Abstract
Expression of the cell adhesion molecule CEACAM1 in melanomas is an independent factor for the risk of metastasis with a predictive value superior to that of tumor thickness. We have previously shown that CEACAM1 co-localizes at the tumor-stroma interface of invading melanoma masses with integrin β3 and that these two adhesion molecules interact via the CEACAM1 cytoplasmic domain. To address the functional consequences of CEACAM1 expression, we investigated invasion and migration of melanocytic and melanoma cells that stably express CEACAM1 using two different in vitro systems. Here, we demonstrate that CEACAM1 expression markedly enhances cell invasion and migration. The enhanced invasion and migration of CEACAM1-transfected cells was dependent on the presence of Tyr-488 within the full-length cytoplasmic CEACAM1 domain. Treatment with anti-CEACAM monoclonal antibodies blocked CEACAM1-enhanced cell invasion and cell migration in a dose-dependent manner. Furthermore, the enhanced invasion and migration of CEACAM1-transfected melanoma cells was blocked by integrin-antagonizing RGD peptides. Expression of integrin β3 induces the up-regulation of CEACAM1 in melanocytic MEL6 cells. These results strengthen the view that CEACAM1 and αvβ3 integrin are functionally interconnected with respect to the invasive growth of melanomas.
In malignant melanoma, dysregulation of cell adhesion molecules is associated with tumor progression.1 In human cutaneous melanoma, the radial growth phase is prognostically favorable, whereas the vertical growth phase often gives rise to metastases.2 The expression of cell adhesion molecules may be either up-regulated or down-regulated when melanomas progress from the radial to the vertical growth phase. Among the most prominent cell adhesion molecules down-regulated during tumor progression is E-cadherin. On the other hand, avβ3 integrin and MCAM are expressed preferentially in the vertical growth phase.3
The prognostic significance of the expression of cell adhesion molecules in human cutaneous melanomas has been substantiated in clinical studies. For example, when avβ3 integrin and/or ICAM-1 are present in tumor tissues, survival rates of patients are significantly lower when compared to patients whose tumors do not express these adhesion molecules.4 These and other correlative data imply that cell adhesion molecules are of functional significance in the progression of human melanomas. This assumption was investigated in experimental studies. The reconstitution of αvβ3 integrin in human melanoma cells induces conversion from the radial to the vertical growth phase as shown in three-dimensional skin reconstructs.5 In in vivo models, both αvβ3 integrin and MCAM lead to increased tumor growth in immunodeficient mice. Tumor growth could be inhibited by agents that functionally interfere with these adhesion molecules.6–8
Recent gene array studies indicate that human melanomas, similar to other human tumors, exhibit considerable heterogeneity with respect to mRNA expression profiles.9 To transfer experimental data into therapeutic applications in patients, it is essential to define the individual contribution of distinct adhesion molecules to the malignant phenotype. Importantly, we showed that expression of CEACAM1, an adhesion molecule of the immunoglobulin superfamily, in human melanomas was associated with a poor prognosis of the patients. Indeed, the predictive value of CEACAM1 expression was superior to that of tumor thickness. Often, the strongest CEACAM1 expression was observed at the invading tumor front.10 These findings suggest that CEACAM1 actively contributes to the progression of malignant melanomas. If this assumption is correct, CEACAM1 may be an additional target to inhibit tumor growth. In this contribution, we provide evidence that the expression of CEACAM1 with its long cytoplasmic domain (CEACAM1-L) in human melanocytes and melanoma cells increases migratory and invasive growth properties in in vitro models. The invasive growth mediated by CEACAM1 could be inhibited by monoclonal CEACAM1 antibodies.
Materials and Methods
Cells and Cell Lines
The human melanoma MV3 and melanocytic MEL6 cell lines were maintained in RPMI 1640 supplemented with 10% heat-inactivated fetal calf serum, 50 U/ml of penicillin, and 50 mg/ml streptomycin at 37°C in 5% CO2 humidified air. The cells were routinely passaged with trypsin-ethylenediaminetetraacetic acid (EDTA) buffer. All tissue culture reagents were purchased from BD Biosciences (Bedford, MA) and Gibco (Invitrogen, Karlsruhe, Germany). All other reagents were purchased from Sigma (Munich, Germany) except for fetal calf serum (PAA Laboratories, Cölbe, Germany).
Antibodies and Purified Adhesive Proteins
The following antibodies were purchased: anti-integrin-β3 CBL 479 (Chemicon, Temecula, CA) and polyclonal anti-integrin-β3 C-20 (Santa Cruz Biotechnology Inc., Santa Cruz, CA). The monoclonal antibody (mAb) 4D1C2 specific for human CEACAM1 was described previously.11,12 The CEACAM mAb 12-140-4 was a kind gift from Øle P. Børmer (Norwegian Radium Hospital, Oslo, Norway). Purified RGD peptides were purchased from Bachem (Weil am Rhein, Germany).
Construction of cDNAs
Cloning of full-length cDNA encoding CEACAM1–4L and generation of point mutants Y488F and Y515F (numbering of amino acid residues refers to the mouse sequence) was performed as described.13–15 The double-tyrosine mutant Y488,515F was generated by the same method starting with a plasmid carrying a single mutation. The short isoform (CEACAM1–4S) was created from human colonic epithelial CEACAM1–4L cDNA using primers corresponding to nucleotides 878 to 903 (upstream primer) and 1354 to 1405 (downstream primer) of the human CEACAM1 coding sequence. Each mutant CEACAM1 construct was verified by dideoxy sequencing and cloned in the expression vector pcDNA 3.1/Zeo (−). Human integrin β3 was amplified from a human cDNA library by polymerase chain reaction and cloned in the EBB expression vector.
Transfection and Selection of Stable Cell Clones
Full-length CEACAM1 and CEACAM mutants cloned in pcDNA vector (4 μg) were mixed with 10 μl of lipofectamine plus (Invitrogen Life Technology) in a final volume of 2 ml of Opti-MEM medium and added to MV3 and MEL6 cells grown to 70% confluence in 100-mm tissue culture plates. After 5 hours fetal calf serum was added to the culture medium. Forty-eight hours after transfection the cells were detached from the dish by treating with trypsin-EDTA followed by washing two times with RPMI 1640 and 10% fetal calf serum. Cells were selected by using selection media (0.3 mg/ml zeocin; (Invitrogen Life Technology). The stable transfected cells were maintained in RPMI 1640 medium containing 0.125 mg/ml of zeocin.
Western Blot Analysis
Total cell lysates (50 μg of protein) were resolved by 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis or sodium dodecyl sulfate-polyacrylamide gel electrophoresis on precast NuPAGE 4 to 10% Bis-Tris gels (Invitrogen Life Technology) and transferred onto nitrocellulose using a semidry transblot apparatus (Biometra, Göttingen, Germany). The membranes were blocked for 1 hour in blocking buffer [phosphate-buffered saline (PBS) containing 5% nonfat dry milk] and incubated overnight with the primary antibody diluted in PBS. After 5 minutes of washing in TBS-Tween (10 mmol/L Tris-HCl, pH 7.5, 150 mmol/L NaCl, 0.1% Tween), the membranes were incubated for 1 hour with goat anti-mouse IgG conjugated to horseradish peroxidase (Dianova, Hamburg, Germany) in PBS. The membranes were then washed in TBS and bound antibody visualized with the enhanced chemiluminescence detection system (Amersham, Freiburg, Germany) according to the manufacturer’s instructions.
In Vitro Invasion Assay
The invasiveness of cells was assayed in the membrane invasion culture system using PET membrane (8-μm pore size) in the upper compartment of a transwell coated with Matrigel (BD BioCoat Matrigel Invasion Chamber; BD Biosciences). Cells were harvested by trypsin-EDTA buffer and washed with RPMI 1640. The cells were then seeded at 2.5 × 104 cells per 500 μl of RPMI 1640 on the Matrigel-coated PET membrane in the upper compartment. The lower compartment was filled with RPMI 1640 medium and the plates were incubated at 37°C for 24 hours. At the end of the incubation time, the upper surface of the membrane was wiped with a cotton-tip applicator to remove nonmigratory cells. Cells that migrated to bottom side of membrane were fixed and stained by Diff-Quick (Dade Behring AG, Düdingen, Switzerland). The cells were quantified by counting nine high-powered fields (460 μm × 700 μm) in the center of each well using a Zeiss microscope. Each measurement was performed in quadruplicate and experiments were repeated five times.
Boyden Chamber Assay
Migration assays were performed as described previously16 in a 96-well microtaxis chamber. Polyester membranes with 8-μm pores (Neuro Probe, Cabin John, MD) were coated with 0.1 mg/ml of collagen I (Collagen Corp., Palo Alto, CA). MV3 and MEL6 cells were harvested using trypsin-EDTA and resuspended in RPMI 1640 containing 0.1% bovine serum albumin. Cells (2.5 × 104) were suspended in 100 μl of serum-free medium containing 0.1% bovine serum albumin and seeded on the upper side of 8-μm-pore filters in the Boyden chamber, and incubated with or without stimuli for 5 hours. Cells on the upper side of the filters were then mechanically removed. Cells on the lower filter side were then fixed and stained with Diff-Quick. The cells were quantified by counting nine high-powered fields (460 μm × 700 μm) in the center of each well using a Zeiss microscope. Each condition was studied in triplicate wells, and each experiment was performed five times.
RNA Preparation and Microarray Analysis
Total RNA was prepared by lysing the cells in Trizol reagent according to the manufacturer’s instructions (Life Technologies Inc., Grand Island, NY). The total RNA was cleaned up using RNeasy mini kit according to the manufacturer’s instructions (Qiagen). For all experiments Affymetrix Human U133 Set GeneChips containing 39,000 genes and expressed sequence tags were used. The targets for Affymetrix DNA microarray analysis were prepared as described by the manufacturer; the amount of total RNA used for the cDNA synthesis was 10 μg for each reaction. GeneChip microarrays were hybridized with the targets for 16 hours at 45°C, washed, and stained using the Affymetrix Fluidics Station according to the GeneChip Expression Analysis Technical Manual. Microarrays were scanned with the Hewlett-Packard Agilent GeneChip scanner, and the signals were processed using the GeneChip expression analysis algorithm (v.2; Affymetrix). To compare samples and experiments, the trimmed mean signal of each array was scaled to a target intensity of 200 for A-arrays. Absolute and comparison analyses were performed with Affymetrix MAS 5.0 and DMT software using default parameters. To assist the identification of genes that were positively or negatively regulated, we selected genes that were increased or decreased at least twofold compared to the baseline.17
Results
Setup of the Experimental System
Several melanoma and melanocytic cell lines were screened for CEACAM1 and integrin β3 expression (not shown). The MV3 melanoma cell line was selected because MV3 cells were completely CEACAM1-negative on protein and RNA level but displayed strong expression of integrin β3. In addition to MV3 cells, cells of the CEACAM1-negative and integrin β3-negative melanocytic cell line MEL6 were studied, either untransfected or CEACAM1-transfected, using invasion and migration assays to delineate the role of CEACAM1 in invasion and migration of melanocytic and melanoma cells.
CEACAM1 Expression Enhances Cell Invasion and Cell Migration
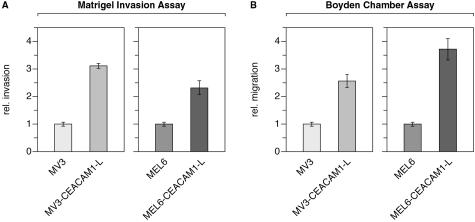
The MV3 and MEL6 cell lines were stably transfected with wild-type CEACAM1 and the invasive capacity was quantified using the Matrigel in vitro invasion assay. The number of cells that invaded through the membrane after incubation for 24 hours was counted. Both cell lines have invasive ability but MV3 cells (mean ± SD, 35 ± 2 cells/field) displayed a higher (more than threefold) motility compared to MEL6 cells (mean ± SD, 10 ± 1.5 cells/field). As shown in Figure 1A, transfection with CEACAM1 exerted a profound effect on cell invasion in both cell lines with considerably higher numbers of invasive cells (>2.5-fold). Using the Boyden chamber assay MV3 cells (mean ± SD, 29 ± 1.5 cells/field) once more displayed a higher (>2.5-fold) motility compared to MEL6 cells (mean ± SD, 11 ± 1 cells/field). As shown in Figure 1B CEACAM1 transfected cells showed a considerably higher (>2.5-fold) motility compared to untransfected cells.
Figure 1.
CEACAM1 expression enhances cell invasion and cell migration. A: Matrigel invasion assay. B: Boyden chamber assay. Cells (2.5 × 104) within passages 6 to 10 were seeded in the upper compartment of transwell chambers containing PET filters precoated with Matrigel (A) or polyester filters precoated with collagen I (B). After 24 hours (invasion assay) or 5 hours (migration assay), under serum-free conditions (RPMI), cells that invaded through the 8-μm pores were stained. The data are means ± SD of five separate experiments. The mean invasion of untransfected MV3 or untransfected MEL6 cells was assigned a value of 1.
Anti-CEACAM Antibodies Inhibit Cell Invasion and Cell Migration
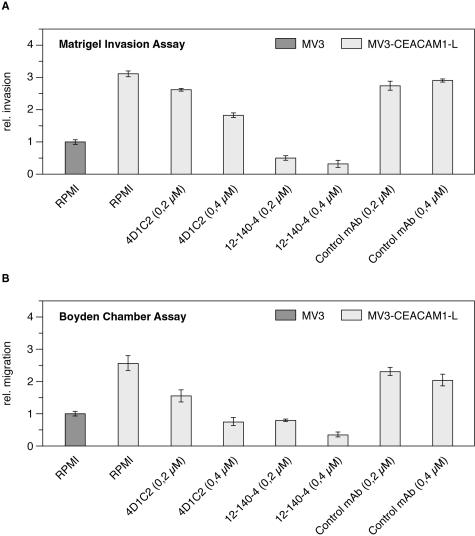
Consistent with a role of CEACAM1 in the regulation of cell invasion we were able to block the enhanced invasive capacity of CEACAM1-transfected MV3 cells in a dose-dependent manner using CEACAM1 mAb 4D1C2. An enhanced blocking effect was observed using equal concentrations of CEACAM mAb 12-140-4, whereas a nonfunctional antibody control mAb did not affect the invasive capacity (Figure 2A). Similar results were observed using the Boyden chamber assay in which both anti-CEACAM antibodies blocked the enhanced migration of CEACAM1-transfected MV3 cells (Figure 2B) in a dose-dependent manner. In conclusion, these data provide evidence that expression of CEACAM1 in melanocytic and melanoma cells enhances their invasive and migratory capacity in two different in vitro systems simulating the extracellular matrix.
Figure 2.
Anti-CEACAM mAb inhibit enhanced CEACAM1-mediated cell invasion and migration. A: Matrigel invasion assay and B: Boyden chamber assay performed as described in Figure 1. Cells that invaded, under serum-free conditions in the absence (RPMI in A and B) or presence of inhibitors, were stained. The data are means ± SD of five separate experiments. The mean invasion of untransfected MV3 cells was assigned a value of 1. Inhibitors were used as follows: 0.2 μmol/L and 0.4 μmol/L 4D1C2 mAb and 0.2 μmol/L and 0.4 μmol/L 12-140-4 mAb as anti-CEACAM1 blocking agents. Control bars are invasion (A) or migration (B) in the presence of 0.2 μmol/L and 0.4 μmol/L nonimmune IgG.
CEACAM1-Enhanced Invasion and Migration Require CEACAM1-Tyr-488
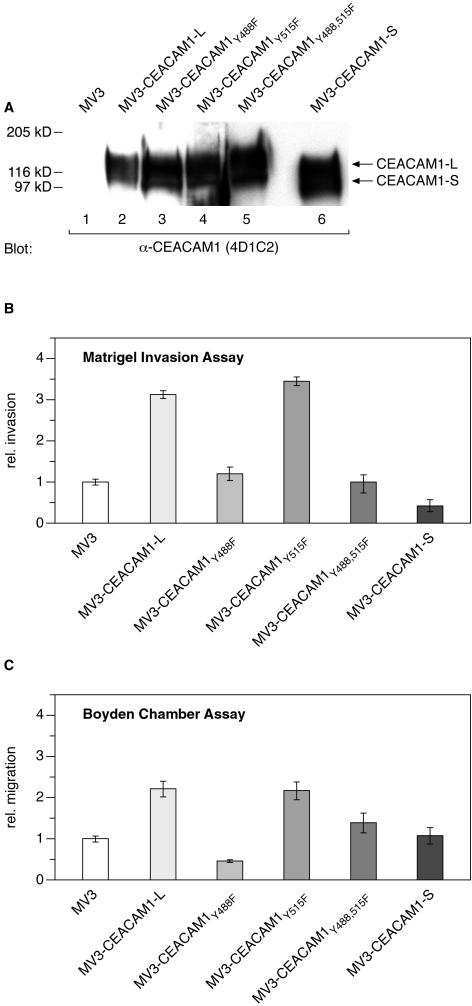
We13–15 and others18,19 have shown that CEACAM1 interacts with several proteins via its cytoplasmic domain. To verify the hypothesis that the intact cytoplasmic domain of CEACAM1 is essential for the observed CEACAM1-enhanced invasion and migration we generated several CEACAM1 mutants differing in the cytoplasmic domain (wild-type, short splice variant, Y488F, Y515F, double-mutant Y488,515F). These constructs were stably transfected into MV3 cells. The CEACAM1 expression levels of the stably transfected cells were determined using Western blot analysis. As shown in Figure 3A, CEACAM1 expression levels in all transfectomas were equivalent.
Figure 3.
CEACAM1-enhanced invasion or migration requires CEACAM1-Tyr-488. A: Western blot analysis of MV3 cells (lane 1) and MV3 cells transfected with wild-type (lane 2) and mutant (lanes 3 to 6) CEACAM1. Cell lysates containing 50 μg of protein each were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and immunoblotted with anti-CEACAM1 mAb 4D1C2. Positions of CEACAM1-L and mutant CEACAM1, containing the short cytoplasmic isoform (CEACAM1-S), are indicated on the right. Matrigel invasion assay (B) and Boyden chamber assay (C) performed as described in Figure 1. The data are means ± SD of five separate experiments. The mean invasion of untransfected MV3 cells was assigned a value of 1.
As shown in Figure 3B enhanced invasion of CEACAM1-transfected cells was dependent on the presence of the intact full-length cytoplasmic domain of CEACAM1 because transfection of MV3 cells with the short isoform of the cytoplasmic CEACAM1 domain had no effect on their invasive capacity. Furthermore, CEACAM1-enhanced invasion of MV3 cells was dependent on the presence of Tyr-488 within its cytoplasmic domain because transfection of mutant Y488F and double-mutant Y488,515F did not display enhanced invasion. On the contrary, presence of Tyr-515 within the cytoplasmic CEACAM1 domain is not required because transfection of MV3 cells with the mutant Y515F did augment cell invasion almost as effectively as transfection with wild-type CEACAM1. As shown in Figure 3C we obtained comparable results using the Boyden chamber assay, which substantiate the functional role of the cytoplasmic CEACAM1 domain, and especially of Tyr-488, in CEACAM1-enhanced invasion and migration. Moreover the presence of Tyr-515 seemed to inhibit migration because transfection of MV3 cells with the mutant Y488F did diminish cell invasion compared to untransfected cells or MV3 cells transfected with the double-mutant Y488,515F.
Integrin-Antagonizing RGD Peptides Block Enhanced Invasion and Migration of CEACAM1-Transfected MV3 Cells
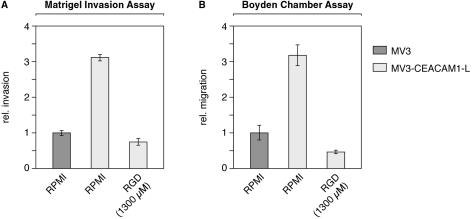
To migrate and invade surrounding tissues, cells need to interact via integrins with several different extracellular matrix components. Therefore, we were interested in whether integrin inhibitors are able to interfere with CEACAM1-enhanced invasion and migration. As shown in Figure 4 the addition of RGD peptides, in a concentration known to antagonize integrin function,20 completely blocked enhanced invasion (Figure 4A) and enhanced migration (Figure 4B) of CEACAM1-transfected MV3 cells.
Figure 4.
CEACAM1-enhanced invasion and migration is blocked by inhibitor of integrin β3 function. A: Matrigel invasion assay and B: Boyden chamber assay performed as described in Figure 1. Cells that invaded, under serum-free conditions in the absence (RPMI) or presence of RGD peptides (1300 μmol/L), were stained. The data are means ± SD of five separate experiments. The mean invasion of untransfected MV3 cells was assigned a value of 1.
Because we have shown15 that CEACAM1 interacts with integrin β3 via the CEACAM1 cytoplasmic domain, dependent on Tyr-488, and that these two adhesion molecules co-localize at the tumor-stroma interface of invading melanoma masses we reasoned a functional role of CEACAM1-integrin β3 interaction in invasion and migration. To further explore the role of CEACAM1-integrin β3 interaction in invasion and migration we used an additional approach with MEL6 cells. These cells express neither CEACAM1 nor integrin β3 and are therefore suitable to study the functional role of both adhesion molecules in this transfection system independently.
CEACAM1 and Integrin β3 Expression Both Enhance Invasion of MEL6 Cells
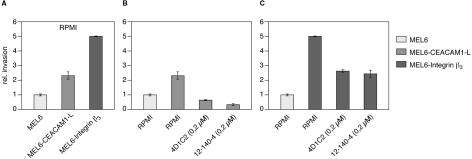
MEL6 cells were stably transfected either with the cDNA for CEACAM1 or for integrin β3 and the invasive capacity of these cells was determined in the Matrigel invasion assay. Consistent with our previous results and the literature4 expression of either CEACAM1 or integrin β3 resulted in enhanced invasion by 2.5- to 5-fold compared to untransfected cells (Figure 5A). This enhanced invasion was blocked by addition of specific CEACAM antibodies (Figure 5B). Unexpectedly, both CEACAM antibodies did reduce the enhanced invasion of integrin β3-transfected MEL6 cells in five independent experiments (Figure 5C).
Figure 5.
Expression of CEACAM1 and integrin β3 in MEL6 cells enhances invasion. A: MEL6 cells or MEL6 cells transfected either with cDNA for CEACAM1 or for integrin β3 were studied using the Matrigel invasion assay as described in Figure 1. B: Invasion of CEACAM1 transfected MEL6 cells in the absence (RPMI) or presence of anti-CEACAM1 inhibitors (0.2 μmol/L 4D1C2 and 0.2 μmol/L 12-140-4). C: Invasion of integrin β3 transfected MEL6 cells in the absence (RPMI) and presence of anti-CEACAM1 inhibitors (0.2 μmol/L 4D1C2 and 0.2 μmol/L 12-140-4). The data are means ± SD of five separate experiments. The mean invasion of untransfected MEL6 cells was assigned a value of 1.
Expression of Integrin β3 Induces the Up-Regulation of CEACAM1
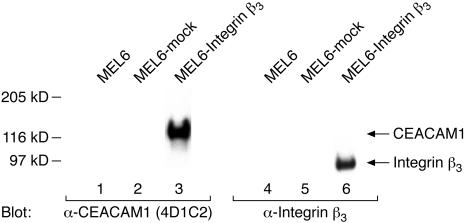
Using Western blot analysis we examined CEACAM1 expression in MEL6 cells. Interestingly, we observed a high level of CEACAM1 expression in MEL6 cells transfected with integrin β3 (Figure 6, lane 3) but no CEACAM1 expression in untransfected MEL6 cells (Figure 6, lane 1) or mock-transfected cells (Figure 6, lane 2). Furthermore, we repeated the transfection procedures and confirmed the CEACAM1 expression in MEL6 cells transfected with integrin β3 on the RNA level using microarrays (not shown) indicating a highly significant detection P value. In addition, the array data demonstrated the presence of integrin αv in MEL6 cells, however, in contrast to CEACAM1, no significant change in the RNA level between the integrin β3-transfected and -untransfected MEL6 cells was observed for integrin αv.
Figure 6.
Expression of integrin β3 induces the up-regulation of CEACAM1. Eighty-μg aliquots of cell lysates of untransfected (lanes 1 and 4), and mock (lanes 2 and 5)- and integrin β3 (lanes 3 and 6)-transfected MEL6 cells were resolved by 7.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted with anti-CEACAM1 mAb 4D1C2 (lanes 1 to 3) or anti-integrin β3 mAb (lanes 4 to 6). The positions of CEACAM1 and integrin β3 are indicated on the right.
Discussion
In previous studies we showed that CEACAM1 is expressed in human melanomas, often at the invading front.15 In melanoma patients, the expression of CEACAM1 is associated with poor prognosis, and CEACAM1 turned out to be a prognostic indicator superior to tumor thickness.10 The results presented here provide a functional basis for these correlative data. We provide evidence that the expression of CEACAM1-L in the CEACAM1-negative MEL6 melanocytic cell line and the CEACAM1-negative MV3 melanoma cell line increases the migratory and invasive growth potentials in in vitro assays.
To our best knowledge, this is the first report that indicates that CEACAM1 actively contributes to tumor progression. Previously, others and we showed that CEACAM1 is down-regulated or dysregulated in carcinomas. For example, in prostate cancer, CEACAM1 is lost during the progression from Gleason stage 3 to 4.21 In colorectal carcinogenesis, a decrease of CEACAM1 mRNA levels is noted already in the adenoma stage.22 However, it is known for some time that the down-regulation of CEACAM1 is not a general phenomenon in epithelial malignancies. For example, in gastric cancer, CEACAM1 is up-regulated.23 Similarly, in non-small cell lung cancer, CEACAM1 is up-regulated24 and an indicator of a poor prognosis.25,26
On a functional level, the differences in CEACAM1 expression in various human malignancies may be explained by the presence of different splice variants. With respect to the cytoplasmic domain, two major isoforms of CEACAM1 can be distinguished, eg, a short version with 10 to 12 amino acids (CEACAM1-S) and a long version with 71 to 73 amino acids (CEACAM1-L).27 The cytoplasmic domain of CEACAM1-L contains two tyrosine residues at positions 488 and 515, respectively. Tyr488 forms part of a perfect immunoreceptor tyrosine-based inhibition motif (ITIM). CEACAM1-S lacks these tyrosine residues. As shown previously, CEACAM1-S, but not CEACAM1-L, mediates the reversion of mammary epithelial cells in Matrigel culture from tumor to normal morphology with respect to gland formation.28 Thus, in glandular tissues, the loss of CEACAM1-S expression may lead to the loss of glandular structures as observed in human prostate cancer. However, in breast cancer cells, the tumor-suppressive effect has been associated with the long cytoplasmic domain that, in contrast to the extracellular CEACAM1 domain, was indispensable for the tumor suppressive effect.29 In mammary epithelial cells transfected with CEACAM1-L, cells undergo apoptosis when transferred from plastic on to Matrigel.28 Although not formally proven, the tumor suppressive effect of the long cytoplasmic domain in experimental tumor models may be based on an increased rate of apoptosis.
In a marked contrast to these results, stable transfection of CEACAM1-L into melanocytes and melanoma cells confers increased migratory and invasive growth potential. The presence of an intact long cytoplasmic domain is a prerequisite for these properties because CEACAM1-S lacked these effects. To further sharpen this contrast, the replacement of the tyrosine residue in position 488 completely abrogated the effect on migration and invasive growth. In contrast, the corresponding tyrosine residue has been shown to be essential for the tumor suppressive effect of CEACAM1-L in vivo, at least in murine colonic carcinoma cells.30 These different effects of CEACAM1 on cell proliferation and cell migration reiterate the increasing body of evidence that cell adhesion molecules act through a web of potential interactions, the actual usage of which varies depending on the cell type and environmental conditions.31
In previous studies, we found that the recombinant cytoplasmic domain of CEACAM1-L specifically associates with the β3 integrin subunit, and that β3 integrin co-localizes with CEACAM1 in melanomas, preferentially at the invasive front.15 A co-localization was also observed in the extravillous intermediate trophoblast, thought to be the invasive component of the trophoblast.32 Because, in tissues other than platelets, the β3 subunit partners with the αv-subunit,33 we suggest that CEACAM1 and αvβ3 integrin interact during the invasive growth of melanomas and trophoblasts. The findings reported here further support the assumption that CEACAM1 interacts with integrins. In the melanoma cell line MV3, the CEACAM1-induced increase in migration and invasive growth properties was blocked completely by RGD peptides. Because this cell line expresses the β3 subunit, the RGD effect may be mediated, either directly or indirectly,34 by αvβ3 integrin. However, it cannot be excluded that the effect of CEACAM1 is mediated by integrins other than β3 integrins.
Our results indicate that the migratory effect of CEACAM1 depends on integrins. In further experiments, we asked if the reverse holds true, eg, if the invasive properties, mediated by integrins, may depend on CEACAM1. Specifically, it was tested if αvβ3 integrin promotes invasive growth properties independent of CEACAM1. Expression of β3 integrin was reconstituted in the CEACAM1-negative melanocytic cell line MEL6. As predicted by results reported previously,34 this integrin promoted migration and invasion that could be blocked by RGD peptides. However, because the expression of β3 integrin stimulated the expression of CEACAM1, CEACAM1 could be targeted by mAbs, which partially reverted the invasive phenotype. These results strengthen the view that CEACAM1 and αvβ3 integrin are functionally interconnected with respect to the invasive growth of melanomas and possibly also of the trophoblast.
The stimulation of migration and invasion may not be the only mechanism by which CEACAM1 promotes progression of melanomas. According to recent results, homophilic interaction of CEACAM1 on melanoma cells and natural killer cells represents a class I MHC-independent inhibitory mechanism of human natural killer cell cytotoxicity.35 Because CEACAM1 supports the progression of melanomas by several mechanisms, patients may benefit from agents that inhibit the functions of CEACAM1.
Acknowledgments
We thank Ole Børmer for the supply of monoclonal antibody 12-140-4 and Ewa Malewski and Katja Redlin for excellent technical assistance.
Footnotes
Address reprint requests to Jens Brümmer, Institut für Klinische Chemie/ Zentrallaboratorien, Zentrum für Klinisch-Theoretische Medizin I, Universitätsklinikum Hamburg-Eppendorf, Martinistrasse 52, 20251 Hamburg, Germany. E-mail: bruemmer@uke.uni-hamburg.de.
Supported by the Deutsche Forschungsgemeinschaft (grant Br 1748/2-1 to J.B. and C.W.) and the Deutsche Krebshilfe (grant 10-2063-Br 2 to J.B. and A.B).
References
- Edward M. Integrins and other adhesion molecules involved in melanocytic tumor progression. Curr Opin Oncol. 1995;7:185–191. doi: 10.1097/00001622-199503000-00015. [DOI] [PubMed] [Google Scholar]
- Buttner P, Garbe C, Bertz J, Burg G, d’Hoedt B, Drepper H, Guggenmoos-Holzmann I, Lechner W, Lippold A, Orfanos CE, Peters A, Rassner G, Stadler R, Stroebel W. Primary cutaneous melanoma. Optimized cutoff points of tumor thickness and importance of Clark’s level for prognostic classification. Cancer. 1995;75:2499–2506. doi: 10.1002/1097-0142(19950515)75:10<2499::aid-cncr2820751016>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Johnson JP. Cell adhesion molecules in the development and progression of malignant melanoma. Cancer Metastasis Rev. 1999;18:345–357. doi: 10.1023/a:1006304806799. [DOI] [PubMed] [Google Scholar]
- Natali PG, Hamby CV, Felding-Habermann B, Liang B, Nicotra MR, Di Filippo F, Giannarelli D, Temponi M, Ferrone S. Clinical significance of alpha(v)beta3 integrin and intercellular adhesion molecule-1 expression in cutaneous malignant melanoma lesions. Cancer Res. 1997;57:1554–1560. [PubMed] [Google Scholar]
- Hsu MY, Shih DT, Meier FE, Van Belle P, Hsu JY, Elder DE, Buck CA, Herlyn M. Adenoviral gene transfer of beta3 integrin subunit induces conversion from radial to vertical growth phase in primary human melanoma. Am J Pathol. 1998;153:1435–1442. doi: 10.1016/s0002-9440(10)65730-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitjans F, Sander D, Adan J, Sutter A, Martinez JM, Jaggle CS, Moyano JM, Kreysch HG, Piulats J, Goodman SL. An anti-alpha v-integrin antibody that blocks integrin function inhibits the development of a human melanoma in nude mice. J Cell Sci. 1995;108:2825–2838. doi: 10.1242/jcs.108.8.2825. [DOI] [PubMed] [Google Scholar]
- Xie S, Luca M, Huang S, Gutman M, Reich R, Johnson JP, Bar-Eli M. Expression of MCAM/MUC18 by human melanoma cells leads to increased tumor growth and metastasis. Cancer Res. 1997;57:2295–2303. [PubMed] [Google Scholar]
- Mills L, Tellez C, Huang S, Baker C, McCarty M, Green L, Gudas JM, Feng X, Bar-Eli M. Fully human antibodies to MCAM/MUC18 inhibit tumor growth and metastasis of human melanoma. Cancer Res. 2002;62:5106–5114. [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Hess AR, Seftor RE. Molecular plasticity of human melanoma cells. Oncogene. 2003;22:3070–3075. doi: 10.1038/sj.onc.1206447. [DOI] [PubMed] [Google Scholar]
- Thies A, Moll I, Berger J, Wagener C, Brummer J, Schulze HJ, Brunner G, Schumacher U. CEACAM1 expression in cutaneous malignant melanoma predicts the development of metastatic disease. J Clin Oncol. 2002;20:2530–2536. doi: 10.1200/JCO.2002.05.033. [DOI] [PubMed] [Google Scholar]
- Wagener C, Clark BR, Rickard KJ, Shively JE. Monoclonal antibodies for carcinoembryonic antigen and related antigens as a model system: determination of affinities and specificities of monoclonal antibodies by using biotin-labeled antibodies and avidin as precipitating agent in a solution phase immunoassay. J Immunol. 1983;130:2302–2307. [PubMed] [Google Scholar]
- Neumaier M, Fenger U, Wagener C. Monoclonal antibodies for carcinoembryonic antigen (CEA) as a model system: identification of two novel CEA-related antigens in meconium and colorectal carcinoma tissue by Western blots and differential immunoaffinity chromatography. J Immunol. 1985;135:3604–3609. [PubMed] [Google Scholar]
- Brummer J, Neumaier M, Gopfert C, Wagener C. Association of pp60c-src with biliary glycoprotein (CD66a), an adhesion molecule of the carcinoembryonic antigen family downregulated in colorectal carcinomas. Oncogene. 1995;11:1649–1655. [PubMed] [Google Scholar]
- Ebrahimnejad A, Flayeh R, Unteregger G, Wagener C, Brummer J. Cell adhesion molecule CEACAM1 associates with paxillin in granulocytes and epithelial and endothelial cells. Exp Cell Res. 2000;260:365–373. doi: 10.1006/excr.2000.5026. [DOI] [PubMed] [Google Scholar]
- Brummer J, Ebrahimnejad A, Flayeh R, Schumacher U, Loning T, Bamberger AM, Wagener C. Cis interaction of the cell adhesion molecule CEACAM1 with integrin beta(3). Am J Pathol. 2001;159:537–546. doi: 10.1016/s0002-9440(10)61725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt NO, Westphal M, Hagel C, Ergun S, Stavrou D, Rosen EM, Lamszus K. Levels of vascular endothelial growth factor, hepatocyte growth factor/scatter factor and basic fibroblast growth factor in human gliomas and their relation to angiogenesis. Int J Cancer. 1999;84:10–18. doi: 10.1002/(sici)1097-0215(19990219)84:1<10::aid-ijc3>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- Rajagopalan D. A comparison of statistical methods for analysis of high density oligonucleotide array data. Bioinformatics. 2003;19:1469–1476. doi: 10.1093/bioinformatics/btg202. [DOI] [PubMed] [Google Scholar]
- Beauchemin N, Kunath T, Robitaille J, Chow B, Turbide C, Daniels E, Veillette A. Association of biliary glycoprotein with protein tyrosine phosphatase SHP-1 in malignant colon epithelial cells. Oncogene. 1997;14:783–790. doi: 10.1038/sj.onc.1200888. [DOI] [PubMed] [Google Scholar]
- Huber M, Izzi L, Grondin P, Houde C, Kunath T, Veillette A, Beauchemin N. The carboxyl-terminal region of biliary glycoprotein controls its tyrosine phosphorylation and association with protein-tyrosine phosphatases SHP-1 and SHP-2 in epithelial cells. J Biol Chem. 1999;274:335–344. doi: 10.1074/jbc.274.1.335. [DOI] [PubMed] [Google Scholar]
- Gehlsen KR, Argraves WS, Pierschbacher MD, Ruoslahti E. Inhibition of in vitro tumor cell invasion by Arg-Gly-Asp-containing synthetic peptides. J Cell Biol. 1988;106:925–930. doi: 10.1083/jcb.106.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch C, Hanssen TA, Wagener C, Obrink B. Down-regulation of CEACAM1 in human prostate cancer: correlation with loss of cell polarity, increased proliferation rate, and Gleason grade 3 to 4 transition. Hum Pathol. 2002;33:290–298. doi: 10.1053/hupa.2002.32218. [DOI] [PubMed] [Google Scholar]
- Nollau P, Scheller H, Kona-Horstmann M, Rohde S, Hagenmuller F, Wagener C, Neumaier M. Expression of CD66a (human C-CAM) and other members of the carcinoembryonic antigen gene family of adhesion molecules in human colorectal adenomas. Cancer Res. 1997;57:2354–2357. [PubMed] [Google Scholar]
- Kinugasa T, Kuroki M, Takeo H, Matsuo Y, Ohshima K, Yamashita Y, Shirakusa T, Matsuoka Y. Expression of four CEA family antigens (CEA, NCA, BGP and CGM2) in normal and cancerous gastric epithelial cells: up-regulation of BGP and CGM2 in carcinomas. Int J Cancer. 1998;76:148–153. doi: 10.1002/(sici)1097-0215(19980330)76:1<148::aid-ijc23>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Ohwada A, Takahashi H, Nagaoka I, Kira S. Biliary glycoprotein mRNA expression is increased in primary lung cancer, especially in squamous cell carcinoma. Am J Respir Cell Mol Biol. 1994;11:214–220. doi: 10.1165/ajrcmb.11.2.8049082. [DOI] [PubMed] [Google Scholar]
- Laack E, Nikbakht H, Peters A, Kugler C, Jasiewicz Y, Edler L, Brummer J, Schumacher U, Hossfeld DK. Expression of CEACAM1 in adenocarcinoma of the lung: a factor of independent prognostic significance. J Clin Oncol. 2002;20:4279–4284. doi: 10.1200/JCO.2002.08.067. [DOI] [PubMed] [Google Scholar]
- Sienel W, Dango S, Woelfle U, Morresi-Hauf A, Wagener C, Brummer J, Mutschler W, Passlick B, Pantel K. Elevated expression of carcinoembryonic antigen-related cell adhesion molecule 1 promotes progression of non-small cell lung cancer. Clin Cancer Res. 2003;9:2260–2266. [PubMed] [Google Scholar]
- Obrink B. CEA adhesion molecules: multifunctional proteins with signal-regulatory properties. Curr Opin Cell Biol. 1997;9:616–626. doi: 10.1016/S0955-0674(97)80114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirshner J, Chen CJ, Liu P, Huang J, Shively JE. CEACAM1-4S, a cell-cell adhesion molecule, mediates apoptosis and reverts mammary carcinoma cells to a normal morphogenic phenotype in a 3D culture. Proc Natl Acad Sci USA. 2003;100:521–526. doi: 10.1073/pnas.232711199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Wood CG, Earley K, Hung MC, Lin SH. Suppression of tumorigenicity of breast cancer cells by an epithelial cell adhesion molecule (C-CAM1): the adhesion and growth suppression are mediated by different domains. Oncogene. 1997;14:1697–1704. doi: 10.1038/sj.onc.1200999. [DOI] [PubMed] [Google Scholar]
- Izzi L, Turbide C, Houde C, Kunath T, Beauchemin N. Cis-determinants in the cytoplasmic domain of CEACAM1 responsible for its tumor inhibitory function. Oncogene. 1999;18:5563–5572. doi: 10.1038/sj.onc.1202935. [DOI] [PubMed] [Google Scholar]
- Schwartz MA, Ginsberg MH. Networks and crosstalk: integrin signalling spreads. Nat Cell Biol. 2002;4:E65–E68. doi: 10.1038/ncb0402-e65. [DOI] [PubMed] [Google Scholar]
- Bamberger AM, Sudahl S, Loning T, Wagener C, Bamberger CM, Drakakis P, Coutifaris C, Makrigiannakis A. The adhesion molecule CEACAM1 (CD66a, C-CAM, BGP) is specifically expressed by the extravillous intermediate trophoblast. Am J Pathol. 2000;156:1165–1170. doi: 10.1016/S0002-9440(10)64985-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2:91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- Markel G, Wolf D, Hanna J, Gazit R, Goldman-Wohl D, Lavy Y, Yagel S, Mandelboim O. Pivotal role of CEACAM1 protein in the inhibition of activated decidual lymphocyte functions. J Clin Invest. 2002;110:943–953. doi: 10.1172/JCI15643. [DOI] [PMC free article] [PubMed] [Google Scholar]