Abstract
The human autosomal-dominant disorder Axenfeld-Rieger syndrome presents with defects in development of the eyes, teeth, and umbilicus. The eye manifests with iris ruptures, irido-corneal adhesions, cloudy corneas, and glaucoma. Transcription factors such as PITX2 and FOXC1 have been found to carry point mutations, causing the disorder. However, for ∼40% of the cases, the pathogenesis is unknown. It has been reported that some mutations in PITX2 increase transactivation, whereas most mutations cause defects in DNA binding or transactivation. It is not known whether up-regulation of PITX2 activity can cause the disorder as well. Here we test this hypothesis directly by overexpressing PITX2A as a transgene in mouse corneal mesenchyme and iris, using keratocan-flanking sequences. The mice presented with corneal opacification, corneal hypertrophy, irido-corneal adhesions, and severely degenerated retina, resembling glaucoma. The corneal hypertrophy also resembles the corneal hypertrophy of Pitx2−/− mice. Control transgenic mice carrying point mutations T68P or K88E in PITX2A were normal. These findings indicate a novel pathogenetic mechanism in which excess corneal and iridal PITX2A cause glaucoma and anterior defects that closely resemble Axenfeld-Rieger syndrome.
Glaucoma is the most common cause of blindness.1 There are many different forms, classified according to, eg, age of onset. Since no drugs are yet available to reverse glaucoma, and because the eye is amenable to local drug delivery, there is significant potential for discovery of novel therapeutics in this area.2 The autosomal-dominant Axenfeld-Rieger syndrome affects the development of eye, tooth, and abdominal organs, and more rarely the heart and pituitary.3,4 The eyes of such patients can have cloudy corneas, iris adhesions to the cornea, iris hypoplasia, apparent ectopic pupils, an anterior and prominent Schwalbe’s line, tearing, and glaucoma. Mutations have been found in the transcription factors PITX2 (4q25) and FOXC1 (6p25), and linkage to as yet unidentified genes on 13q14 and 16q24.5 There are Rieger syndrome patients with balanced translocations, for example t(4;16) and t(4;11).6,7
For ∼40% of the cases of Axenfeld-Rieger syndrome investigated, no causative gene defect has been found. Mice disrupted in one allele for PITX2 display several phenotypes reminiscent of the human disorder, and the homozygous mice die prenatally of developmental defects of the heart.8–11 Notably, the ocular phenotype in the Pitx2−/− mice is corneal mesenchymal hypertrophy and increased corneal thickness.9,11 Pitx2 is also involved in the Nodal/Sonic hedgehog pathway, which determines left/right polarity of mesoderm-derived organs such as heart, gut, and stomach.12,13 In addition, Pitx2 is important for hindlimb development,14 and has been suggested to regulate terminal neuronal differentiation in the developing mouse thalamus and midbrain.15 Most mutations in human patients decrease DNA binding, transactivation, or both, when tested in cell culture or in vitro.3,5 Among the mutations found in PITX2 that cause Axenfeld-Rieger syndrome, there is one, V45L, with increased transactivation properties.16 This led the authors to postulate that excess PITX2 may also cause glaucoma and/or Axenfeld-Rieger syndrome.16
Mouse keratocan 5′-flanking sequences has previously been shown to direct β-galactosidase activity to periocular tissues from 13.5 days post coitum (E13.5), and to corneal mesenchyme from E14.5 to adult stages.17 At the mid-embryonic stage E14.5, native Pitx2 protein corneal expression is peaking, later to be progressively reduced in corneal structures, but remaining strong in the forming iris at E18.5 just before birth.18 Here we used the keratocan-flanking sequences to drive PITX2 expression in transgenic mice. The mice display with cloudy corneas, tearing, retinal degeneration, and irido-corneal adhesions. Together, these data demonstrate a novel pathogenetic mechanism in which gain-of-function alterations of PITX2 activity can cause defects similar to Rieger anomaly and glaucoma.
Materials and Methods
Transgenic Construct
A 4.7-kb fragment with 3.2-kb mouse keratocan (Ktcn) gene upstream regulatory region, 183-bp noncoding exon 1, 1.4-kb intron 1, and 6 noncoding bp of exon 2 was amplified from plasmid mGKtcn Bam/R16.6kb17 using primers (MWG Biotech, Edersberg, Germany): Kt-1: 5′-CGTCGGTACCATCCATCCCATAATCAACTTCCAAAC-3′,with a KpnI site in the 5′-end and Kt-2: 5′-CGTCGTCGACATTACAGCACCTGTGGAATAAGCAAC-3′, with a SalI site in the 5′-end. Advantage2 polymerase mix (Clontech/BD Biosciences, Bedford, MA) was used. Polymerase chain reaction (PCR) profile: 94°C for 30 seconds, 68°C for 4 minutes, for 7 cycles, 94°C for 30 seconds, 58°C for 1 minute, 68°C for 4 minutes for 27 cycles, 68°C for 4 minutes. A 917-bp cDNA for human PITX2A was amplified from total human cDNA (Clontech/BD Biosciences) using primers described19 with SalI in the 5′ end and NotI in the 3′ end. A 433-bp rabbit β-globin poly(A) signal sequence was PCR amplified from plasmid pCAGGS20 using primers polyAup1: 5′-GTACTGCAGATGCGGCCGCCTGGCTCACAAATACCAC-3′, with a NotI site in the 5′-end, and polyAlow-1: 5′-CGTCGGTACCTCAGGAGAGGAGGAAAAATCTG-3′, with a KpnI site in the 5′-end. The construct was joined in the order Ktcn promoter-PITX2A cDNA-rabbit polyA signals by ligation, and amplified using primers Kt-3: 5′-CGTCGGTACCCCCATAATCAACTTCCAAACGCTGAC-3′, and polyAlow-1:5′-CGTCGGTACCTCAGGAGAGGAGGAAAAATCTG-3′, yielding KpnI sites in both ends. The 6.1-kb product was gel purified, digested with KpnI, and ligated to a pBluescript vector. After sequencing, the construct was excised using KpnI, gel purified, and used for pronuclear injections. Point mutations T68P and K88E were generated in the PITX2A cDNA using PCR mutagenesis.21 Constructs carrying the point mutations in the PITX2A reading frame were made as for Ktcn-PITX2, resulting in constructs/mice Ktcn-K88E and Ktcn-T68P.
Mice
The ethics committee for animal research of Lund’s Tingsrätt has approved of our experiments. Staff at the Karolinska Institute Transgenic Core, Stockholm, performed pronuclear injections. The purified DNA construct was injected into fertilized mouse eggs at the one-cell stage. After injection, eggs were transferred into oviducts of pseudo pregnant female mice. The resulting F2 CBA × C57BL/6 mice were weaned, and sent to the BMC animal facilities at Lund University, Lund, were they were housed and treated according to guidelines of national and local animal ethics guidelines.
Genotyping
A 2.83-kb BglII fragment, unique for mice harboring the transgenic construct, was detected using a labeled PITX2a cDNA fragment in Southern blotting. By PCR using primers Kpa-1: 5′-GCCTGGATGGGAAGTTGATACTCT-3′, for intron 1 of Ktcn, and Kpa-5: 5′-CGAAGCCATTCTTGCATAGCTC-3′, for PITX2A, a 476-bp fragment, unique for mice harboring the transgenic construct, could be detected. PCR profile: 94°C for 30 seconds, 60°C for 1 minute, 68°C for 1 minute for 35 cycles.
Determination of Chromosomal Integration Site
We used a Genome Walker kit (Clontech/BD Biosciences). Briefly, mouse genomic DNA was digested with restriction enzymes and adaptors ligated to the ends, enabling regions adjacent to our construct to be amplified and sequenced.
Reverse Transcriptase-PCR
Isolation of total RNA from eyes was with the Trizol (InVitrogen, Carlsbad, CA) and RNeasy MinElute Clean-up (Qiagen, Valencia, CA) kits. The RNA was treated with RQ1 RNase-free DNase (Promega, Madison, WI), extracted with phenol/chloroform, and precipitated. Reverse transcription was performed using SuperscriptII (Gibco/BRL/InVitrogen, Carlsbad, CA) and a mix of random hexamer and oligo-dT primers, and according to the recommendations of the manufacturer. PITX2A cDNA was amplified using primers 5′-CGTCGTCGACTAACGGGGAAATGGAG-3′ and 5′-GTACTGCAGATGCGGCCGCAGCATAATTCCCAGTC-3′.19 Ribosomal protein Rps18 was amplified using 5′-GGGCTGGAGAACTCACGGAGGAT-3′ and 5′-GGCCCAGAGACTCATTTCTTCTT-3′, 350 bp; cyclinD2: 5′-GGGTAAGCTGAAGTGGAACCTG, and 5′-CCCGAATGGCTTCCTCACAGGT-3′, 450 bp. PCR profile: 10 cycles of 94°C for 30 seconds, 68°C for 90 seconds, 18 cycles of 94°C for 30 seconds, 60°C for 1 minute, 68°C for 1 minute.
Histochemistry and Immunohistochemistry
Eight-μm sections of frozen tissue were prepared and stained with PITX2 antibody or stained with hematoxylin and eosin (H&E) as described.18 For staining of MKi-67, we used Ki67 Ab-4 (Lab Vision/Neomarkers, Fremont, CA). The keratocan antibody has been described.22 The secondary antibody was goat anti-rabbit Alexafluor 488 (Molecular Probes, Eugene, OR). Slides were mounted in Permafluor (Beckman-Coulter, Fullerton, CA). For whole-mount staining, mouse eyes were fixed and permeabilized in 2% paraformaldehyde/2% Triton X-100 in phosphate-buffered saline (PBS) at 20°C for 10 minutes, washed in PBS and 50 mmol/L glycine/PBS, blocked in 3% bovine serum albumin/PBS at 4°C for 1 hour, and incubated overnight with the P2R10 PITX2 antibody.18 Washing was in 10 ml of PBS for 2 × 2 hours at 4°C. The secondary antibody was goat anti-rabbit Alexafluor 488. Washed whole mounts were viewed in the microscope. We used at 20° Zeiss Axioplan 2 microscope (Carl Zeiss AG, Göttingen, Germany), ×2.5/0.075, ×20/0.75, and ×40/0.95 lenses, a Hamamatsu ORCA-ER digital camera (Bridgewater, NJ), and Openlab 3.1.2 (Improvision, Coventry, UK) acquisition software.
Visual Tests
We used a visual tracking drum to evaluate overall eye performance in mice.23 Briefly, mice were put, unrestrained, on a small platform surrounded by a large vertically rotating drum, the inside of which was coated with black and white vertical stripes. Slow rotation (2 rpm) of the drum elicits a slow, synchronized head movement reflex in mice with normal vision. We used stripe width (4 mm) and radial distance (16 cm) corresponding to 1.4° to 2° (depending on how mice position themselves on the platform). Mice with normal visual acuity can resolved an angle of 1° readily.23,24 The visual tests were performed four times for each mouse (twice to the left and twice to the right) per occasion. We scored mice that failed all four subtests as having poor vision, and mice that passed at least three of the four subtests as having normal vision.
Terminal dUTP Nick-End Labeling (TUNEL) Assay
Testing apoptotic activity was performed on frozen, unfixed, 8-μm sections using a TUNEL assay with the In situ cell death detection kit, fluorescein (Roche, Indianapolis, IN), according to the recommendations of the manufacturer. Slides were mounted in Permafluor (Beckman-Coulter) and viewed in a fluorescence microscope.
Transmission Electron Microscopy
Eyes of 1-month-old wild-type and Ktcn-PITX2 mice were dissected, and then opened in the posterior end using a scalpel blade. Eyes were fixed for 3 hours with 2.5% glutaraldehyde, rinsed in Sörensen’s phosphate buffer, postfixed in 1% OsO4, and embedded in Epon (Resolution Performance Products, Houston, TX). Ultra-thin sections were stained with uranyl acetate and lead citrate. The samples were examined by transmission electron microscopy (Philips CM10, Bothell, WA).
Results
Generation of Transgenic Mice Overexpressing PITX2A Selectively in Corneal Mesenchyme
To achieve cornea-specific expression of PITX2A in mice as a transgene, human PITX2A cDNA was placed under control of the mouse Ktcn 5′-flanking sequences17 followed by rabbit β-globin polyA signal sequences20 (Figure 1A). The construct was termed Ktcn-PITX2. Two additional constructs were made, similar to Ktcn-PITX2, but carrying point mutations in the PITX2A reading frame originally isolated from human patients with Rieger syndrome, T68P and K88E.19,25 The resulting constructs and mice were termed Ktcn-T68P and Ktcn-K88E, respectively (Figure 1A). To analyze whether the transgenic construct was present in the genomic DNA, we isolated DNA from mouse tails for genotyping. Genotyping examples are presented in Figure 1B. We also analyzed eye RNA from 1-month-old Ktcn-PITX2 and wild-type mice for levels of PITX2 mRNA to detect overexpressing strains by semiquantitative reverse transcriptase (RT)-PCR (Figure 1B). PITX2A mRNA was drastically elevated in transgenic eyes. Control ribosomal RNA RPS18 was evenly expressed in both wild-type and transgenic eyes. A suggested PITX2 target gene in heart development, cyclin D2,26 was evenly expressed in both Ktcn-PITX2 and wild-type eyes. Procollagen lysyl hydroxylase 2 (Plod2), another suggested target gene,27 was slightly down-regulated in this assay. Of the 59 Ktcn-PITX2 mice obtained, 13 carried the transgene as determined by Southern blotting or PCR genotyping. Of the 29 Ktcn-T68P mice obtained, 5 carried the transgene, and of the 64 Ktcn-K88E mice obtained, 8 carried the transgene. Most lines expressed PITX2A in adult eye as determined by RT-PCR or immunofluorescence, and were similar in visible phenotypes. We herein report on the analyses of two lines of Ktcn-PITX2, A and B, which were identical in phenotypes and expression patterns. We determined the site of chromosomal integration for these lines. In line A, multiple copies were integrated on chromosome 10B1, in an intergenic region in the C57BL/6J genome, build 32.1, 380 kb from LOC234373 (similar to glutathione S-transferase Mu 3, mRNA accession number: XM_137047), and 570 kb from LOC382361 (similar to Gag, mRNA accession number: XM_356461). In line B, multiple copies were integrated on chromosome 12B3, in intron 12 of Akap6 (protein kinase A anchor protein 6, mRNA accession number: NM_198111).
Figure 1.
A: Ktcn-PITX2 transgenic construct. The mouse keratocan (Ktcn) 5′ flank, promoter, noncoding exon 1, intron 1, and 6 bp of noncoding exon 2 are depicted together in the white box. The human PITX2A reading frame (gray box) and rabbit poly-A signals (black box) were fused to the Ktcn sequences. Key restriction enzyme sites are marked above the construct. Start and stop codons for PITX2A are marked below. The horizontal arrows depict location of PCR primers used for genotyping. The map is not to scale. The Ktcn-T68P and Ktcn-K88E constructs are shown below, with sites for point mutations introduced marked with vertical arrows. B: Genotyping and RT-PCR. Typical results from genotyping Ktcn-PITX2 transgenic (t) and wild-type (w) mouse tail DNA, using PCR (left), or Southern blotting (middle). Right: Results from RT-PCR of 1-month-old Ktcn-PITX2 mouse eyes.
Ktcn-PITX2 Mice Have Clouded Corneas, Degraded Retinas, and Loss of Vision
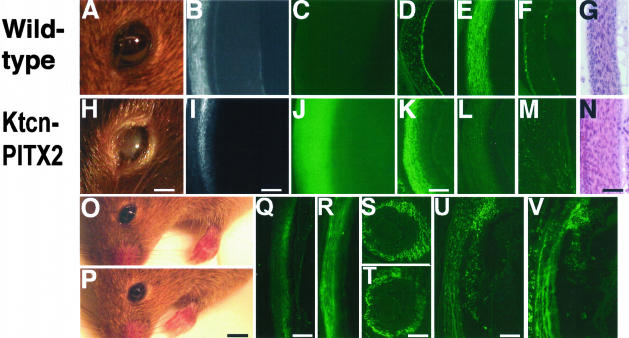
The eyes of 1-month-old Ktcn-PITX2 mice were opaque and tearing to varying degrees (Figure 2H), similar to human patients with glaucoma or cataracts. When viewed sideways in a light microscope, the mutant eyes displayed clouded, grainy corneas that occluded light from passing through, whereas the wild-type mice had clear corneas (Figure 2, I and B). The expression difference of PITX2 protein between wild-type and Ktcn-PITX2 corneas was dramatic. This was evident both for the 1-month-old eyes (Figure 2, C and J) as well as for 5-day-old eyes (Figure 2, D and K). Keratocan (Ktcn) protein, a marker for normal corneal identity, was lower in mutants compared to wild type (Figure 2, L and E). Proliferation, as indicated by Ki67 protein,28 was higher in mutant corneas (Figure 2, M and F). The Ktcn-PITX2 corneas were hypertrophic (Figure 2N) to varying degrees (see also Figure 4). The Ktcn-PITX2 mice also presented with a foreleg phenotype, because of a brief embryonic pulse of Ktcn promoter expression in foreleg mesenchyme. This foreleg phenotype is still under investigation and will be presented elsewhere. The control transgenic mice Ktcn-K88E and Ktcn-T68P appeared completely normal in all aspects (Figure 2, O and P). It has been shown previously that both mutated proteins T68P and K88E can be expressed stably in cell culture.19,25 We also noted overexpression of the mutated PITX2 proteins in corneas of the Ktcn-K88E and Ktcn-T68P mice (Figure 2, Q and R).
Figure 2.
Transgenic mice corneas become thick and cloudy. A–C: Wild-type 1-month-old mouse eyes; H–J: Ktcn-PITX2 1-month-old mouse eyes; D–G: wild-type 5-day-old corneas; K–N: Ktcn-PITX2 5-day-old corneas. A and H, Digital photographs; B and I, light microscopy photographs; C and J, whole-mount immunofluorescence of PITX2; D and K, immunofluorescence of sections with PITX2 antibody; E and L, immunofluorescence of sections with Ktcn antibody; F and M, immunofluorescence of sections with Ki-67 antibody; G and N, H&E-stained sections. O: Two-month-old Ktcn-K88E mouse; P, 2-month-old Ktcn-T68P mouse. Q and R: PITX2 immunostainings of 1-month-old cornea sections. Q, Ktcn-K88E; R, Ktcn-T68P. S and T: PITX2 immunostainings of E11.5 eyes. S, wild-type; T, Ktcn-PITX2. U and V: PITX2 immunostainings of E17.5 corneas and iris. U, wild-type; V, Ktcn-PITX2. Scale bars: 2 mm (H); 100 μm (I); 50 μm (K); 60 μm (N); 5 mm (P); 300 μm (Q); 150 μm (T); 200 μm (U).
Figure 4.
Severe irido-corneal phenotype of newborn Ktcn-PITX2 mice. A–F: Wild-type 5-day-old littermate; G–M: Transgenic Ktcn-PITX2 mouse. A, G, M: H&E staining; B and H: immunofluorescence of PITX2; C, D, I, J: immunofluorescence of MKi-67; E and K: immunofluorescence of Ktcn; F and L: TUNEL assay of retinas. Scale bar, 40 μm.
Expression in our PITX2 constructs is driven by the Ktcn promoter, which engages at E13.5 in the eye.17 Thus, we examined PITX2 expression in wild-type and Ktcn-PITX2 eyes at E11.5, before the Ktcn promoter is active; and at E17.5, when the Ktcn promoter has been activated. The results indicate that PITX2 is expressed in periocular mesenchyme in E11.5 wild-type and Ktcn-PITX2 mice at approximately equal levels (Figure 2, S and T). We observed a slight increase in PITX2 expression in both cornea and iris for the E17.5 Ktcn-PITX2 mice.
We tested the visual capacities of transgenic and wild-type mice by using a visual tracking drum.23 The results indicate that 60% of the young (1 to 4 months old) transgenic mice had poor vision (Table 1). With increasing age (6 to 8 months) vision deteriorated rapidly and 85% had poor vision. This can be compared to healthy mice in which only 5% of the 6 to 8 months old had impaired vision. The Ktcn-K88E and Ktcn-T68P mice had normal vision by visual tracking drum tests (not shown). The loss of vision of the Ktcn-PITX2 mice can in part be attributed to the clouded corneas (Figure 2). When we studied the retinas of wild-type and Ktcn-PITX2 mice, we saw a dramatic decrease in retinal thickness, from 150 μm in 3-month-old wild-type mice (Figure 3A) to 50 μm in 3-month-old transgenic mice (Figure 3B). All retinal cell layers appeared reduced in size. The outer nuclear cell layer was reduced by 50% in the transgenic mice. The inner retinal cell layer was reduced by 50 to 67%. The cell density of the retinal ganglion cells was also dramatically reduced. The retinal damage progressed with age. In some 6-month-old transgenic eyes the entire retina was degraded, and only the retinal pigment epithelium remained (Figure 3C). The severity also varied between the eyes of individual Ktcn-PITX2 transgenic mice, but both eyes were always affected. We studied the mechanism of cell death by using a TUNEL test, and saw dramatic incidence of TUNEL-positive nuclei in all cell types in the degenerating Ktcn-PITX2 retinas (Figure 3E), whereas no or very little TUNEL staining was evident in the wild-type retinas (Figure 3D). We detected compression or occlusion of some 5-day-old irido-corneal angles (Figure 3G and Figure 4M). Possibly these early angle malformations causes the later retinal dysgenesis (Figure 3, B and C) of the Ktcn-PITX2 mice.
Table 1.
Visual Tracking Drum Tests
| Age (months) | Wild type
|
Ktcn-PITX2
|
P value* | ||
|---|---|---|---|---|---|
| Normal | Poor | Normal | Poor | ||
| 1–4 | 10 | 1 | 7 | 11 | 0.0080 |
| 6 | 15 | 0 | 1 | 5 | 0.0003 |
| 8 | 5 | 1 | 1 | 6 | 0.0291 |
| Total | 30 | 2 | 9 | 22 | <0.0001 |
Two-tailed P value from Fischer’s exact test with 95% confidence interval.
Figure 3.
Dramatic retinal degeneration in Ktcn-PITX2 retinas. Wild-type 3-month-old (A), Ktcn-PITX2 3-month-old (B), and Ktcn-PITX2 8-month-old (C) mice retinas were sectioned and stained with H&E. Arrows indicate retinal ganglion cells. D: Wild-type retina TUNEL assay; E: Ktcn-PITX2 retina TUNEL assay. F and G: H&E-stained sections of irido-corneal angles. F, Wild type, 9 days old, arrow indicates angle; G, Ktcn-PITX2, 9 days old. Scale bars: 20 μm (A); 150 μm (G).
The Ktcn-PITX2 Mice Display Hypertrophic Cornea and Iris
All Ktcn-PITX2 mice displayed hypertrophic, or thick, corneas of varying degrees of severity (Figure 2, G and N). Some of the transgenic embryos in both lines A and B presented early with a severe ocular phenotype (Figure 4). Compared to a 5-day-old wild-type littermate (Figure 4A), the transgenic corneal mesenchyme was massively hypertrophic, and involuted toward the similarly hypertrophic iris (Figure 4, G and M). The iris in central and peripheral parts adhered to the cornea, and appeared fused to each other (Figure 4, G and M). The expression of PITX2 protein was evident in corneal mesenchyme, and in iris, and more intense in the cornea for the transgene (Figure 4H) than for the wild-type (Figure 4B). We also studied expression of the MKi-67 protein, a nuclear proliferation marker.28 In wild-type cornea (Figure 4C), no or very weak staining was detected, whereas in wild-type retina, widespread, intense staining was seen (Figure 4D). For the Ktcn-PITX2 mice, the inverse pattern applied; strong staining of MKi-67 in cornea (Figure 4I), but weak staining in retina (Figure 4J). Keratocan protein expression was high and uniform in wild-type cornea (Figure 4E), but patchy and weaker in transgenic cornea (Figure 4K). TUNEL staining in retinas at this early stage were negative (Figure 4, F and L), and there were no signs of retinal degeneration (not shown).
Ktcn-PITX2 Corneas Have Severely Disrupted Collagen Matrix Structure
We previously identified the PLOD (procollagen lysyl hydroxylase) family as likely cognate downstream target genes of PITX2 in the eye.27 PLOD is an enzyme that affects the stability of collagen crosslinking.29,30 To examine whether dysregulation of PITX2 caused defects in collagen fibril organization, we studied Ktcn-PITX2 and wild-type corneas of 1-month-old mice by electron microscopy (Figure 5). The Ktcn-PITX2 corneal mesenchyme was hypertrophic (Figures 2 and 4), and the corneal epithelium in 5-day-old Ktcn-PITX2 mice was also hypertrophic (Figure 4). In contrast, the 1-month-old Ktcn-PITX2 corneal epithelium appeared five times thinner than wild type (Figure 5, A and B). The Ktcn-PITX2 mid-corneal collagen superstructure was severely disrupted (Figure 5C). This can be compared with the characteristic collagen fibrillar structure of wild-type corneas (Figure 5D). In addition, the fibroblast cells that normally are thin and elongated (Figure 5, A and D) were in Ktcn-PITX2 corneas relaxed and expanded and making contacts with other nearby fibroblasts (Figure 5C). We noted a slight down-regulation in 1-month-old Ktcn-PITX2 mice by semiquantitative RT-PCR for one PITX2 target gene, Plod2 (Figure 1), also visible at day 5 (not shown).
Figure 5.
Ktcn-PITX2 corneas display disrupted collagen morphology. A–D: Electron microscopy. A, D: Wild-type corneas. B, C: Ktcn-PITX2 corneas. Scale bars: 10 μm (A, C, D); 2 μm (B).
Discussion
The Axenfeld-Rieger syndrome is inherited as autosomal dominant, resulting from either dominant-negative or haploinsufficient loss-of-function of PITX2, or of FOXC1 or other, in 40% of the cases of unknown causes.3,4 Here we have created a novel mouse model that resembles the ocular manifestation of Axenfeld-Rieger syndrome, based on a gain-of-function mechanism represented by higher-than-normal transcriptional activity of PITX2. Several lines of evidence support the idea of a gain-of-function mechanism contributing to the Axenfeld-Rieger syndrome: first, the Ktcn-PITX2 mice and Axenfeld-Rieger syndrome patients present with irido-corneal adhesions, corneal clouding, and glaucoma, with large variations of severity. This suggests that many of the ocular defects of the human disorder can be mimicked by PITX2A overexpression in mice. Second, the Ktcn-PITX2 mice and the Pitx2−/− mice present with corneal hypertrophy.9,11 Megalocornea, enlarged corneas, is occasionally seen in Axenfeld-Rieger patients.31 This implies that at least two different pathways involving Pitx2 exist that produce corneal hypertrophy; lack of Pitx2 or excess Pitx2. Third, there is one Rieger syndrome mutation, V45L, which has increased transactivation properties,16 supporting a gain-of-function pathogenesis. Mice hypomorphic for PITX2, that is, expressing slightly lower than normal levels of the mRNA/protein, also display serious malformations.11 Thus, minor defects, up or down, in PITX2 gene expression or function seem to have dire consequences for eye development (Figure 6). We think it likely that a fraction of patients with the Axenfeld-Rieger syndrome spectrum of disorders have genetic defects in the proximal promoter or distal regulatory elements of PITX2, causing up-regulation of normal PITX2 transcription levels. Alternatively, factors are mutated that normally suppress or control the level of PITX2 transcription. Any of these variants would produce gain-of-function alterations of PITX2, which is at least possible to fit within the 40% of unexplained causatives of Axenfeld-Rieger syndrome. It is well known that gain-of-function mutations in genes for transcription factors can cause inherited disorders. Examples include MSX2 in Boston-type craniosynostosis,32–34 TCF2/HNFβ in disorders of the kidney and pancreas,35,36 and NFKBIA in X-linked anhidrotic ectodermal dysplasia with immunodeficiency.37 A gain-of-function mechanism has also, based on a mouse model, been proposed for PROP1 in pituitary endocrinopathies.38
Figure 6.
Model for gain-or loss-of-function in Rieger syndrome. Known heterozygous defects in the PITX2 gene can cause eye disorders. The human and mouse homozygotes are early embryonic lethal. These mice develop hypertrophic corneas. Heterozygous mice develop phenotypes resembling human patients, even the hypomorphic variants, with PITX2 expression levels between heterozygous and normal. The Rieger-syndrome-causing mutation V45L elevates PITX2 transactivating activity. The Ktcn-PITX2 mice overexpress PITX2 in the cornea and iris, resulting in hypertrophic corneas like the mouse Pitx2 homozygotes, and Rieger syndrome-like effects including irido-corneal adhesions and retinal degeneration/glaucoma.
Human patients with Axenfeld-Rieger syndrome are typically heterozygous for point mutations in the PITX2 reading frame. Because PITX2 binds and transactivates DNA as a homodimer in in vitro and in cell culture experiments, the equal gene dosage of normal and defective protein then contributes, by different mechanisms, to the disease.3,25 When overexpressed, we postulated that T68P or K88E mutant forms of PITX2A would form an excess of defective protein. The mirrored expression engineered here, such that the Ktcn 5′-flank directs expression of our constructs to times in development when native PITX2 is down-regulated, would minimize the chances of disruptive heterozygous dimer formation. This seems to have been the case, because both the Ktcn-T68P and Ktcn-K88E mice were normal in all tests used.
Our Ktcn-PITX2 transgenic construct directs expression to the corneal mesenchyme as shown here, and also early periocular mesenchyme, as described for the β-galactosidase fusion of the keratocan 5′ flank.17 There is a native background of iris expression of Pitx2 even at late embryonic stages.18 For the severe phenotype of Ktcn-PITX2, we detected a dramatically increased cell mass of the iris, in addition to the increased cell mass of the corneal mesenchyme. This is most likely explained by Ktcn-driven expression in the early periocular mesenchyme, which later develops into corneal and iridal structures.
When Pitx2 is overexpressed in HeLa cell cultures without native Pitx2 expression, cell spreading and associated genes are induced.39 In our model, hypertrophic growth is induced in corneal mesenchyme, and in iris. This is linked to a subsequent degradation of the retina, most likely via the aberrant irido-corneal angles and ocular overpressure. In the more severe hypertrophic form of Ktcn-PITX2, the retina is also detached by the physical intrusion of the irido-corneal cell mass. In both the milder and more severe form of Ktcn-PITX2 corneal hypertrophy, we detected higher proliferation in corneas, and lower in retinas, by studying MKi-67 protein expression. By contrast, another proliferative marker, cyclin D2, was not elevated. Cyclin D2 has been identified as a PITX2 target gene in heart,23,40 but its expression was not affected in the stages of Ktcn-PITX2 mice eyes we tested. We previously identified PLOD as a likely PITX2 target gene family.27 The PLOD enzymes are crucial for maintaining collagen crosslinking stability.29,30 The fact that the corneal collagen fibrillar structure is severely damaged in the Ktcn-PITX2 mice could be interpreted as PITX2 overactivity affecting PLOD function. We detected a slight reduction of Plod2 mRNA in Ktcn-PITX2 mice by RT-PCR, which might be part of the explanation. For more exact confirmation of various PITX2 target genes, an inducible Ktcn-PITX2 transgene with many immediate time samples would be required. The keratocan protein is a marker for corneal mesenchyme cells.17 Keratocan was expressed at lower levels than normal in the Ktcn-PITX2 mouse corneas. This gives molecular support for the histological observations of loss of corneal mesenchymal identity. We could also see dramatic aberrations in mature corneas by electron microscopy. Because ablation of the Ktcn gene22 results in modest corneal hypotrophy, thinning of the corneas, and very subtle disruptions of collagen superstructure, we do not believe genomic insertion of our transgenic construct has affected native Ktcn expression.
In summary, our mouse model shows that high levels of PITX2 can invoke Rieger syndrome-like conditions. This suggests that gain-of-function of PITX2 may also cause Rieger syndrome in human patients. It will therefore be important to study whether there are human disease-causing gain-of-function mutations in genes that code for factors that regulate PITX2 expression or perhaps in regulatory regions of the PITX2 gene.
Acknowledgments
We thank Dr. Jun-Ichi Miyazaki for providing the plasmid pCAGGS; Dr. Johannes Wilbertz and staff at the Karolinska Institute Transgenic Core, Stockholm; Dr. Lena Persson-Feld and staff at Lund University Animal Care facilities; Eric Carlemalm and staff at the Electron Microscopy core, Lund; and Drs. T. van Veen, P. Ekblom, M. Ekblom, and M. Durbeej-Hjalt for helpful discussions.
Footnotes
Address reprint requests to Tord A. Hjalt, Lund University, Department of Cell and Molecular Biology, Section for Cell and Developmental Biology, BMC B12, Tornavägen 10, SE-22184 Lund, Sweden. E-mail: tord.hjalt@medkem.lu.se.
Supported by the Swedish Research Council (K2002-32X-14190-01A, K2004-31X-14190-03A, and 621-2001-2007 to T.A.H.), Kungliga Fysiografiska Sällskapet (to T.A.H.), Magnus Bergvalls Stiftelse (to T.A.H.), Crafoordska Stiftelsen (to T.A.H.), Åke Wibergs Stiftelse (to T.A.H.), and the National Institutes of Health (grant RO1 EY12486 to C.-Y.L.).
References
- Quigley HA. Number of people with glaucoma worldwide. Br J Ophthalmol. 1996;80:389–393. doi: 10.1136/bjo.80.5.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AF, Yorio T. Ophthalmic drug discovery. Nat Rev Drug Disc. 2003;2:448–459. doi: 10.1038/nrd1106. [DOI] [PubMed] [Google Scholar]
- Amendt BA, Semina EV, Alward WLM. Rieger syndrome: a clinical, molecular, and biochemical analysis. Cell Mol Life Sci. 2000;57:1652–1666. doi: 10.1007/PL00000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage PJ, Suh H, Camper SA. The bicoid-related Pitx gene family in development. Mamm Genome. 1999;10:197–200. doi: 10.1007/s003359900970. [DOI] [PubMed] [Google Scholar]
- Lines MA, Kozlowski K, Walter MA. Molecular genetics of Axenfeld-Rieger malformations. Hum Mol Genet. 2002;11:1177–1184. doi: 10.1093/hmg/11.10.1177. [DOI] [PubMed] [Google Scholar]
- Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, Datson NA, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel U, Carey JC, Murray JC. Cloning and characterization of a novel bicoid-related homeobox gene, RIEG, involved in Rieger syndrome. Nat Genet. 1996;14:392–399. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- Trembath DG, Semina EV, Jones DH, Patil SR, Qian Q, Amendt BA, Russo AF, Murray JC. Analysis of two translocation breakpoints and identification of a negative regulatory element in patients with Rieger’s syndrome. Birth Defects Res Part A Clin Mol Teratol. 2004;70:82–91. doi: 10.1002/bdra.10154. [DOI] [PubMed] [Google Scholar]
- Campione M, Steinbeisser H, Schweickert A, Deissler K, van Bebber F, Lowe LA, Nowotschin S, Viebahn C, Haffter P, Kuehn MR, Blum M. The homeobox gene Pitx2: mediator of asymmetric left-right signaling in vertebrate heart and gut looping. Development. 1999;126:1225–1234. doi: 10.1242/dev.126.6.1225. [DOI] [PubMed] [Google Scholar]
- Lu M-F, Pressman C, Dyer R, Johnson RL, Martin JF. Function of Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature. 1999;401:276–278. doi: 10.1038/45797. [DOI] [PubMed] [Google Scholar]
- Lin CR, Kioussi C, O’Connell S, Briata P, Szeto D, Liu F, Izpisua-Belmonte JC, Rosenfeld MG. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401:279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- Gage PJ, Suh H, Camper SA. Dosage requirement of Pitx2 for development of multiple organs. Development. 1999;126:4643–4651. doi: 10.1242/dev.126.20.4643. [DOI] [PubMed] [Google Scholar]
- Hamada H, Meno C, Watanabe D, Saijoh Y. Establishment of vertebrate left-right asymmetry. Nat Rev Genet. 2002;3:103–113. doi: 10.1038/nrg732. [DOI] [PubMed] [Google Scholar]
- Tamura K, Yonei-Tamura S, Belmonte JC. Molecular basis of left-right asymmetry. Dev Growth Differ. 1999;41:645–656. doi: 10.1046/j.1440-169x.1999.00465.x. [DOI] [PubMed] [Google Scholar]
- Marcil A, Dumontier E, Chamberland M, Camper SA, Drouin J. Pitx1 and Pitx2 are required for development of hindlimb buds. Development. 2003;130:45–55. doi: 10.1242/dev.00192. [DOI] [PubMed] [Google Scholar]
- Martin DM, Skidmore JM, Philips ST, Vieira C, Gage PJ, Condie BG, Raphael Y, Martinez S, Camper SA. PITX2 is required for normal development of neurons in the mouse subthalamic nucleus and midbrain. Dev Biol. 2004;267:93–108. doi: 10.1016/j.ydbio.2003.10.035. [DOI] [PubMed] [Google Scholar]
- Priston M, Kozlowski K, Gill D, Letwin K, Buys Y, Levin AL, Walter MA, Heon E. Functional analyses of two newly identified PITX2 mutants reveal a novel molecular mechanism for Axenfeld-Rieger syndrome. Hum Mol Gen. 2001;10:1631–1638. doi: 10.1093/hmg/10.16.1631. [DOI] [PubMed] [Google Scholar]
- Liu C-Y, Arar H, Kao C, Kao WW-Y. Identification of a 3.2 kb 5′-flanking region of the murine keratocan gene that directs β-galactoside expression in the adult corneal stroma of transgenic mice. Gene. 2000;250:85–96. doi: 10.1016/s0378-1119(00)00165-7. [DOI] [PubMed] [Google Scholar]
- Hjalt TA, Semina EV, Amendt BA, Murray JC. The Pitx2 protein in mouse development. Dev Dyn. 2000;218:95–200. doi: 10.1002/(SICI)1097-0177(200005)218:1<195::AID-DVDY17>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Amendt BA, Sutherland LB, Semina EV, Russo AF. The molecular basis of Rieger syndrome. J Biol Chem. 1998;273:20066–20072. doi: 10.1074/jbc.273.32.20066. [DOI] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Horton RM, Ho SN, Pullen JK, Hunt HD, Cai Z, Pease LR. Gene splicing by overlap extension. Methods Enzymol. 1993;217:270–279. doi: 10.1016/0076-6879(93)17067-f. [DOI] [PubMed] [Google Scholar]
- Liu CY, Birk DE, Hassell JR, Kane B, Kao WW. Keratocan-deficient mice display alterations in corneal structure. J Biol Chem. 2003;278:21672–21677. doi: 10.1074/jbc.M301169200. [DOI] [PubMed] [Google Scholar]
- Thaung C, Arnold K, Jackson IJ, Coffey PJ. Presence of visual head tracking differentiates normal sighted from retinal degenerate mice. Neurosci Lett. 2002;325:21–24. doi: 10.1016/s0304-3940(02)00223-9. [DOI] [PubMed] [Google Scholar]
- Artal P, Herreros de Tejada P, Munoz Teda C, Green DG. Retinal image quality in the rodent eye. Vis Neurosci. 1998;15:597–605. doi: 10.1017/s0952523898154020. [DOI] [PubMed] [Google Scholar]
- Saadi I, Semina EV, Amendt BA, Harris DJ, Murphy KP, Murray JC, Russo AF. Identification of a dominant negative homeodomain mutation in Rieger syndrome. J Biol Chem. 2001;276:23034–23041. doi: 10.1074/jbc.M008592200. [DOI] [PubMed] [Google Scholar]
- Kioussi C, Briata P, Baek SH, Rose DW, Hamblet NS, Herman T, Ohgi KA, Lin C, Gleiberman A, Wang J, Brault V, Ruiz-Lozano P, Nguyen HD, Kemler R, Glass CK, Wynshaw-Boris A, Rosenfeld MG. Identification of a Wnt/Dvl/b-Catenin-Pitx2 pathway mediating cell-type-specific proliferation during development. Cell. 2002;111:673–685. doi: 10.1016/s0092-8674(02)01084-x. [DOI] [PubMed] [Google Scholar]
- Hjalt TA, Amendt BA, Murray JC. PITX2 regulates procollagen lysyl hydroxylase (PLOD) gene expression: implications for the pathology of Rieger syndrome. J Cell Biol. 2001;152:545–552. doi: 10.1083/jcb.152.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes J, Li L, Schlueter C, Duchrow M, Wohlenberg C, Gerlach C, Stahmer I, Kloth S, Brandt E, Flad HD. Immunobiochemical and molecular biologic characterization of the cell proliferation-associated nuclear antigen that is defined by monoclonal antibody Ki-67. Am J Pathol. 1991;138:867–873. [PMC free article] [PubMed] [Google Scholar]
- Kivirikko KI, Myllylä R, Pihlajaniemi T. Hydroxylation of proline and lysine residues in collagens and other animal and plant proteins. Harding JJ, Crabba MJC, editors. Boca Raton: CRC Press; Focus on Post-Translational Modifications of Proteins. 1992:pp 1–51. [Google Scholar]
- Kivirikko KI, Pihlajaniemi T. Collagen hydroxylases and the protein disulfide isomerase subunit of prolyl 4-hydroxylases. Adv Enzymol Relat Areas Mol Biol. 1998;72:325–398. doi: 10.1002/9780470123188.ch9. [DOI] [PubMed] [Google Scholar]
- Traboulsi EI. Traboulsi EI, editor. New York: Oxford University Press; Genetic Diseases of the Eye. 1998:pp 81–98. [Google Scholar]
- Jabs EW, Muller U, Li X, Ma L, Luo W, Haworth IS, Klisak I, Sparkes R, Warman ML, Mulliken JB, Snead ML, Maxson R. A mutation in the homeodomain of the human MSX2 gene in a family affected with autosomal dominant craniosynostosis. Cell. 1993;75:443–450. doi: 10.1016/0092-8674(93)90379-5. [DOI] [PubMed] [Google Scholar]
- Liu YH, Kundu R, Wu L, Luo W, Ignelzi MA, Jr, Snead ML, Maxson JR RE. Premature suture closure and ectopic cranial bone in mice expressing Msx2 transgenes in the developing skull. Proc Natl Acad Sci USA. 1995;92:6137–6141. doi: 10.1073/pnas.92.13.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Golden S, Wu L, Maxson R. The molecular basis of Boston-type craniosynostosis: the pro148-to-his mutation in the N-terminal arm of the MSX2 homeodomain stabilizes DNA binding without altering nucleotide sequence preferences. Hum Mol Genet. 1996;5:1915–1920. doi: 10.1093/hmg/5.12.1915. [DOI] [PubMed] [Google Scholar]
- Bingham C, Ellard S, Allen L, Bulman M, Shepherd M, Frayling T, Berry PJ, Clark PM, Lindner T, Bell GI, Ryffel GU, Nicholls AJ, Hattersley AT. Abnormal nephron development associated with a frameshift mutation in the transcription factor hepatocyte nuclear factor-1-beta. Kidney Int. 2000;57:898–907. doi: 10.1046/j.1523-1755.2000.057003898.x. [DOI] [PubMed] [Google Scholar]
- Wild W, Pogge von Strandmann E, Nastos A, Senkel S, Lingott-Frieg A, Bulman M, Bingham C, Ellard S, Hattersley AT, Ryffel GU. The mutated human gene encoding hepatocyte nuclear factor 1-beta inhibits kidney formation in developing Xenopus embryos. Proc Natl Acad Sci USA. 2000;97:4695–4700. doi: 10.1073/pnas.080010897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois G, Smahi A, Reichenbach J, Doffinger R, Cancrini C, Bonnet M, Puel A, Chable-Bessia C, Yamaoka S, Feinberg J, Dupuis-Girod S, Bodemer C, Livadiotti S, Novelli F, Rossi P, Fischer A, Israel A, Munnich A, Le Deist F, Casanova J-L. A hypermorphic I-kappa-B-alpha mutation is associated with autosomal dominant anhidrotic ectodermal dysplasia and T cell immunodeficiency. J Clin Invest. 2003;112:1108–1115. doi: 10.1172/JCI18714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman LJ, Watkins-Chow DE, Brinkmeier ML, Raetzman LT, Radak AL, Lloyd RV, Camper SA. Persistent Prop1 expression delays gonadotrope differentiation and enhances pituitary tumor susceptibility. Hum Mol Genet. 2001;10:1141–1153. doi: 10.1093/hmg/10.11.1141. [DOI] [PubMed] [Google Scholar]
- Wei Q, Adelstein RS. Pitx2a expression alters actin-myosin cytoskeleton and migration of HeLa cells through Rho GTPase signalling. Mol Biol Cell. 2002;13:683–697. doi: 10.1091/mbc.01-07-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briata P, Llengo C, Corte G, Moroni C, Rosenfeld MG, Chen C-Y, Gherzi R. The Wnt/b-catenin-PITX2 pathway controls the turnover of Pitx2 and other unstable mRNAs. Mol Cell. 2003;12:1201–1211. doi: 10.1016/s1097-2765(03)00407-6. [DOI] [PubMed] [Google Scholar]