Haag B, III, Johnson RS, Kahn CR. Alternative pathway of insulin signalling in mice with targeted disruption of the IRS-1 gene. Nature. 1994;372:186–190. doi: 10.1038/372186a0. [DOI] [PubMed] [Google Scholar]
- Tamemoto H, Kadowaki T, Tobe K, Yagi T, Sakura H, Hayakawa T, Terauchi Y, Ueki K, Kaburagi Y, Satoh S, Sekihara H, Yoshioka S, Horikoshi H, Furuta Y, Ikawa Y, Kasuga M, Yazaki Y, Aizawa S. Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Nature. 1994;372:182–186. doi: 10.1038/372182a0. [DOI] [PubMed] [Google Scholar]
- Withers DJ, Gutierrez JS, Towery H, Burks DJ, Ren JM, Previs S, Zhang Y, Bernal D, Pons S, Shulman GI, Bonner-Weir S, White MF. Disruption of IRS-2 causes type 2 diabetes in mice. Nature. 1998;391:900–904. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Hanada T, Tokuhisa T, Kosai K, Sata M, Kohara M, Yoshimura A. Activation of STAT3 by the hepatitis C virus core protein leads to cellular transformation. J Exp Med. 2002;196:641–653. doi: 10.1084/jem.20012127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28:412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- Simmonds P, Holmes EC, Cha TA, Chan SW, McOmish F, Irvine B, Beall E, Yap PL, Kolberg J, Urdea MS. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993;74:2391–2399. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- Aoyagi K, Ohue C, Iida K, Kimura T, Tanaka E, Kiyosawa K, Yagi S. Development of a simple and highly sensitive enzyme immunoassay for hepatitis C virus core antigen. J Clin Microbiol. 1999;37:1802–1808. doi: 10.1128/jcm.37.6.1802-1808.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedossa P, Poynard T. An algorithm for the grading of activity in chronic hepatitis C. The METAVIR Cooperative Study Group. Hepatology. 1996;24:289–293. doi: 10.1002/hep.510240201. [DOI] [PubMed] [Google Scholar]
- Kawaguchi T, Sakisaka S, Sata M, Mori M, Tanikawa K. Different lobular distributions of altered hepatocyte tight junctions in rat models of intrahepatic and extrahepatic cholestasis. Hepatology. 1999;29:205–216. doi: 10.1002/hep.510290115. [DOI] [PubMed] [Google Scholar]
- Kawaguchi T, Sakisaka S, Mitsuyama K, Harada M, Koga H, Taniguchi E, Sasatomi K, Kimura R, Ueno T, Sawada N, Mori M, Sata M. Cholestasis with altered structure and function of hepatocyte tight junction and decreased expression of canalicular multispecific organic anion transporter in a rat model of colitis. Hepatology. 2000;31:1285–1295. doi: 10.1053/jhep.2000.7435. [DOI] [PubMed] [Google Scholar]
- Nagayama K, Kurosaki M, Enomoto N, Maekawa SY, Miyasaka Y, Tazawa J, Izumi N, Marumo F, Sato C. Time-related changes in full-length hepatitis C virus sequences and hepatitis activity. Virology. 1999;263:244–253. doi: 10.1006/viro.1999.9924. [DOI] [PubMed] [Google Scholar]
- Zannis VI, Breslow JL, SanGiacomo TR, Aden DP, Knowles BB. Characterization of the major apolipoproteins secreted by two human hepatoma cell lines. Biochemistry. 1981;20:7089–7096. doi: 10.1021/bi00528a006. [DOI] [PubMed] [Google Scholar]
- Nakabayashi H, Taketa K, Yamane T, Miyazaki M, Miyano K, Sato J. Phenotypical stability of a human hepatoma cell line, HuH-7, in long-term culture with chemically defined medium. Gann. 1984;75:151–158. [PubMed] [Google Scholar]
- Kawaguchi T, Takenoshita M, Kabashima T, Uyeda K. Glucose and cAMP regulate the L-type pyruvate kinase gene by phosphorylation/dephosphorylation of the carbohydrate response element binding protein. Proc Natl Acad Sci USA. 2001;98:13710–13715. doi: 10.1073/pnas.231370798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi T, Osatomi K, Yamashita H, Kabashima T, Uyeda K. Mechanism for fatty acid “sparing” effect on glucose-induced transcription: regulation of carbohydrate-responsive element-binding protein by AMP-activated protein kinase. J Biol Chem. 2002;277:3829–3835. doi: 10.1074/jbc.M107895200. [DOI] [PubMed] [Google Scholar]
- Kawaguchi T, Veech RL, Uyeda K. Regulation of energy metabolism in macrophages during hypoxia. Roles of fructose 2,6-bisphosphate and ribose 1,5-bisphosphate. J Biol Chem. 2001;276:28554–28561. doi: 10.1074/jbc.M101396200. [DOI] [PubMed] [Google Scholar]
- Honda A, Hatano M, Kohara M, Arai Y, Hartatik T, Moriyama T, Imawari M, Koike K, Yokosuka O, Shimotohno K, Tokuhisa T. HCV-core protein accelerates recovery from the insensitivity of liver cells to Fas-mediated apoptosis induced by an injection of anti-Fas antibody in mice. J Hepatol. 2000;33:440–447. doi: 10.1016/s0168-8278(00)80280-9. [DOI] [PubMed] [Google Scholar]
- Moriya K, Fujie H, Shintani Y, Yotsuyanagi H, Tsutsumi T, Ishibashi K, Matsuura Y, Kimura S, Miyamura T, Koike K. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat Med. 1998;4:1065–1067. doi: 10.1038/2053. [DOI] [PubMed] [Google Scholar]
- Mangia A, Schiavone G, Lezzi G, Marmo R, Bruno F, Villani MR, Cascavilla I, Fantasia L, Andriulli A. HCV and diabetes mellitus: evidence for a negative association. Am J Gastroenterol. 1998;93:2363–2367. doi: 10.1111/j.1572-0241.1998.00688.x. [DOI] [PubMed] [Google Scholar]
- Zignego AL, Ferri C, Giannini C, Monti M, La Civita L, Careccia G, Longombardo G, Lombardini F, Bombardieri S, Gentilini P. Hepatitis C virus genotype analysis in patients with type II mixed cryoglobulinemia. Ann Intern Med. 1996;124:31–34. doi: 10.7326/0003-4819-124-1_Part_1-199601010-00006. [DOI] [PubMed] [Google Scholar]
- Andreone P, Gramenzi A, Cursaro C, Bernardi M, Zignego AL. Monoclonal gammopathy in patients with chronic hepatitis C virus infection. Blood. 1996;88:1122. [PubMed] [Google Scholar]
- Sanyal AJ. Insulin resistance and tissue repair: a “fato-logical” phenomenon. Gastroenterology. 2003;125:1886–1889. doi: 10.1053/j.gastro.2003.10.021. [DOI] [PubMed] [Google Scholar]
- Svegliati-Baroni G, Ridolfi F, Di Sario A, Casini A, Marucci L, Gaggiotti G, Orlandoni P, Macarri G, Perego L, Benedetti A, Folli F. Insulin and insulin-like growth factor-1 stimulate proliferation and type I collagen accumulation by human hepatic stellate cells: differential effects on signal transduction pathways. Hepatology. 1999;29:1743–1751. doi: 10.1002/hep.510290632. [DOI] [PubMed] [Google Scholar]
- Rui L, Yuan M, Frantz D, Shoelson S, White MF. SOCS-1 and SOCS-3 block insulin signaling by ubiquitin-mediated degradation of IRS1 and IRS2. J Biol Chem. 2002;277:42394–42398. doi: 10.1074/jbc.C200444200. [DOI] [PubMed] [Google Scholar]
- Zhang JG, Farley A, Nicholson SE, Willson TA, Zugaro LM, Simpson RJ, Moritz RL, Cary D, Richardson R, Hausmann G, Kile BJ, Kent SB, Alexander WS, Metcalf D, Hilton DJ, Nicola NA, Baca M. The conserved SOCS box motif in suppressors of cytokine signaling binds to elongins B and C and may couple bound proteins to proteasomal degradation. Proc Natl Acad Sci USA. 1999;96:2071–2076. doi: 10.1073/pnas.96.5.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessin JE, Saltiel AR. Signaling pathways in insulin action: molecular targets of insulin resistance. J Clin Invest. 2000;106:165–169. doi: 10.1172/JCI10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprez J, Vertommen D, Alessi DR, Hue L, Rider MH. Phosphorylation and activation of heart 6-phosphofructo-2-kinase by protein kinase B and other protein kinases of the insulin signaling cascades. J Biol Chem. 1997;272:17269–17275. doi: 10.1074/jbc.272.28.17269. [DOI] [PubMed] [Google Scholar]
- Aytug S, Reich D, Sapiro LE, Bernstein D, Begum N. Impaired IRS-1/PI3-kinase signaling in patients with HCV: a mechanism for increased prevalence of type 2 diabetes. Hepatology. 2003;38:1384–1392. doi: 10.1016/j.hep.2003.09.012. [DOI] [PubMed] [Google Scholar]
- Feinstein R, Kanety H, Papa MZ, Lunenfeld B, Karasik A. Tumor necrosis factor-alpha suppresses insulin-induced tyrosine phosphorylation of insulin receptor and its substrates. J Biol Chem. 1993;268:26055–26058. [PubMed] [Google Scholar]
- Yu C, Chen Y, Cline GW, Zhang D, Zong H, Wang Y, Bergeron R, Kim JK, Cushman SW, Cooney GJ, Atcheson B, White MF, Kraegen EW, Shulman GI. Mechanism by which fatty acids inhibit insulin activation of insulin receptor substrate-1 (IRS-1)-associated phosphatidylinositol 3-kinase activity in muscle. J Biol Chem. 2002;277:50230–50236. doi: 10.1074/jbc.M200958200. [DOI] [PubMed] [Google Scholar]
- Oncul O, Top C, Cavuplu T. Correlation of serum leptin levels with insulin sensitivity in patients with chronic hepatitis-C infection. Diabetes Care. 2002;25:937. doi: 10.2337/diacare.25.5.937. [DOI] [PubMed] [Google Scholar]