Abstract
The tet(M) gene encodes a protein which is related to tetracycline ribosomal protection, one of the mechanisms of tetracycline resistance. A tet(M) gene that is 100% homologous to the tet(M) gene of Staphylococcus aureus has been found in a clinical isolate of Acinetobacter baumannii, which also carries the tet(A) gene encoding a tetracycline efflux pump.
Acinetobacter baumannii is an opportunistic nosocomial pathogen whose importance has steadily risen due to its facility to develop resistance to a great variety of antimicrobial agents, including tetracyclines (13). Tetracyclines are bacteriostatic antibiotics which inhibit protein synthesis by preventing the attachment of aminoacyl-tRNA to the ribosomal acceptor side (4). To date, the main mechanisms responsible for tetracycline resistance have been identified as (i) expression of efflux pumps and (ii) ribosomal protection. tet(M) is one of the genes related to the ribosomal protection function and, together with the tet(O) gene, is the most extensively characterized of the whole group. Moreover, this gene has generally been associated with conjugative chromosomal elements which code for their own transfer, such as Tn916 and Tn1545 (4).
To our knowledge, the only data published on the mechanisms of resistance to tetracycline in A. baumannii were those of Guardabassi et al. (5), who found the Tet A and Tet B determinants in clinical and aquatic strains of this microorganism. In their article, the authors also reported on the search for the presence of other determinants, including Tet M. Unfortunately, they did not succeed in this effort. In spite of the description of different tetracycline determinants in A. baumannii, little is known about the mechanisms in Acinetobacter that are responsible for tetracycline resistance. Thus, the aim of the present study was to analyze the molecular mechanisms of resistance to tetracycline in 15 clinical isolates of A. baumannii and to characterize the Tet M determinant found in one of the studied strains.
To achieve this aim, a total of 15 epidemiologically unrelated isolates of A. baumannii were recovered from different biological samples, mainly respiratory secretions, and submitted to the Clinical Laboratory of Microbiology of the Hospital Clinic of Barcelona, Spain (14). These isolates were identified as A. baumannii by using standard biochemical procedures according to the criteria of Bouvet and Grimont (3) and by amplified ribosomal DNA restriction analysis (ARDRA) (12). The MICs of tetracycline and minocycline were determined by using Mueller-Hinton medium (2) with the Etest (AB Biodisk, Sölna, Sweden) in accordance with the manufacturer's instructions. The MICs of tetracycline were also determined by a microdilution method, in accordance with the guidelines established by the National Committee for Clinical Laboratory Standards (8). The amplification of 2,039 bp corresponding to the whole gene of tet(M) in A. baumannii was established several times by PCR with the primers specific for this gene, namely, tetM1 (5′-TGGGCTTTTGAATGGAGGAA-3′) and tetM2 (5′-ATCTCCTCCTTTACACTTTA-3′), under the conditions previously described (9), except that the annealing temperature was 50 instead of 55°C. The prevalence of this gene was determined by PCR by analyzing the 15 epidemiologically unrelated clinical isolates of A. baumannii (14). Moreover, the presence of the tet(A) gene was established in these strains by using the conditions described by Guardabassi et al. (5). The PCR products of both the tet(A) and tet(M) genes were recovered from the agarose gel and purified with the Concert rapid purification system according to the manufacturer's instructions (Gibco BRL, Life Technologies Inc., Gaithersburg, Md.). The sample was then directly processed for DNA sequencing by using the dRhodamine terminator cycle sequencing kit and was analyzed with an automatic DNA sequencer (ABI PRISM 377; Perkin-Elmer, Emeryville, Calif.). The DNA sequencing for the tet(M) gene was repeated twice in order to avoid the effects of contamination.
Cloning and transformation procedures were performed with a TA cloning kit dual promoter (Invitrogen, Groningen, The Netherlands). The dot blot analysis was performed as described previously (10).
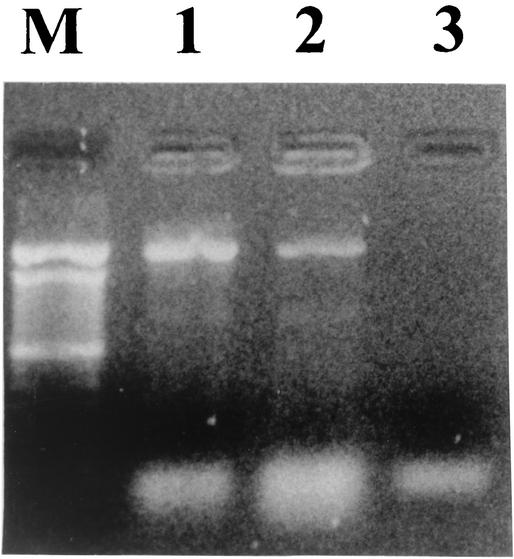
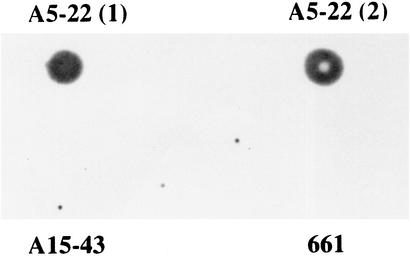
After performing dot blotting, amplification, and sequencing of the tet(M) gene, we found that the tet(M) gene was present in one clinical isolate of A. baumannii, A5-22 (Fig. 1 and 2), and that this gene was 100% homologous to the tet(M) gene identified in Staphylococcus aureus strain Mu50 (EMBL accession number AP003359) (6). Since our strain also carried a Tet A determinant (A. Ribera, I. Roca, J. Ruiz, I. Gibert, and J. Vila, submitted for publication), the tet(M) gene from the A. baumannii strain was cloned in a vector and transformed in an Escherichia coli DH5α strain to demonstrate the activity of Tet M by itself. The MICs of tetracycline and minocycline (Table 1) were 512 and 8 μg/ml, respectively, for the A. baumannii strain and 128 and 8 μg/ml, respectively, for the transformed E. coli strain, while the MICs of these antibiotics for the E. coli wild-type strain were 0.75 and 1.5 μg/ml, respectively. The differences between the MICs of tetracycline for the A. baumannii strain and the transformed strain could be explained by additional mechanisms of tetracycline resistance in the A. baumannii strain, such as the efflux determinant Tet A among others, which is able to pump tetracycline but not minocycline out of the cell (4).
FIG. 1.
PCR amplification of the tet(M) gene. Lane M, 100-bp DNA ladder(GIBCO-BRL, Life Technologies); lane 1, A. baumannii strain A5-22; lane 2, E. coli DH5α transformed strain; lane 3, E. coli DH5α wild-type strain.
FIG. 2.
Dot blot analysis using an amplified tet(M) product as the probe. The experiment was done with two different genomic DNA extracts from strain A5-22 and with the genomic DNA extract from strains 661 and A15-43, which contained the tet(A) gene but did not possess the tet(M) gene. The dot blot was positive for the two genomic DNA extracts of strain A5-22 and was negative for the other strains.
TABLE 1.
MICs of tetracycline and minocycline for the A. baumannii strain and for both transformed and wild-type E. coli strains
| Strain | MIC (μg/ml)
|
|
|---|---|---|
| Tetracycline | Minocycline | |
| A5-22 | 512 | 8 |
| E. coli (wild type) | 1 | 1 |
| E. coli (transformed)a | 128 | 8 |
Transformed with the plasmid pCR-II in which the tet(M) gene from the A. baumannii strain (A5-22) was cloned.
Previous experiments suggest that tetracycline resistance is inducible, although it is unclear whether this is due to the tet(A) gene, the tet(M) gene, or to other tetracycline resistance mechanisms.
Therefore, our finding is another example where a tet gene, such as tet(M), that has been identified as having a gram-positive origin has been identified in a gram-negative bacterium, in this case, A. baumannii. It is now known that the number of gram-negative bacteria carrying tet genes as well as other types of genes with gram-positive origins is increasing (1, 4, 7).
The results of the prevalence study (Table 2) showed a low frequency of the tet(M) gene in the clinical isolates (1 out of 15 isolates, or 6.6%). In contrast, the results suggested a higher prevalence of the tet(A) gene among these clinical isolates (6 out of 15 isolates, or 40%); these results are consistent with those of Guardabassi et al. (5), who found that tet(A) and tet(B) are the genes responsible for tetracycline resistance that are most frequently encountered in clinical isolates of A. baumannii. It is known that Tet A confers resistance to tetracycline but not to minocycline and that Tet B confers resistance to both antibiotics (4). Therefore, our results support these data, as the tet(A) gene was identified in the strains resistant to tetracycline but not to minocycline, while the gene was not found in the strains resistant to both antibiotics. These strains may possess the Tet B determinant, which would confer resistance to both antibiotics. Another possibility may be the presence of mutations in the 16S rRNA gene (11) in these strains, since they have recently been identified as conferring resistance to tetracyclines in Helicobacter pylori. However, further studies are necessary to confirm this hypothesis.
TABLE 2.
MICs of tetracycline and minocycline for 15 strains of A. baumannii and results of the PCR amplification of the tet(A) and tet(M) genes
| Strain | MIC (μg/ml)
|
PCR result
|
||
|---|---|---|---|---|
| Tetracycline | Minocycline | tet(A) | tet(M) | |
| A5-22 | 512 | 8 | + | + |
| 623 | 512 | 1 | + | − |
| F13 | 512 | 2 | + | − |
| 661 | 256 | 1 | + | − |
| A15-43 | 512 | 1 | + | − |
| AC236 | 256 | 1 | + | − |
| 6948V | 512 | 32 | − | − |
| 5985V | >512 | 32 | − | − |
| 215I | 512 | 32 | − | − |
| 709-R | >512 | 32 | − | − |
| 6F | >512 | 32 | − | − |
| O14-47 | >512 | 32 | − | − |
| L30 | 3 | 0.25 | − | − |
| 46I | 16 | 0.50 | − | − |
| 93 | 3 | 0.125 | − | − |
In the present study, the identification of the tet(M) gene in a clinical isolate of A. baumannii that also carries the tet(A) gene is a novel finding. The fact that the gene is 100% homologous to the tet(M) gene of S. aureus suggests a horizontal transference of genetic material between gram-positive and gram-negative bacteria.
Acknowledgments
We thank Stuart B. Levy and his laboratory staff (Department of Molecular Biology and Microbiology, Center for Adaptation Genetics and Drug Resistance, Tufts University School of Medicine, Boston, Mass.) for their advice regarding tetracycline resistance and for their generosity.
This study was supported by a grant from Fondo de Investigaciones Sanitarias (FIS 00/0997), Spain. A.R. has a fellowship from the Ministerio de Educación y Ciencia, Spain.
REFERENCES
- 1.Alonso, A., P. Sanchez, and J. L. Martínez. 2000. Stenotrophomonas maltophilia D457R contains a cluster of genes from gram-positive bacteria involved in antibiotic and heavy metal resistance. Antimicrob. Agents Chemother. 44:1778-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atlas, R. M. 1993. Handbook of microbiological media. CRC Press, Inc., London, United Kingdom.
- 3.Bouvet, P. J. M., and P. A. D. Grimont. 1986. Taxonomy of the genus Acinetobacter with the recognition of Acinetobacter baumannii sp. nov., Acinetobacter haemolyticus sp. nov., Acinetobacter johnsonii sp. nov., Acinetobacter junii sp. nov., and emended descriptions of Acinetobacter calcoaceticus and Acinetobacter lwoffii. Int. J. Syst. Bacteriol. 36:228-240. [Google Scholar]
- 4.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guardabassi, L., L. Dijkshoorn, J. M. Collard, J. E. Olsen, and A. Dalsgaard. 2000. Distribution and in-vitro transfer of tetracycline resistance determinants in clinical and aquatic Acinetobacter strains. J. Med. Microbiol. 49:929-936. [DOI] [PubMed] [Google Scholar]
- 6.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. Kobayashi, T. Tanaka, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 7.Luna, V. A., S. Cousin, Jr., W. L. H. Whittington, and M. C. Roberts. 2000. Identification of the conjugative mef gene in clinical Acinetobacter junii and Neisseria gonorrhoeae isolates. Antimicrob. Agents Chemother. 44:2503-2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.National Committee for Clinical Laboratory Standards. 2001. Performance standards for antimicrobial susceptibility testing. Approved standard M100-S11. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 9.Ruiz, J., M. M. Navia, C. Casals, J. M. Sierra, M. T. Jiménez de Anta, and J. Vila. Integron mediated antibiotic multiresistance in Acinetobacter baumannii clinical isolates. Clin. Microbiol. Infect. in press. [DOI] [PubMed]
- 10.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 11.Trieber, C. A., and D. E. Taylor. 2002. Mutations in the 16S rRNA genes of Helicobacter pylori mediate resistance to tetracycline. J. Bacteriol. 184:2131-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaneechoutte, M., L. Dijkshoorn, I. Tjernberg, A. Elaichouni, P. de Vos, G. Claeys, and G. Verschraegen. 1995. Identification of Acinetobacter genomic species by amplified ribosomal DNA restriction analysis. J. Clin. Microbiol. 33:11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vila, J. 1998. Mechanisms of antimicrobial resistance in Acinetobacter baumannii. Rev. Med. Microbiol. 9:87-97. [Google Scholar]
- 14.Vila, J., A. Ribera, F. Marco, J. Ruiz, J. Mensa, and M. T. Jiménez de Anta. 2002. Activity of clinafloxacin, compared with other four quinolones, against Acinetobacter baumannii clinical isolates with known mechanisms of quinolone resistance. J. Antimicrob. Chemother. 49:471-477. [DOI] [PubMed] [Google Scholar]