Abstract
Cluster designation (CD) antigens are cell surface markers that can be used to identify constituent cell populations of an organ. We have previously determined the CD phenotype of normal prostate parenchymal cells (Am J Pathol 2002, 160:37–43) and are now extending this analysis to prostate cancer. Since expression of CD antigens is associated with cellular differentiation, cancer cells may differ from their normal counterpart in their CD profile. Compared with luminal secretory cells, prostate adenocarcinoma cells are frequently negative for CD10 and CD13, express increased levels of the cell activation molecule CD24, and decreased levels of the apoptosis-associated multifunctional enzyme CD38. Expression of CD57, CD63, CD75s, CD107a, CD107b, CD164, and CD166 by cancer cells is similar to that of secretory cells. Prostate basal epithelial cells do not express the CD antigens characteristic of prostate secretory cells; and the basal cell CD markers, CD29, CD44, CD49b, CD49f, CD104, and nerve growth factor receptor (NGFR) are not expressed by cancer cells. The preferential expression of secretory cell-associated CD markers by prostate cancer cells suggests a closer lineage relationship between cancer cells and secretory cells than basal cells. Although the above cancer CD phenotype was the most frequently seen, some prostate cancers contained populations of CD10- and/or CD13-positive cells, and CD57-negative cells. Furthermore, the cancer phenotype of tumor metastasis is different. Despite its low frequency in primary tumors, CD10 is expressed by virtually all of the nodal metastases of prostate cancer. In addition, stromal fibromuscular cells associated with primary prostate cancer differ from stromal cells in benign prostate tissue by an increased level of expression of the cell activation molecule, CD90. In summary, our data show that the CD marker expression profile of prostate cancer cells most closely resembles that of secretory prostate epithelial cells and that some prostate cancers consist of heterogeneous cell populations as distinguished by CD-marker expression profiles.
Prostate cancer is the most frequent cancer in men, and the second leading cause of cancer mortality in the US.1 However, the prevalence of prostate cancer is at least fivefold greater than the frequency with which it causes morbidity. Parameters that can stratify patients for type of therapy based on likelihood of tumor progression are clinical stage, serum PSA, and histological differentiation, conventionally reported as the Gleason grade.2 Although these parameters predict outcome for populations of patients, they are only weak predictors of the course of the disease in the individual patient. Thus, better tissue markers are needed that can supplement Gleason grade and be applied either to biopsies to stratify for primary therapy or to radical prostatectomy samples to stratify for adjuvant therapy.
Cell and tissue markers that may be informative on the outcome of prostate cancer include Ki67 fraction, hepsin, PIM-1, EZH2, etc.3–5 In general, the predictive power of these markers has not been demonstrated in multivariate analyses. In addition, a useful tumor marker is one that would also serve as a target for therapy. One of the efforts of our prostate cancer genomics program is to identify these markers.6 While this approach has great potential, useful reagents, once identified, require time for development and validation. We propose that a panel of commercially available, well-characterized antibodies to cell surface molecules could be used to identify cancers with distinct clinical behaviors.
Cluster designation (CD) antigens, of which there are now more than 200, were first defined on leukocytes. These markers have been proven to be very useful in lineage study of hematopoietic cell types (http://ncbi.nlm.nih.gov/prow/). We have completed a study immunophenotyping the prostate, in which CD molecules specific for the different cell types were identified7 (http://scgap.systemsbiology.net/figures/CD_specificity.php). Since expression of CD molecules is linked to physiological changes,8,9 tumor cells may have distinct complements of CD molecules that differ from those of normal cells. Furthermore, these differentially expressed molecules may have functional significance. We have observed distinct CD profiles in the prostate cancer cell lines LNCaP, PC3, and DU145,10 which differ in their malignant property. Since prostate cancer is a heterogeneous disease with respect to tumor behavior, we questioned whether patterns of CD expression could also be used to identify clinically distinctive cancer cell types in both primary tumors and metastases.11 If true, the behavior of a tumor might be predicted by its cell CD phenotype. Furthermore, CD molecules common to all prostate cancer cell types might serve as therapeutic targets for cell killing. This study was undertaken to identify CD molecules that are differentially expressed in prostate cancers.
Materials and Methods
Prostate Tissues and Lymph Node Metastases
Tissue blocks from about 80 radical prostatectomy and 25 lymph node metastasis were used for this study. To maximize yield of tumor and minimize cell degradation, we used the following protocol. Immediately upon receipt of the radical prostatectomy in the frozen section room of the Department of Pathology, serial 3-mm thick transverse sections were cut. Quadrants were prepared from each transverse section after submitting the apex and base for permanent sections. The posterior tissue quadrants from alternate 3-mm thick transverse sections were embedded in OCT and frozen. The frozen tissue blocks were assigned an anonymized code and accessioned into our tumor bank. Those frozen blocks containing tumor foci were labeled as cancer tissue sample (CP) while those containing no tumor were labeled as sample of normal prostate tissue (NP). For flow cytometry analysis to assess the feasibility of sorting cancer cell populations, only tumors with a relatively large volume (at least 1 ml) in a single focus were chosen. In nearly all cases the block selected for staining contained the largest amount of tumor. Tissue enriched for cancer, in which at least 85% of the cells in the corresponding frozen section were cancer cells and weighing at least 100 mg, was dissected from the opposing aspect of the non-fixed section that was adjacent to the block that was frozen. For immunohistochemistry, tissue blocks (containing cancer) were randomly selected from the tissue bank. Each specimen had a numeric code with a letter code indicating the site origin of the block (right apex, left mid, etc). Multiple 5-μm thin serial sections were sliced from each tissue block and fixed in cold acetone. Lymph node metastasis specimens (the numerals, if present, after the letter code indicate a particular node in a group of several) were similarly processed. We used frozen sections for these studies since a majority of the CD antibodies that we use do not immunoreact with antigens in formalin-fixed, paraffin-embedded tissue.
Immunohistochemistry
CD expression was probed principally by immunohistochemistry, and all monoclonal CD antibodies (mouse IgG1, IgG2a, IgG2b, IgM, and rat IgG2b) were obtained from BD-PharMingen (San Diego, CA). Isotype-specific CD-antigen irrelevant antibody clones MOPC21 (IgG1) and G155–178 (IgG2a) were used as negative controls. Immunolocalization was done using an indirect avidin-biotin-peroxidase method. The most used primary antibodies: CD10, CD13, CD24, CD26, CD29, CD38, CD44, CD45, CD49a, CD49b, CD49f, CD53, CD55, CD57, CD63, CD69, CD71, CDw75 (updated to CD75s), CD90, CD104, CD107a, CD107b, CD164, CD166, and nerve growth factor receptor (NGFR) were used at concentrations listed in Table 1, based on a protocol as described previously.7 The titers were selected to minimize non-specific staining. The positive tissue controls are those prostate parenchymal cells that we have identified as expressing each respective antigen.7 The secondary antibodies were either biotinylated anti-mouse IgG (BA-2000, Vector Labs, Burlingame, CA) which also reacts with rat IgG antibodies), or anti-mouse IgM (BA-2020) for the CD57 and CD75s antibodies. Reaction product was detected by incubating sections in a solution of avidin-biotin-peroxidase, followed by a solution of the chromogen diaminobenzidine. The sections were counterstained with hematoxylin. Specificity of labeling was confirmed by omission of the primary antibody.
Table 1.
Anti-CD Antibodies
| Antibody | Clone | Isotype | Concentration |
|---|---|---|---|
| CD10 | HI10a | mouse IgG1 | 4.6 ng/μl |
| CD13 | WM15 | mouse IgG1 | 0.032 μl 50 tests/ml/μl |
| CD24 | ML5 | mouse IgG2a | 4.6 ng/μl |
| CD26 | M-A261 | mouse IgG1 | 0.03 μl 50 tests/ml/μl |
| CD29 | MAR4 | mouse IgG1 | 3.1 ng/μl |
| CD38 | HIT2 | mouse IgG1 | 4 ng/μl |
| CD44 | G44-26 | mouse IgG2b | 0.8 ng/μl |
| CD45 | HI30 | mouse IgG1 | 6 ng/μl |
| CD49a | SR84 | mouse IgG1 | 0.036 μl 50 tests/ml/μl |
| CD49b | 12F1-H6 | mouse IgG2a | 3.3 ng/μl |
| CD49f | GoH3 | rat IgG2a | 3.9 ng/μl |
| CD53 | HI29 | mouse IgG1 | 8 ng/μl |
| CD55 | IA10 | mouse IgG2a | 6.7 ng/μl |
| CD57 | NK-1 | mouse IgM | 6.2 ng/μl |
| CD63 | H5C6 | mouse IgG1 | 0.025 μl 50 tests/ml/μl |
| CD69 | FN50 | mouse IgG1 | 8 ng/μl |
| CD71 | M-A712 | mouse IgG2a | 4 ng/μl |
| CDw75 | LN1 | mouse IgM | 5.7 ng/μl |
| CD90 | 5E10 | mouse IgG1 | 2.7 ng/μl |
| CD104 | 439-9B | rat IgG2b | 3.6 ng/μl |
| CD105 | 266 | mouse IgG1 | 6 ng/μl |
| CD107a | H4A3 | mouse IgG1 | 2 ng/μl |
| CD107b | H4B4 | mouse IgG1 | 0.006 μl 50 tests/ml/μl |
| CD164 | 103B2 | mouse IgG3 | 7.5 ng/μl |
| CD166 | 3A6 | mouse IgG1 | 0.03 μl 50 tests/ml/μl |
| NGFR | C40-1457 | mouse IgG1 | 0.04 μl 50 tests/ml/μl |
The immunohistochemistry staining level was categorized as: intense, in which the immunoreaction deposit was distinctly more optically dense than background and tissue that did not express the antigen; equivocal, in which the immunoreaction deposit was either similar enough in optical density to the background and/or to tissue that did not express the antigen, or was so focal, ie, < 5% of cells, that there was reasonable uncertainty regarding whether the cells expressed the antigen; and none, in which there was either no immunoreaction deposit or that the reaction product was no more optically dense than background.
Cell-Type Analysis by Flow Cytometry
Matched tumor (CP) and non-tumor (NP) samples, obtained as described above, were digested with type I collagenase (GIBCO-BRL, Rockville, MD) to prepare populations of single cells.11,12 The cells were resuspended in 50-μl aliquots of 0.1% bovine serum albumin-Hanks’ balanced salt solution (BSA-HBSS). Aliquots of CD antibodies (≤0.1 μg) were added to the cell suspensions. The CD antibodies are conjugated to either R-phycoerythrin (PE) or fluorescein isothiocyanate (FITC) fluorescent molecules. After labeling, the cells were resuspended and fixed in a solution of 2% formalin in HBSS for flow analysis. Omission of the primary antibody or irrelevant isotype-specific fluorochromated antibodies were used as negative controls to delineate the autofluorescent (negative) cell population. Events that exhibited fluorescence outside the ranges delineated by the negative controls were scored as positive as described previously.10 The percentage of labeled cells for each CD antibody was determined. Since the CP samples did not contain pure populations of cancer cells, but contained some normal epithelial cells and stromal cells of various types, the percentage of labeled cells in CP was normalized to the percentage of CD57-positive cells. We chose CD57 as the normalization cell phenotype since CD57 is expressed by cancer cells as well as by normal secretory cells.
Results
Luminal Secretory-Cell CD Markers in Cancer
For the majority of specimens analyzed, the cancer cell profile was CD10−/CD13−/CD24↑/CD26+/CD38↓/CD57+/CD75s+/CD107b+ (where ↑ denotes increased and ↓ denotes decreased immunostaining intensity) as shown in Figure 1. For these specific markers, the secretory cell profile is predominantly CD10+/CD13+/CD24+/CD26+/CD38+/CD57+/CD75s+/CD107b+, while the basal cell profile is CD10−/CD13−/CD24−/CD26−/CD38−/CD57−/CD75s−/CD107b−. Other luminal CD markers found in cancer were CD63, CD107a, CD164, and CD166, while basal CD markers including CD29, CD44, CD49b, CD49f, CD104, and NGFR were not expressed by cancer cells (data not shown, but available at our database: http://scgap.systemsbiology.net/). This database also contains information regarding heterogeneity of immunostaining of the histologically normal epithelium. The significance of this heterogeneity is unclear to us at present. Thus, cancer cells differed from their normal luminal cell counterpart in the absence of expression of CD10 and CD13. In addition, CD24 immunoreactivity was more intense and CD38 immunoreactivity less intense in cancer cells compared with normal cells.
Figure 1.
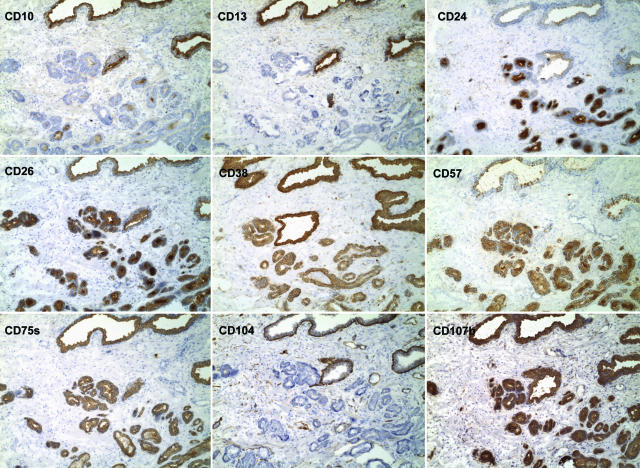
CD phenotype of prostate cancer. Shown are the staining results of CD10, CD13, CD24, CD26, CD38, CD57, CD75s, CD104, and CD107b on serial sections of specimen 01–064D. In this specimen, the tumor glands in the lower right of the field are negative for CD10 (the photomicrographs are identified by the CD molecules), CD13 and CD104; positive for CD24, CD26, CD38, CD57, CD75s, and CD107b. The tumor glands are visualized by their intense staining for CD24 compared to the larger benign glands. In contrast, tumor glands are stained weaker for CD38 compared to benign glands. They are also more uniformly stained for CD57, however, normal glands elsewhere in this specimen were well stained by CD57. CD104 stains the basal cell layer of benign glands, which is absent in cancer glands. Magnification is ×100.
This profile of immunohistochemistry-defined CD expression by cancer correlated with expression patterns obtained by flow analysis of cells prepared from dissected samples. The percentage of CD10- and CD13-positive cells was decreased in the cancer sample shown in Figure 2. There was no appreciable decrease in the CD26- and CD107b-positive cell fractions, in agreement with the immunohistochemistry results. The percentage of cells expressing basal cell markers CD29, CD44, CD55, and CD104 was decreased in CP compared with NP, consistent with our observation that cancer cells do not express basal cell CD markers. The fraction of cells expressing CD49a, which is uniformly expressed by stromal fibromuscular cells, was decreased to some extent. This finding is consistent with the interpretation that the glands of cancer are usually more densely packed than are normal glands. As a consequence of glandular crowding, the percentage of stromal cells in cancer-containing tissue is reduced. There were no sizeable differences in the fraction of CD53, CD69, and CD71 cells. The first two are markers of lymphoid cells, which appeared to be equally represented in CP and NP. CD71 is reactive to multiple cell types. Because we did not know which cell types were being labeled in flow analysis (for example, CD13 stains not only luminal cells, but also endothelial cells, nerve elements, and leukocytes7), no attempt was made to refine this analysis other than to show that immunoreactivity was concordant between immunohistochemistry and flow cytometry.
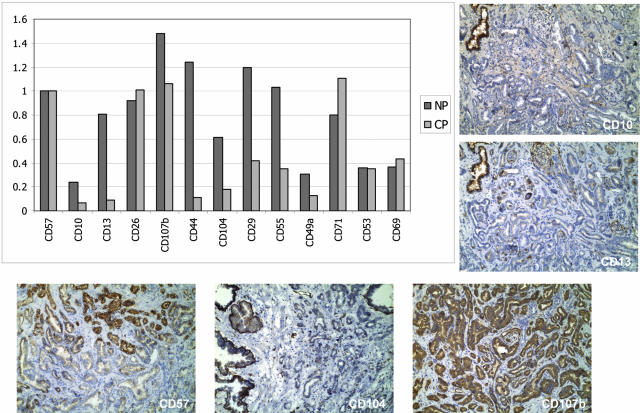
Figure 2.
Flow analysis of cancer. For the purpose of comparison, the number of positive events to each CD specificity (x axis) is adjusted to the percentage of CD57+ cells in either NP (13%) or CP (57.6%) (y axis), and tabulated. The finding that the cancer of specimen 99–010 is predominantly CD57+ was shown by immunohistochemistry of frozen blocks coded as 99–010E and 99–010F, although the tissue sample analyzed by flow cytometry was not the same as that represented in the tissue blocks. Staining results for CD10, CD13, CD104 and CD107b are also shown. CD57, CD10, CD13, CD26, and CD107b are reactive to luminal cells; CD44, CD104, CD29, and CD55 are reactive to basal cells; CD29, CD55, and CD49a are reactive to stromal fibromuscular cells; CD71 is reactive to multiple cell types; while CD53 and CD69 are reactive to white blood cells. Note the decreased positivity for CD10 and CD13 in CP. The CD44 and CD104 results are indicative of an under-representation of basal cells.
Our analysis reported here concerns cancer and a systematic analysis of high-grade prostatic intraepithelial neoplasia (HGPIN) was not carried out. We reported earlier a lower intensity of CD10 staining in HGPIN compared to normal epithelium.13
Cancer CD Phenotypes
In addition to the CD10−/CD13−/CD24↑/CD26+/CD38↓/CD57+/CD75s+/CD107b+ phenotype, other cancer phenotypes were found. These included tumors that were scored as CD10+, CD13+, and/or CD57−. The tumor shown in Figure 3A contained cancer cells that were CD10+ and CD13+. In a number of cases, two populations of the same tumor were found to express different CD profiles, which were not apparent by routine staining. In one example, one tumor focus was characterized by the more common CD13−/CD57+ phenotype and the other by a variant CD13+/CD57− phenotype (data not shown). The following two examples are shown to illustrate the staining heterogeneity for a high Gleason-grade tumor and a low Gleason-grade tumor. In Figure 3B, a staining gradient separated two foci of cancer cells graded Gleason 4. One focus was characterized as CD10−/CD13−/CD24+/CD26+/CD38+/CD57+/CD75s+ and the other as CD10+/CD13−/CD24±/CD26±/CD38−/CD57−/CD75s+ (both were CD104−). In Figure 3C, the two cancer phenotypes graded Gleason 3 were not as clearly segregated. One type was characterized as CD10−/CD13−/CD57− while the other was CD10+/CD13+/CD57+; both were CD24+/CD26+/CD38+/CD75s+/CD104−. Tumor specimens containing aggregates of cancer cells with a non-glandular (Gleason pattern 4) histology constituted about 20% of the samples studied, and the example shown in Figure 4 was scored as CD10−/CD13−/CD24+/CD26+/CD38−/CD57+/CD75s+/CD104−/CD107b+. CD10+ or CD13+ cancer cells were also seen in other such examples (data not shown).
Figure 3.
(Continues)
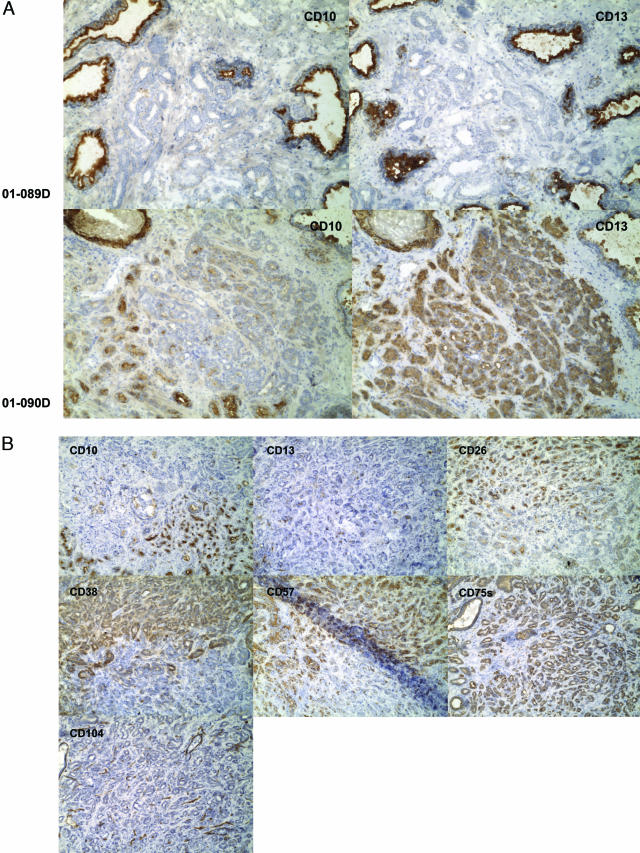
Figure 3.
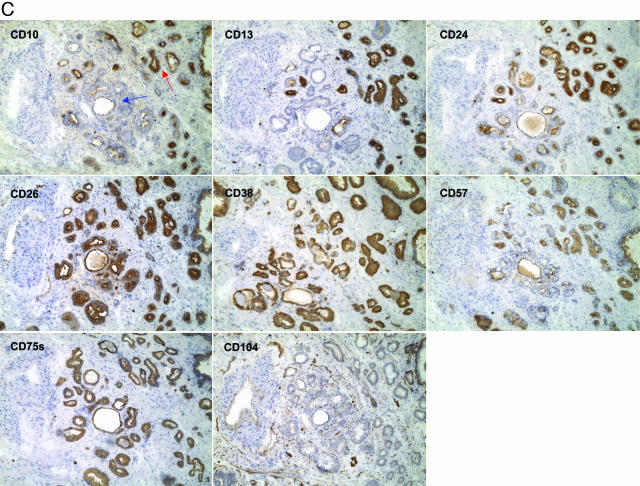
Tumor heterogeneity. A: CD10+/CD13+ tumor glands. Two specimens, 01–089D and 01–090D, are contrasted. In the top panels, the cancer glands (middle) of 01–089D are not stained for CD10 and CD13 (whereas surrounding benign glands are stained). In the bottom panels, some of the small cancer glands in 01–090D are stained for CD10, and all for CD13. The cancer glands are negative for CD104 (not shown). Magnification is ×100. B: Adjacent, differentially stained tumor cell populations of high Gleason grade. Serial sections of specimen 01–081D were stained with the antibodies identified in the individual micrographs. Two tumor CD phenotypes are evident. Both types are positive for CD75s and negative for CD104 (stained are basal cells of benign glands, endothelial cells of blood vessels, and nerve cells in this field). The two types are otherwise indistinguishable by histomorphology. The Gleason score given for this specimen is 4 + 4. Magnification is ×100. C: Adjacent differentially stained tumor glands of low Gleason grade. One type of tumor glands is scored as CD10−/CD13−/CD57− (blue arrow) while the other is scored as CD10+/CD13+/CD57+ (red arrow) in specimen 01–076D. Both cancer gland types are CD24+/CD26+/CD38+/CD75s+/CD104−. The Gleason score given for this specimen is 3 + 3. Magnification is ×100.
Figure 4.
CD phenotype of a non-glandular pattern tumor. The Gleason score given for specimen 98–366D is 4 + 5, and the tumor mass (arrowed in the CD10 panel, note benign glands are stained) are CD10−/CD13−/CD24+/CD26+/CD38−/CD57+/CD75s+/CD104−/CD107b+. Except for the complete absence of CD38 staining, the phenotype is like that of the specimen shown in Figure 1. Magnification is ×100.
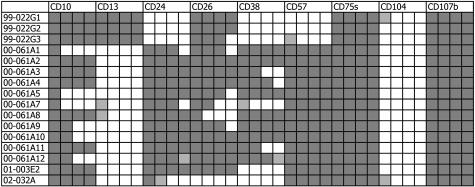
The immunostaining results of the first 31 tumor specimens analyzed are presented in Figure 5. The tumors were all negative for CD104. Expression of CD10, CD13, and CD38 was variable, whereas expression of CD24, CD26, CD57, CD75s, and CD107b was less variable. With regard to CD10, CD13, and CD57 expression among a cohort of 16 cases that were screened, six phenotypes could be assigned. There were nine typed as CD10−/CD13−/CD57+, two each as CD10−/CD13+/CD57+ and CD10+/CD13−/CD57+, and one each as CD10+/CD13+/CD57+, CD10+/CD13+/CD57−, and CD10−/CD13+/CD57−. Overall, CD10+ cancer cells were detected at a frequency of 27%, CD13+ cells at 32%, and CD57+ cells at 80% (and CD24+ cells at 84%, CD26+ cells at 90%, CD38+ cells at 45%, CD75s+ cells at 100%, CD104+ cells at 0%, and CD107b+ cells at 98%) among the tumor foci.
Figure 5.
CD reactivity of prostate tumors. Thirty-one tumor specimens listed in the first column were scored for the expression of the CD markers listed. The subsequent columns indicate the immunoreactivity of the cancer to the CD antibodies indicated. Percentages of cancer showing positive immunostaining are presented in quartile with each filled box representing an increment of 25%. Unfilled boxes indicate no staining. Darker and lighter shading of the boxes are used to indicate increased intensity of staining in cancer versus non-cancer, as for CD24, and decreased intensity of staining, as for CD38, respectively. 99–022H1 and H2 are two separate tumor foci of the same specimen. For comparison, the CD phenotypes (as determined by flow cytometry) of cancer cell lines LNCaP, PC3, DU145, and CL1 are shown at the bottom. The lighter shaded boxes for CD24 in DU145 and CL1, and for CD104 in LNCaP are used to represent a labeling percentage of less than 25%. Because cultured cells were detached by trypsin before analysis epitopes of molecules like GPI-anchored CD24 might be affected by the enzyme treatment.
In this first cohort, a probable association between CD10+ cancer cells and higher-grade tumors could be detected with a P value for the Fisher’s exact test of association of 0.09. Most Gleason 3 tumors were CD10−.To validate this finding a larger cohort of 80 specimens was analyzed for cancer-CD10 staining as presented in Figure 6. Each specimen was scored for its component of Gleason 3, 4, and 5 patterns and the percentage of staining in each pattern was estimated. For data presentation, the specimens were sorted by their overall percentage of CD10 staining. It is evident that many of the tumors with only Gleason pattern 3 showed no staining. There were also many tumors containing at least 5% CD10+ cancer cells. Two cases (because the patients were surgically treated for their metastases) of cancer recurrence (indicated by bold in the first column) contained a significant proportion of CD10+ cancer cells in the primary tumors. Of those cases staged N+, almost all contained CD10+ cells (≥10%) except cases 02–007 and 99–068. However, the particular section (labeled C) of 02–007 showed only a Gleason 3 pattern while the overall Gleason score was 4 + 3 (therefore the Gleason pattern 4 in a different region of the prostate might contain CD10+ cells). Similarly, the sections of 99–068 scored (Gleason 3 and 4) as well as a T4 case, 99–002, showed no Gleason 5 pattern, however, the overall scores were 4 + 5 and 3 + 5, respectively. Thus, sampling may account for these discrepancies of association between CD10 and advanced disease. This sampling problem can be illustrated with 01–182B that showed 90% Gleason 3, 10% Gleason 4; and its sister block 01–182D (representing a different tumor locus of the same gland) that showed 70% Gleason 3, 25% Gleason 4, and 5% Gleason 5 cells. For case 99–022, which was shown to have positive nodes, the cancer was recorded as stage T2a. The TNM staging for the cases was based on the 2002 version.14 In cases 01–079, 00–175, and 98–009 where tumor volume was relatively small (<1 ml) the CD10+ cancer cells might be insignificant. The TNM stages for cases with only CD10− cells are distributed in the following way: one T2a, one T2b, 16 T2c, three T3a, one T3b, and one T3c. T2c was scored for case 02–034, even though it was a 5.2-ml predominantly pattern 5 tumor the percentage of CD10+ cells was only 5%. These examples illustrate the challenging aspect in this type of analysis. Nevertheless, our data showed the following trend: median percentage of tumor cells positive for CD10 was 0% for Gleason 6, 13% for Gleason 7, and 10% for Gleason >7; 5% for stage <T3, 15% for >T2, and 20% for N+. A more detailed statistical analysis of this data set with additional informative results will be communicated in another report.
Figure 6.
CD10 expression and Gleason components. The specimens in the first column are listed in the order of increasing percentage of CD10-positive cancer cells. The prostate cancer cases are characterized by percentages of Gleason 3, 4, and 5 components and the percentages of CD10+ cells in each component. For example, the tumor of 01–182D (first entry) is composed of 70% Gleason pattern 3, of which 0% is CD10-positive; 25% Gleason pattern 4, of which 0% is CD10-positive; and 5% Gleason pattern 5, of which 0% is CD10-positive. The tumor of 01–135C (36th entry) is composed of 50% Gleason pattern 5, of which 5% is CD10-positive; 40% Gleason pattern 4, of which 5% is CD10-positive; 10% Gleason pattern 5, of which 0% is CD10-positive. Its total positive percentage is 50 × 5/100 + 40 × 5/100 = 4.5%. The two cases highlighted in bold (98–395D and 97–233F) are known disease recurrence. The tumor volume is in ml. Some cases, like 99–155 (22.75% CD10-positive cancer cells) and 98–348 (55%), support the contention that presence of CD10+ cancer cells indicate likelihood of disease progression (as indicated by node involvement). Others, like 99–068 and 02–007, were assigned N+ yet the specimens 99–068D and 02–007C contained no CD10-positive cancer cells. However, our sampling only analyzed a portion of the tumor for many of these cases (see text). na, not available.
Cancer Cell Types of Lymph Node Metastases
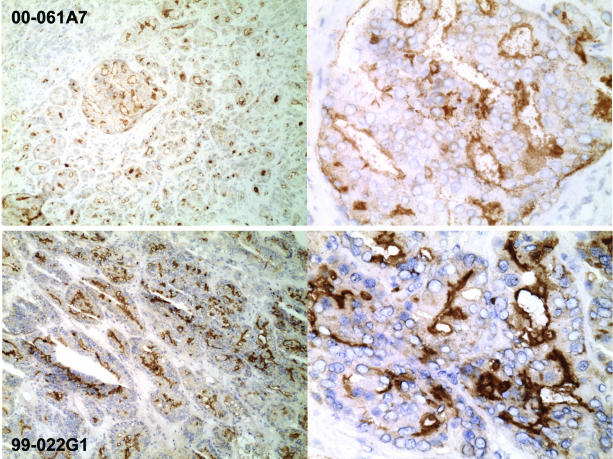
One striking difference between primary tumors and lymph node metastases was the observation that most cancer cells in the node metastases expressed CD10 (Figure 7), in contrast to the finding of CD10+ cancer cells in a minority of primary tumors. With the exception of CD10, expression of the other CD molecules was not significantly different between primary tumors (Figure 5) and metastases (Figure 8). Note that the cancer cells of node specimens 99–022G1, G2, and G3 could have originated from cancer cells in the corresponding primary tumor focus in 99–022H1 rather than from 99–022H2 (Figure 5) based on the presence of the same CD24−/CD38−/CD57− cancer cell type in 99–022H1. A comparative flow analysis was carried out on samples obtained from a second matched prostate cancer and node metastasis (specimen 99–042). The prostate tumor tissue was shown to contain ∼1% CD10+ cells and 40% CD107b+ cells; while the node was shown to contain 16% CD10+ cells and 15% CD107b+ cells. Assuming that CD107b+ cells represented cancer epithelial cells, the percentage of CD10+ cells increased from a small fraction (∼1%) in the prostate tumor to 100% in the lymph node (CD45+ lymphocytes, if present, were negative for CD10, see also Figure 9).
Figure 7.
Lymph node metastases. Prostate cancer cells in the lymph nodes were identified by CD107b (and CD75s staining; and lymphocytes by CD45 staining). CD10+ cancer cells are evident in the two specimens, 00–061A7 and 99–022G1, shown. Staining appears to be localized to the apical surface of gland-like luminal space. CD10− cancer cells are also evident in 00–061A7 (upper left). Magnification is ×100 (left) and ×400 (right).
Figure 8.
CD reactivity of lymph node metastases. Sixteen specimens from four (available) different surgical cases (99–022, 00–061, 01–003, and 02–032) are listed. As in Figure 5, percentages of cancer showing positive immunostaining are presented in quartile with each filled box representing an increment of 25%. Unfilled boxes indicate no staining. The lighter shaded boxes indicate equivocal staining results. Note that the node patterns of 99–022G1, G2, G3 are similar to that of some tumor cells in the corresponding prostate, 99–022H1.
Figure 9.
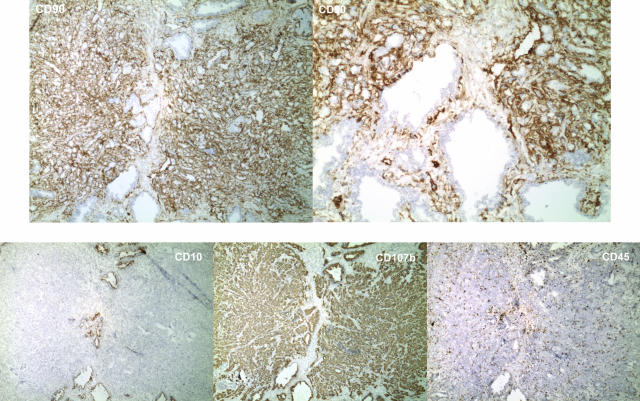
CD90 reactivity of cancer-associated stroma. The stromal fibromuscular cells of the two cancer foci found in specimen 01–178F are stained more intensely for CD90 than stromal fibromuscular cells of non-cancer tissue. Shown also are the staining results for CD10, CD45, and CD107b. The finding that staining is not due to infiltrating lymphocytes that are CD90+ is indicated by the low number of CD45-positive cells in the tumor foci. Both tumor and benign glands are stained for CD107b, tumor cells are negative for CD10. Magnification is ×40 except top right, which is ×100.
Cancer-Associated CD Expression Changes in the Stromal Compartment
In addition to both qualitative and quantitative CD expression changes in the epithelial cancer cells, some quantitative changes were detected in the stromal fibromuscular cells. CD90 (Thy-1) is a marker for stromal cells in the prostate,7 and the stromal cells of tumors were stained more intensely than those of benign tissue (Figure 9). This increased CD90 staining appeared to be a common feature for nearly every tumor specimen analyzed. In fact, the pronounced CD90 staining could serve to delineate tumor foci in tissue sections, as this staining difference did not appear to extend beyond the tumor area. An increased staining of cancer area was also seen with CD105 (endoglin/TGFβR), although it was difficult to distinguish whether the stained cells represented an increased number of endothelial cells of blood vessels15 (data not shown).
Discussion
Based on the utility of CD phenotypes in leukemia classification, we were interested in whether prostate cancer could also be CD phenotyped, whether different cancer CD phenotypes could be correlated to disease outcomes, and whether the compositional difference of CD cancer cell types could be a basis of tumor heterogeneity. A cohort of over 80 prostatectomy specimens was selected for this study. Since conditions of primary antibody immunoreactivity restricted us to using frozen samples we were limited to using samples collected as frozen tissue within the past 4 years. The clinical parameters of these specimens were available in a secured, restricted-access database. The prostatic epithelium contains three epithelial cell types: luminal secretory and basal cells that constitute the principal populations plus a small population of neuroendocrine cells.16 A number of experimental observations would suggest that cancer cells are derived from the luminal or some luminal-like intermediate cell type.17 In support of that, many CD molecules of luminal cells are expressed by cancer cells whereas basal markers are generally not expressed by cancer cells. Among the luminal markers, cancer cells of most primary tumors are positive for CD24, CD26, CD38, CD57, CD75s, and CD107b but negative for CD10 and CD13. Tumors characterized by a non-glandular pattern, despite having a higher Gleason score, are not remarkably different from glandular tumors (of a lower score) with regard to the CD phenotype. In fact, two morphologically distinct tumor types have been reported to have similar clinical outcomes.18 The prostate cancer CD phenotype is, however, unlike that of LNCaP (CD10+/CD13−/CD24−/CD26−/CD38−/CD57−/CD75s−), PC3 (CD10−/CD13+/CD24−/CD38−/CD44+/CD57−/CD75s−/CD104+), or DU145 (CD10−/CD13−/CD26−/CD38−/CD44+/CD57−/CD75s−/CD104+),10 three cell lines that are used as prostate cancer models. This is not unexpected since these cell lines were established from metastatic lesions.
Lack of CD10 and CD13 expression in cancer cells suggests that their absence might play a role in the process that gives rise to transformed cells. According to the hypothesis that equates oncogeny to partially-blocked ontogeny,19 cancer cells may appear as a result of a block in the differentiation of an intermediate cell type characterized by CD10−/CD13− in the epithelial cell lineage. CD10, which is neutral endopeptidase or the common acute lymphoblastic leukemia antigen (CALLA) expressed by normal and neoplastic cell types,20 and CD13, which is aminopeptidase N,21 are involved in the processing of diverse regulatory peptides (CD26 is another such enzyme). These activities release amino acid residues from the N-terminus with broad substrate specificity. The malignant transformation process may thus involve aberrant signaling by peptide molecules that are the substrates of CD10 and CD13. Possible substrates include bombesin, endothelin-1, neurotensin, enkephalins, and substance P for CD10,22,23 neurotensin, enkephalins, bradykinins, and dynorphin-related peptides for CD13.24 Some of these are likely products of the neuroendocrine cell type.25 Loss of CD10 expression was previously reported to be associated with androgen-independent growth of prostate cancer cells,26 however, its loss in primary tumors cannot be attributed to androgen. Down-regulation of CD10 might be an early event in prostate cancer with decreased staining evident in the presumed premalignant HGPIN,13 and lower CD10 expression is detectable by DNA array analysis.4,27 CD10 is also down-regulated in some renal cell cancers.28 Smoking reduces CD10 activity of bronchial epithelial cells, and this might predispose to lung cancer.29 Diminished CD13 reactivity in prostate cancer was also previously reported.30 On the other hand, virtually all cancer cells exhibit intense staining of CD24 compared to luminal cells. CD24 is a GPI-anchored glycoprotein in B-cell proliferation and maturation (implying a cell activation function), and CD24 is also found in breast cancer.31 Though variable, cancer cells show a weaker staining for CD38. CD38 acts as an NAD glycohydrolase in T cells, and its expression is known to be down-regulated in prostate cancer.32 Low CD38 is also known to parallel a lower propensity of (hematopoietic) cells to undergo apoptosis,33 which is the presumed fate of normal luminal cells. By inference, prostate cancer cells with lower CD38 are less responsive to apoptotic signals. Staining for CD57, CD75s, and CD107b shows little variability, CD75s and CD107b, in particular, are expressed by nearly all cancer cell types in primary tumors. CD57 is an natural killer (NK) cell marker, CD75s functions in intercellular adhesion,34 and CD107b is a glycosylated lysosome-associated membrane protein LAMP2 (LAMP1 is CD107a and LAMP3 is CD63) found in activated lymphocytes and platelets.35
In addition to the predominant cancer CD phenotype, other cancer cell types were found that are CD10+, CD13+, or CD57−. This variance is also reflected in the different CD phenotypes of prostate cancer cell lines and xenografts. Are they distinct cancer cell types of different malignant behavior that contribute to tumor heterogeneity? Do they have independent origin or are they related in cancer progression? Possible lineages based on their frequencies could be CD10−/CD13−/CD57+ → CD10+/CD13−/CD57+ → CD10+/CD13+/CD57+ → CD10+/CD13+/CD57− in one; and CD10−/CD13−/CD57+ → CD10−/CD13+/CD57+ → CD10−/CD13+/CD57− in another. Whether these cell types are in fact related by lineage remains to be determined. Answers to the above questions are crucial to the potential application of CD typing in prostate cancer stratification. Comparative analysis of cancer cell-type composition between primary tumors and metastases may therefore be informative. Whereas the predominant cell type in primary tumors is CD10−, node metastases in our study invariably contained CD10+ cancer cells. LNCaP and xenograft LuCaP 35, both established from lymph node metastases, are typed CD10+ (10 and unpublished data). CD10+ cells were detected in one of the corresponding prostates available for comparative immunohistochemistry (specimen 99–022), and it is quite possible that the CD10+ cancer cells could originate from the primary tumor rather than from clones newly arisen in the nodes. It appears then that CD10+ cancer cells in primary tumors would indicate a likelihood of node metastasis. The two studies using tissue microarrays, however, reported no statistical significant link between CD10 expression and disease progression,27,36 though a detailed CD expression pattern presented by Zellweger et al37 in a more recent meeting abstract showed the following: 100% in benign prostatic hypertrophy (BPH), 65% in prostatic intraepithelial neoplasia (PIN), 35% in localized cancer, 50% in hormone refractory cancer, and 40% in metastases. In our series, CD10+ cancer cells were often found in Gleason ≥4 tumors, though not all such tumors examined contained CD10+ cancer cells in agreement with the other studies. In another recently reported series, there was generally no CD10 staining in low Gleason grade tumors but staining in a subset of Gleason pattern 4 and 5 tumors.38 The presence of CD10+ cancer cells in nodes would suggest a role for CD10 in the metastatic process (although CD10 has been shown to inhibit cell migration39). Indeed, CD10 expression appears to correlate with liver metastasis of colorectal adenocarcinoma, in which tumors with higher CD10 positivity are associated with metastasis.40 CD10 antigen is also up-regulated in melanoma metastasis. In that study, there were cases of primary tumors and metastases containing CD10+ cancer cells, and a patient with a CD10− tumor who survived for 19 years versus one with a CD10+ tumor who died within 2 years.41 Our ongoing analysis showed a high frequency of CD10+ prostate tumors in cases that failed as indicated by a rising serum PSA level (M. Dall’Era, Department of Urology, University of Washington, Seattle, WA, personal communication). Our data, however, cannot determine if CD10+ cancer cells are derived from CD10− cancer cells or from CD10+ normal cells. Besides CD10, other CD molecules may also be involved in disease progression, CD13+ cancer cells might also be metastatic because CD13 antiserum can inhibit invasion of metastatic cells.42 Non-small cell lung cancer that contains CD13+ cancer cells has a worse prognosis than that without, or with only CD10+ cells.43 CL1, a hormone-insensitive, highly tumorigenic cell line derived from in vitro selection of LNCaP cells in androgen-depleted media,44 differs in its CD expression from LNCaP. While LNCaP is characterized by CD10 positivity, CL1 is characterized by positivity of CD13 and CD44 with little of CD10.13 Although CD13 is a marker of luminal cells there is a noticeable shift to the expression of basal markers such as CD44, CD55, and CD104 in the more aggressive CL1 cells (CD104 in the non-lymph node-derived PC3 and DU145 cells as shown in Figure 5). This is in agreement with our previous analysis on the LuCaP xenografts in intact versus castrated animals.45 In summary, these observations show that gene expression (of CD molecules) is modulated variably in cancer progression as exemplified by CD10. CD10 is present in luminal secretory cells, absent in cancer cells of primary tumors, present in cancer cells of lymph nodes, and absent in androgen-independent cancer cells as reported by Nanus and co-workers.26 This illustrates that a gene product may serve different functions depending on the cell type in which it is expressed.
In addition, tumor-associated changes were also observed in stromal mesenchyme cells as indicated by the increased staining for CD90. Among the functions attributed to CD90 are co-stimulation of lymphocytes and inhibition of stem cell proliferation and differentiation. Its cell stimulatory function is similar to that of CD24, expression of which is increased in the neighboring cancer epithelial cells. Prostate functioning and homeostasis are dependent on reciprocal stromal/epithelial interaction.46 Epithelial development is influenced by stromal cell-derived growth factors, and stromal cells serve as an intermediary in the transmission of androgen-induced stimuli to epithelial cells.47 It has been experimentally shown that carcinoma-associated stromal cells differ in their influence on epithelial cells.48 Therefore, it is not unexpected that gene expression changes are present in these cells as well. In cancer foci, the peri-glandular stromal cells are more fibroblastic than myoid,49 showing a loss of androgen receptor expression50 as well as changes in the expression of several other genes (such as the loss of the smooth muscle marker, calponin51).
CD molecules that are found in all prostate cancer cell types can in principle be exploited as targets for specific cell killing. Candidate CD molecules include CD47 (integrin-associated protein IAP), CD63, CD75s, CD107a, CD107b, CD147 (neurothelin), CD164 (sialomucin), and CD166 (activated leukocyte cell adhesion molecule ALCAM). A therapeutic strategy may entail the generation of immunotoxins by recombinant DNA techniques of single-chain antibody variable (scFv) domains coupled to toxin molecules.52 The toxin-antibody conjugate can be designed in such a way as to target cancer cells that co-express these CD molecules while sparing normal cells that do not co-express them.
Note Added in Proof
In a study by Osman I et al [Neutral endopeptidase protein expression and prognosis in localized prostate cancer. Clin Cancer Res 2004, 10:4096–4100], complete loss of CD10 expression was reported to be associated with PSA relapse. CD10 immunohistochemistry was scored for primary tumors (but no lymph node metastases) of 223 patients, represented by an African-American majority. Since loss of CD10 expression was seen in a majority of patients, one would expect a correspondingly large proportion of patients to fail. Perhaps of significance in this data set was the increased percentage of CD10+ tumors in African-American patients: 38.2% CD10 positive to 33.3% for Caucasian patients, 19.8% CD10 heterogeneous to 7.6% for Caucasian patients, and 42% CD10 negative to 59.1% for Caucasian patients. The higher positive rate and much lower negative rate for African-American patients might be used to explain the worse disease course seen for this patient group (if CD10+ tumor is associated with poor outcome as suggested by our data).
Acknowledgments
We thank Susan Saiget of BD-PharMingen for her invaluable contribution to this work, Tracy Sherertz for photography, Sarah Hawley for statistical analysis, and Beatrice Knudsen for critical comments, suggestions, and improvements.
Footnotes
Address reprint requests to Alvin Y. Liu, Department of Urology, Box 356510, University of Washington, Seattle, WA 98195. E-mail: aliu@u.washington.edu.
Supported by CaPCURE Foundation, grants CA85859, CA98699, and DK63630.
Presented in part at the Annual Meeting of the US-Canadian Academy of Pathology, 2000.
References
- Greenlee RT, Murray T, Bolden S, Wingo PA. Cancer Statistics 2000. CA Cancer J Clin. 2000;50:7–33. doi: 10.3322/canjclin.50.1.7. [DOI] [PubMed] [Google Scholar]
- Kronz JD, Silberman MA, Allsbrook WC, Bastacky SI, Burks RT, Cina SJ, Mills SE, Ross JS, Sakr WA, Tomaszewski JE, True LD, Ulbright TM, Weinstein MW, Yantiss RK, Young RH, Epstein JI. Pathology residents’ use of a Web-based tutorial to improve Gleason grading of prostate carcinoma on needle biopsies. Hum Pathol. 2000;31:1044–1050. doi: 10.1053/hupa.2000.16278. [DOI] [PubMed] [Google Scholar]
- Khoo VS, Pollack A, Cowen D, Joon DL, Patel N, Terry NHA, Zagars GK, von Eschenbach AC, Meistrich ML, Troncoso P. Relationship of Ki-67 labeling index to DNA-ploidy, S-phase fraction, and outcome in prostate cancer treated with radiotherapy. Prostate. 1999;41:166–172. doi: 10.1002/(sici)1097-0045(19991101)41:3<166::aid-pros3>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Liu AY, Nelson PS, van den Engh G, Hood L. Human prostate epithelial cell-type cDNA libraries and prostate expression patterns. Prostate. 2002;50:92–103. doi: 10.1002/pros.10036. [DOI] [PubMed] [Google Scholar]
- Liu AY, True LD. Characterization of prostate cell types by CD cell surface molecules. Am J Pathol. 2002;160:37–43. doi: 10.1016/S0002-9440(10)64346-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AY, Peehl DM. Characterization of cultured human prostatic epithelial cells by cluster designation antigen expression. Cell Tissue Res. 2001;305:389–397. doi: 10.1007/s004410100419. [DOI] [PubMed] [Google Scholar]
- Liu AY, LaTray L, van den Engh G. Changes in cell surface molecules associated with in vitro culture of prostatic stromal cells. Prostate. 2000;45:303–312. doi: 10.1002/1097-0045(20000901)44:4<303::aid-pros7>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Liu AY. Differential expression of cell surface molecules in prostate cancer cells. Cancer Res. 2000;60:3429–3434. [PubMed] [Google Scholar]
- Liu AY, True LD, LaTray L, Ellis WJ, Vessella RL, Lange PH, Higano CS, Hood L, van den Engh G. Analysis and sorting of prostate cancer cell types by flow cytometry. Prostate. 1999;40:192–199. doi: 10.1002/(sici)1097-0045(19990801)40:3<192::aid-pros7>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Liu AY, True LD, LaTray L, Nelson PS, Ellis WJ, Vessella RL, Lange PH, Hood L, van den Engh G. Cell-cell interaction in prostate gene regulation and cytodifferentiation. Proc Natl Acad Sci USA. 1997;94:10705–10710. doi: 10.1073/pnas.94.20.10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedland SJ, Seligson DB, Liu AY, Pantuck AJ, Paik SH, Horvath S, Wieder JA, Zisman A, Nguyen D, Tso C, Palotie AV, Belldegrun AS. Loss of CD10 (neutral endopeptidase) is a frequent and early event in human prostate cancer. Prostate. 2003;55:71–80. doi: 10.1002/pros.10202. [DOI] [PubMed] [Google Scholar]
- Greene FL, Page DL, Fleming ID, Fritz AG, Balch CM, Haller DG, Morrow M. AJCC Cancer Staging Handbook. New York, NY: Springer-Verlag; 2002 [Google Scholar]
- Wikström P, Lissbrant IF, Stattin P, Egevad L, Bergh A. Endoglin (CD105) is expressed on immature blood vessels and is a marker for survival in prostate cancer. Prostate. 2002;51:268–275. doi: 10.1002/pros.10083. [DOI] [PubMed] [Google Scholar]
- Bonkhoff H, Stein U, Remberger K. Multidirectional differentiation in the normal, hyperplastic, and neoplastic human prostate: simultaneous demonstration of cell-specific epithelial markers. Hum Pathol. 1994;25:42–46. doi: 10.1016/0046-8177(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Okada H, Tsubura A, Okamura A, Senzaki H, Naka Y, Komatz Y, Morii S. Keratin profiles in normal/hyperplastic prostates and prostate carcinoma. Virchows Arch A Pathol Anat. 1992;421:157–161. doi: 10.1007/BF01607049. [DOI] [PubMed] [Google Scholar]
- Rubin MA, de La Taille A, Bagiella E, Olsson CA, O’Toole KM. Cribriform carcinoma of the prostate and cribriform prostatic intraepithelial neoplasia. Am J Surg Pathol. 1998;22:840–848. doi: 10.1097/00000478-199807000-00006. [DOI] [PubMed] [Google Scholar]
- Potter VR. Phenotypic diversity in experimental hepatomas: the concept of partially blocked ontogeny. Br J Cancer. 1978;38:1–23. doi: 10.1038/bjc.1978.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipp MA, Richardson NE, Sayre PH, Brown NR, Masteller EL, Clayton LK, Ritz J, Reinherz EL. Molecular cloning of the common acute lymphoblastic leukemia antigen (CALLA) identifies a type II integral membrane protein. Proc Natl Acad Sci. 1988;85:4819–4823. doi: 10.1073/pnas.85.13.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Look AT, Ashmun RA, Shapiro LH, Peiper SC. Human myeloid plasma membrane glycoprotein CD13 (gp150) is identical to aminopeptidase N. J Clin Invest. 1989;83:1299–1307. doi: 10.1172/JCI114015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson JB, Carducci MA. Small bioactive peptides and cell surface peptidases in androgen-independent prostate cancer. Cancer Invest. 2000;18:87–96. doi: 10.3109/07357900009023066. [DOI] [PubMed] [Google Scholar]
- Joshi DD, Dang A, Yadav P, Qian J, Bandari PS, Chen K, Donnelly R, Castro T, Gascon P, Haider A, Rameshwar P. Negative feedback on the effects of stem cell factor on hematopoiesis is partly mediated through neutral endopeptidase activity on substance P: a combined functional and proteomic study. Blood. 2001;98:2697–2706. doi: 10.1182/blood.v98.9.2697. [DOI] [PubMed] [Google Scholar]
- Moody TW, Mayr CA, Gillespie T, Davis TP. Neurotensin is metabolized by endogenous proteases in prostate cancer cell lines. Peptides. 1998;19:253–258. doi: 10.1016/s0196-9781(97)00306-9. [DOI] [PubMed] [Google Scholar]
- di Sant’Agnese PA. Neuroendocrine cells of the prostate and neuroendocrine differentiation in prostatic carcinoma: a review of morphologic aspects. Urology. 1998;51(Suppl 5A):121–124. doi: 10.1016/s0090-4295(98)00064-8. [DOI] [PubMed] [Google Scholar]
- Papandreou CN, Usmani B, Geng Y, Bogenrieder T, Freeman R, Wilk S, Finstad CL, Reuter VE, Powell CT, Scheinberg D, Magill C, Scher HI, Albino AP, Nanus DM. Neutral endopeptidase 24.11 loss in metastatic human prostate cancer contributes to androgen-independent progression. Nat Med. 1998;4:50–57. doi: 10.1038/nm0198-050. [DOI] [PubMed] [Google Scholar]
- Best CJM, Leiva IM, Chuaqui RF, Gillespie JW, Duray PH, Murgai M, Zhao Y, Simon R, Kang JJ, Green JE, Bostwick DG, Linehan WM, Emmert-Buck MR. Molecular differentiation of high- and moderate-grade human prostate cancer by cDNA microarray analysis. Diagn Mol Pathol. 2003;12:63–70. doi: 10.1097/00019606-200306000-00001. [DOI] [PubMed] [Google Scholar]
- Gohring B, Holzhausen HJ, Meye A, Heynemann H, Rebmann U, Langner J, Riemann D. Endopeptidase 24.11/CD10 is down-regulated in renal cell cancer. Int J Mol Med. 1998;2:409–414. doi: 10.3892/ijmm.2.4.409. [DOI] [PubMed] [Google Scholar]
- Shipp MA, Tarr GE, Chen C, Switzer SN, Hersh LB, Stein H, Sunday ME, Reinherz EL. CD10/neutral endopeptidase 24.11 hydrolyzes bombesin-like peptides and regulates the growth of small cell carcinomas of the lung. Proc Natl Acad Sci USA. 1991;88:10662–10666. doi: 10.1073/pnas.88.23.10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenrieder T, Finstad CL, Freeman RH, Papandreou CN, Scher HI, Albino AP, Reuter VE, Nanus DM. Expression and localization of aminopeptidase A, aminopeptidase N, and dipeptidyl peptidase IV in benign and malignant human prostate tissue. Prostate. 1997;33:225–232. doi: 10.1002/(sici)1097-0045(19971201)33:4<225::aid-pros1>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Fogel M, Friederichs J, Zeller Y, Husar M, Smirnov A, Roitman L, Altevogt P, Sthoeger ZM. CD24 is a marker for human breast carcinoma. Cancer Lett. 1999;143:87–94. doi: 10.1016/s0304-3835(99)00195-0. [DOI] [PubMed] [Google Scholar]
- Kramer G, Steiner G, Födinger D, Fiebiger E, Rappersberger C, Binder S, Hofbauer J, Marberger M. High expression of a CD38-like molecule in normal prostatic epithelium and its differential loss in benign and malignant disease. J Urol. 1995;154:1636–1641. [PubMed] [Google Scholar]
- Chow KU, Boehrer S, Bojunga J, Stieler M, Rummel MJ, Fauth F, Schneider B, Martin H, Hoelzer D, Weidmann E, Mitrou PS. Induction of apoptosis by cladribine (2-CdA), gemcitabine and other chemotherapeutic drugs on CD34+/CD38+, and CD34+/CD38− hematopoietic progenitor cells: selective effects of doxorubicin and 2-CdA with protection of immature cells. Leuk Lymphoma. 2002;43:377–384. doi: 10.1080/10428190290006198. [DOI] [PubMed] [Google Scholar]
- Epstein AL, Marder RJ, Winter JN, Fox RI. Two new monoclonal antibodies (LN-1, LN-2) reactive in B5 formalin-fixed, paraffin-embedded tissues with follicular center and mantle zone human B lymphocytes and derived tumors. J Immunol. 1984;133:1028–1036. [PubMed] [Google Scholar]
- Mane SM, Marzella L, Bainton DF, Holt VK, Cha Y, Hildreth JEK, August JT. Purification and characterization of human lysosomal membrane glycoproteins. Arch Biochem Biophys. 1989;268:360–378. doi: 10.1016/0003-9861(89)90597-3. [DOI] [PubMed] [Google Scholar]
- Zellweger T, Ninck C, Mirlacher M, Annefeld M, Glass AG, Gasser TC, Mihatsch MJ, Gelmann EP, Bubendorf L. Tissue microarray analysis reveals prognostic significance of syndecan-1 expression in prostate cancer. Prostate. 2003;55:20–29. doi: 10.1002/pros.10209. [DOI] [PubMed] [Google Scholar]
- Zellweger T, Niuck C, Mirlacher M, Koivisto PA, Bloch M, Mihatsch MJ, Gasser TC, Bubendorf L. Expression patterns of potential therapeutic targets in prostate cancer. J Urol. 2003;169 5:213. doi: 10.1002/ijc.20615. [DOI] [PubMed] [Google Scholar]
- Tawfic S, Niehans GA, Manivel JC. The pattern of CD10 expression in selected pathologic entities of the prostate gland. Hum Pathol. 2003;34:450–456. doi: 10.1016/s0046-8177(03)00077-7. [DOI] [PubMed] [Google Scholar]
- Sumitomo M, Shen R, Walburg M, Dai J, Geng Y, Navarro D, Boileau G, Papandreou CN, Giancotti FG, Knudsen B, Nanus DM. Neutral endopeptidase inhibits prostate cancer cell migration by blocking focal adhesion kinase signaling. J Clin Invest. 2000;106:1399–1407. doi: 10.1172/JCI10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T, Takata M, Tustsumi S, Nishiyama K, Taguchi K, Nagai E, Tsuneyoshi M. Phenotypic expression of gastrointestinal differentiation markers in colorectal adenocarcinomas with liver metastasis. Pathology. 2002;34:556–560. doi: 10.1080/0031302021000035965-4. [DOI] [PubMed] [Google Scholar]
- Kanitakis J, Narvaez D, Claudy A. Differential expression of the CD10 antigen (neutral endopeptidase) in primary versus metastatic malignant melanomas of the skin. Melanoma Res. 2002;12:241–244. doi: 10.1097/00008390-200206000-00007. [DOI] [PubMed] [Google Scholar]
- Saiki I, Fujii H, Yoneda J, Abe F, Nakajima M, Tsuruo T, Azuma I. Role of aminopeptidase N (CD13) in tumor-cell invasion and extracellular matrix degradation. Int J Cancer. 1993;54:137–143. doi: 10.1002/ijc.2910540122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuhara T, Adachi M, Hashida H, Ishida H, Taki T, Higashiyama M, Kodama K, Tachibana S, Sasaki S, Miyake M. Neutral endopeptidase/CD10 and aminopeptidase N/CD13 gene expression as a prognostic factor in non-small cell lung cancer. Jpn J Thorac Cardiovasc Surg. 2001;49:489–496. doi: 10.1007/BF02919543. [DOI] [PubMed] [Google Scholar]
- Tso C, McBride WH, Sun J, Patel B, Tsui K, Paik SH, Gitlitz B, Caliliw R, van Ophoven A, Wu L, deKernion J, Belldegrun A. Androgen deprivation induces selective outgrowth of aggressive hormone-refractory prostate cancer clones expressing distinct cellular and molecular properties not present in parental androgen-dependent cancer cells. Cancer J. 2000;6:220–233. [PubMed] [Google Scholar]
- Liu AY, Corey E, Bladou F, Lange PH, Vessella RL. Prostatic cell lineage markers: emergence of BCL2+ cells of human prostate cancer xenograft LuCaP 23 following castration. Int J Cancer. 1996;65:85–89. doi: 10.1002/(SICI)1097-0215(19960103)65:1<85::AID-IJC15>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Hayward SW, Haughney PC, Rosen MA, Greulich KM, Weier HU, Dahiya R, Cunha GR. Interactions between adult human prostatic epithelium and rat urogenital sinus mesenchyme in a tissue recombination model. Differentiation. 1998;63:131–140. doi: 10.1046/j.1432-0436.1998.6330131.x. [DOI] [PubMed] [Google Scholar]
- Shima H, Tsuji M, Elfman F, Cunha GR. Development of the male urogenital epithelia elicited by soluble mesenchymal factors. J Androl. 1995;16:233–241. [PubMed] [Google Scholar]
- Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, Cunha GR. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999;59:5002–5011. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward SW, Grossfeld GD, Tlsty TD, Cunha GR. Genetic and epigenetic influences in prostatic carcinogenesis. Int J Oncol. 1998;13:35–47. [PubMed] [Google Scholar]
- Olapade-Olaopa EO, MacKay EH, Taub NA, Sandhu DPS, Terry TR, Habib FK. Malignant transformation of human prostatic epithelium is associated with the loss of androgen receptor immunoreactivity in the surrounding stroma. Clin Cancer Res. 1999;5:569–576. [PubMed] [Google Scholar]
- Tuxhorn JA, Ayala GE, Smith MJ, Smith VC, Dang TD, Rowley DR. Reactive stroma in human prostate cancer: induction of myofibroblast phenotype and extracellular matrix remodeling. Clin Cancer Res. 2002;8:2912–2923. [PubMed] [Google Scholar]
- Colcher D, Bird R, Roselli M, Hardman KD, Johnson S, Pope S, Dodd SW, Pantoliano MW, Milenic DE, Schlom J. In vivo tumor targeting of a recombinant single-chain antigen-binding protein. J Natl Cancer Inst. 1990;82:1191–1197. doi: 10.1093/jnci/82.14.1191. [DOI] [PubMed] [Google Scholar]