Abstract
Based on the RNAi technique, we have developed a new approach that generates transgenic animals capable of mimicking human genetic diseases. The new system is a combination of siRNA with Cre-loxP and tetracycline-on. It has the characteristics of being stable, inheritable, and inducible, with the siRNA able to be transcribed tissue specifically. To support the ability of this new method to generate a model for a disease, we created an ABCA1-deficient mouse line that mimics Tangier disease under controlled conditions. Thus, it should now be possible to rapidly establish human genetic diseases as a whole animal model without the use of embryonic stem cell and gene targeting. This system also provides a tool for pathological and pharmacological studies of aspects peculiar to particular human genetic diseases.
Mutant mice lacking specific genes have become a very useful model for studying gene function. Classical approaches to creating animal models for human genetic diseases such as gene knockout and using the Cre-loxP systems may possibly have a lethal impact during embryogenesis, if the target gene is crucial during development. This prevents some abnormal phenotypes from reaching maturity. The RNA interference (RNAi) approach that uses double-strand RNA of 21–23 nucleotides to mediate a sequence-specific post-transcriptional gene silencing has been adopted as a tool and used in a variety of functional genomic projects in a wide range of species.1,2 The application of short double-strand RNA has successfully eliminated the interferon response and non-specific mRNA degradation induced by the anti-sense long double-strand RNA (>500 nucleotides) in the cytoplasm.3 Now, RNAi with short nucleotides has been adapted for high-throughput use in the transient knockdown of gene expression in cell lines and animals.4–7 However, the promoters used in the RNAi approach, such as U6 and H1 RNA polymerase III (pol III), are continually active in all of the tissues and cannot be applied to generate tissue-specific gene knockdown.
Recently, two lentivirus-based RNAi systems were used to generate transgenic animals with specific gene knockdown.8,9 Likewise, the lentivirus-based system is again active throughout embryo development and cannot be used to affect important genes crucial to embryogenesis. In addition, the lentivirus-based system with the pol III promoter expresses siRNA in all tissues and thus lacks tissue specificity. Subsequently, by replacing the pol III promoter with pol II, the RNAi approach was used to generate partial tissue-specific gene knockdown.10 The system named pDECAP has been applied to ski-deficient mice. However, overexpressed siRNA from the pol II promoter was exhibited in all pol II-related tissues. To overcome these limitations encountered in the production of specific gene-knockdown animals, a spatiotemporally controlled adult somatic mutagenesis system is required. In this paper, we describe, for the first time, the development of a stable, inheritable, and inducible gene-knockdown system named the SIRIUS-Cre system that combines three approaches including the siRNA for specific gene-knockdown, Cre-loxP for tissue-specific expression, and tetracycline-on for inducible expression. The transgenic mice were created through the mating of two specific parental lines that contain a specific siRNA of interest gene and tissue-specific recombinase under tetracycline control. We have successfully adapted the system to mimic Tangier disease, a genetic disease that is due to the mutation of ABCA1 gene,11 and have shown that embryonic lethality has been avoided.
Materials and Methods
Plasmid Construction and in Vivo Verification
The SIRIUS-ABCA1 vector expresses an enhanced green fluorescent protein (EGFP) cDNA under control of the cytomegalovirus (CMV) promoter and also the hairpin oligo of ABCA1 siRNA under the control of the mouse U6 RNA polymerase III promoter after recombining two loxP sites by Cre recombinase. This vector was constructed using the following steps. First, an U6 promoter, which contains a Mlu I cloning site at the 3′ end was inserted into the BclI site of the pLEGFP-C1 vector (BD Biosciences, San Jose, CA). Second, two loxP oligonucleotides were inserted at the XhoI, ApaI, and Mlu I restriction sites down- and up-stream of the EGFP cluster. The sequence of the loxP is 5′-ATA ACT TCG TAT AGC ATA CAT TAT ACG AAG TTA T-3′. Third, an oligonucleotide encoding a hairpin siRNA against ABCA1, being an inverted repeat separated by a 9-nt (TTC AAG AGA) spacer, was inserted beyond the loxP using the BamHI and NotI cloning sites. The sequence of the oligonucleotide encoding the ABCA1 siRNA hairpin is 5′-GAA CCT CAC TTT CAG AAG ATT CAA GAG ATC TTC TGA AAG TGA GG-3′. The resulting vector was named SIRIUS-ABCA1. The specific gene-knockdown effect of the ABCA1 siRNA oligonucleotide was examined by comparing the reduction in ABCA1 for siRNA and non-silencing oligo-transfected cells. The non-silencing siRNA oligo (4611, Ambion Inc, Austin, TX) and ABCA1 siRNA oligos were separately transfected into HepG2 cytoplasm and the ABCA1 was detected by RT-PCR. The reduction in ABCA1 only occurred displayed in the ABCA1 siRNA oligo-transfected cells (data not shown).
The AlbTet-Cre vector, which contains a modified liver-specific human albumin promoter with TRE, Cre recombinase, and tetracycline silencer-rtTA, was constructed. The vector expressed the Cre recombinase under the control of albumin promoter and doxycycline (tetracycline analog). This vector was constructed utilizing the following steps. First, full-length cDNA of the Cre gene was inserted into BglII site of the pLHCX vector (BD Biosciences). Next, Mlu I sites and ApaI sites were created upstream of the Cre gene, and a modified albumin promoter was inserted into Mlu I and ApaI sites of the pLHCX vector. Then, a fragment encoding TRE (tetracycline responsible element) was created downstream of albumin promoter12 for conditional and tissue-specific control. Finally, the tetracycline silencer-rtTA was inserted at the 3′ end of the CMV promoter using the HindIII and HpaI sites. The resulting vector was named AlbTet-Cre. The sequence of TRE is 5′-CTC CCT ATC AGT TGA TAG AGA AA-3′. Additional details of the cloning strategy are available from the authors on request. The recombinant reSIRIUS-ABCA1 plasmid, when created in vivo, will have two distinct features; it will be smaller in size due to loxP recombination caused by the Cre recombinase and it would have lost fluorescence for a similar reason. Cre recombinase is produced under albumin promoter control through doxycycline induction.
Cells of the hepatoma cell line HepG2 were transfected 24 hours after passage using the manufacturer’s protocol (siPORT amine transfection agent; Ambion Inc.). Cellular EGFP expression was examined visually under a fluorescent microscope. As proof of recombination, the DNA of the transfected cells was extracted and then amplified with specific primers (p1581: 5′-TTA TCC AGC CCT CAC TCC TTC T-3′ and p4214: 5′-CAT TAA GGG ATC AGT TAT CTA G-3′) for PCR analysis.
Transgenic Animals
Fertilized eggs of adult ICR mice (Inbr 29. Albino. Origin: BLU:Ha (ICR) outbred stock from the Institute of Cancer Research in 1958, to Arbor Scientific Co. Ltd., to University of British Columbia, Vancouver in 1977, followed by sib mating) over 4 weeks old were collected after super-ovulation as previously described.13 Eluted eggs were maintained in Dulbecco’s phosphate-buffered salines (DPBS) medium in an atmosphere of 5% CO2 at 37°C. The DNAs of both transgene vectors were cut with specific restriction enzymes (ScaI for SIRIUS-ABCA1 and AflIII for AlbTet-Cre) after purification by CsCl gradient centrifugation. The DNA was diluted to 5 ng/ul in sterile MilliQ water and injected into eggs. These zygotes were re-implanted into pseudo-pregnant foster mothers, and the offspring were screened for the presence of transgenes by Southern blotting and PCR. Genomic DNA was isolated from the tail blood of the transgenic animals by proteinase K digestion followed by phenol extraction and ethanol precipitation. Southern blotting demonstrated that all transgenes were integrated into the genome of animals tested (data not shown). PCR analyses were used to identify transgenic mice using specific primers (p1581 and p4214 primers for SIRIUS-ABCA1 and the CreF and CreD primers for AlbTet-Cre). The primer sequences of CreF and CreD are 5′-ATG GCA CCC AAG AAG AAG AGG A-3′ and 5′-CTA ATC GCC ATC TTG CAG CAG G-3′, respectively (Figure 1). Four F0-S transgenic mice successfully passed the transgene through their germ line. Transgenic expression of EGFP under CMV control was also shown by the fluorescent pattern under an anatomical microscope. Six F0-A AlbTet-Cre transgenic mice were constructed and confirmed in a similar manner. These two founders successfully passed this transgene to their offspring too. All transgenic mice developed and bred normally. Next, these two lines of transgenic mice were mated to produce offspring (F1-SA), which simultaneously contain both the SIRIUS-ABCA1 and AlbTet-Cre transgenes. In the first experiment, four F1-SA mice were obtained that successfully carried transgenes in their genomic DNA as detected by Southern blotting (data not shown), PCR, and the presence of fluorescence. Then, the F1-SA transgenic animals (1-week-old) were supplied with doxycycline hydrochloride for 4 days via breast milk (2 mg/ml doxycycline). To enhance the induction, 4 mg/ml doxycycline in 0.5 ml of normal saline was injected intraperitoneally. These treatments of doxycycline were repeated twice at 24-hour intervals.
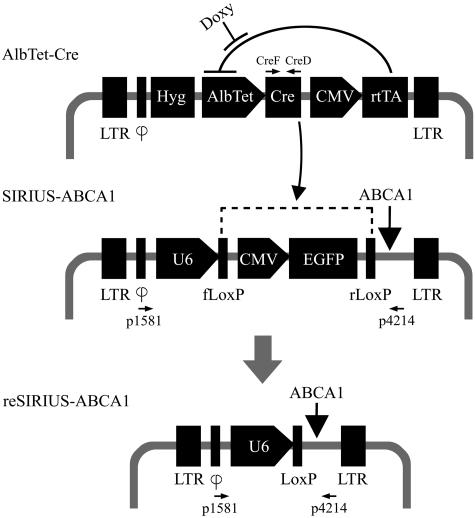
Figure 1.
Construction and expression of SIRIUS-Cre system. Schematic diagrams of SIRIUS-ABCA1, AlbTet-Cre, and the resulting reSIRIUS-ABCA1 constructions, all vectors contain LTR (long terminal repeat) and ϕ (virus packaged-signal) fragments. HepG2 cells were co-transfected with plasmids containing SIRIUS-ABCA1 or AlbTet-Cre. After doxycycline treatment and the subsequent recombination, reSIRIUS-ABCA1 was generated. The rtTA, reverse tet-controlled transcriptional activator, binds the TRE (tet-responsive element) in the presence of doxycycline and activates transcription of Cre recombinase under AlbTet promoter controlling. Cre recombinase recognizes loxP sites in the SIRIUS-ABCA1, facilitating precise recombination (dotted line) and activates transcription of ABCA1 (ATP-binding cassette subfamily A member 1) siRNA. These neomycin and hygomycin resistance genes were inserted in SIRIUS-ABCA1 and AlbTet-Cre vectors, respectively. Primers used in this study include CreF, CreD, p1581, and p4214 and are indicated.
ABCA1-Related Examinations
In the ABCA1 siRNA assay, RNAs from cells and different tissues of transgenic F1-SA were separated on a 15% denaturing gel with 7 mol/L urea after doxycycline treatment and then were electro-transferred to a nylon membrane for Northern blotting. Synthetic DNA oligonucleotides (5′-TCT CTT GAA TCT TCT GAA AGT GAG GTT C-3′) with a complementary sequence to the ABCA1 dsRNA hairpin were labeled with [γ-32P-ATP] at 3′ end and used as a probe for the dsRNA hairpin detection. Hybridization was performed at 65°C in ExpressHyb hybridization solution (BD Biosciences) and washed with 2X SSC plus 1% sodium dodecyl sulfate at 50°C. For endogenous ABCA1 expression, 10 micrograms of RNA isolated from liver was separated on a gel and transferred to a membrane. A specific-ABCA1 oligo-probe (5′-ACA CCT GGA GAG AAG CTT TCA ACG AGA CTA ACC-3′) was end-labeled with [γ-32P-ATP] and used to detect the expression of ABCA1 by Northern blotting. In the Western blotting assay, the ABCA1 antibody was purchased from Santa Cruz, Inc. (sc-5491, Santa Cruz, CA). Cell and tissue lysates were prepared and aliquots of the extracts containing equal amounts of protein were analyzed by immunoblotting. Additionally, tissues taken for lipid analysis were frozen in OCT (optimal cutting temperature) medium. Frozen sections (5 μm in thickness) were stained with Oil Red O (Sigma, St. Louis, MO) for cholesterol display and were counter-stained with hematoxylin.
Plasma Cholesterol Measurement
Eight-week-old F1-SA transgenic mice were fed with doxycycline in drinking water (2 mg/ml). Blood was isolated from the heart after sacrifice and spun at 3000 rpm for 15 minutes at 4°C to isolate plasma. The total cholesterol was determined by enzymatic methods using Kodak Ektachem-700 analyzer with Clinical Chemistry Slides (Eastman Kodak, Rochester, NY) and expressed as mg/dl. In principle, cholesterol ester in plasma is hydrolyzed and then oxidized to generate hydrogen peroxide. This hydrogen peroxide generates a colored product that is produced by its reaction with a triarylimidazole leuco dye. Values of cholesterol were recorded and are based on analysis of individual animals and two animals were induced for every time point.
Results
The method of producing these transgenic mice was via mating of one mouse, which carries the siRNA/loxP plasmid, with another mouse, which carries expression of Cre recombinase in specific tissues. The regulation of the RNAi in the offspring of transgenic mice was strictly through the tetracycline-on system. On induction by tetracycline or doxycycline, the tissue-specific Cre recombinase were produced and in turn mediated the recombination of two loxP sites that lead to the switching on of the siRNA transcription cassette in the same cell. Using ABCA1 (ATP-binding cassette subfamily A member 1) as a target gene of the study, two new vectors, SIRIUS-ABCA1 and AlbTet-Cre, the combination of which efficiently blocks the ABCA1 expression in liver under strict and spatiotemporal control, were created. The SIRIUS-ABCA1 vector (Figure 1) was designed to express ABCA1 siRNA under the control of U6 promoter after the recombination of the two different loxP sites by Cre recombinase from AlbTet-Cre. EGFP was incorporated to produce the visual fluorescence effect. AlbTet-Cre (Figure 1) was designed to initially express two products, rtTA and Cre recombinase, under separate promoter controls, a modified liver-specific human albumin promoter with TRE and a constitutive CMV promoter to drive Cre and rtTA, respectively.
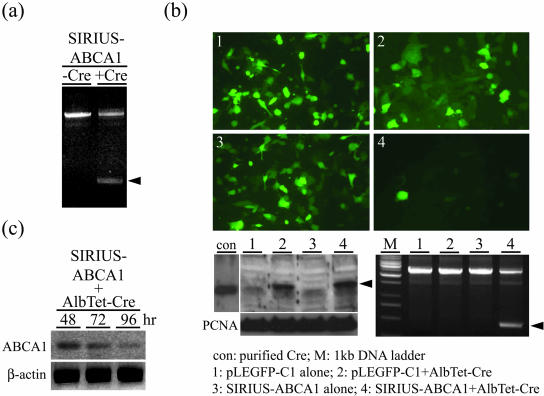
To determine the effect of the Cre recombinase on SIRIUS-ABCA1, in vitro and in vivo recombination assays were first carried out and the results are shown in Figure 2, a to c. Firstly, the SIRIUS-ABCA1 plasmid was treated with purchased Cre recombinase and the recombination of SIRIUS-ABCA1 was confirmed by PCR using specific primers (p1581 and p4214 primers). Without Cre recombinase treatment, where loxP recombination did not occur and a 2.6-kb PCR fragment was detected. The fragment was reduced to less than 0.5-kb after Cre recombinase treatment where the CMV promoter and EGFP were knocked out (Figure 2a). Both PCR fragments are close to estimated sizes from the original vector (SIRIUS-ABCA1) and the Cre recombinase-treated vector (reSIRIUS-ABCA1). In addition, cells of HepG2 displayed down-regulation of GFP expression when co-transfected with SIRIUS-ABCA1 and AlbTet-Cre and after treatment with doxycycline (2 μg/ml) in culture medium (Figure 2b-4). Cells co-transfected with pLEGFP in which loxP was not incorporated, however, did not show any fluorescence reduction (Figure 2b-1). The disappearance of fluorescence was attributed to the elimination of the CMV and EGFP fragments by the Cre recombinase-mediated loxP recombination. Western blotting, as shown in Figure 2b, lower left panel, demonstrated the production of Cre recombinase in cells transfected by AlbTet-Cre where only cells transfected with AlbTet-Cre (Figure 2b, lower left panel, lanes 2 and 4) produced Cre recombinase. The expressed Cre recombinase displayed its effect on loxP recombination when tested by genomic-PCR (bottom right panel) following the same protocol as used in Figure 2a. Subsequently, to obtain evidence that expression of ABCA1 was decreased in these cells, Western blotting using the ABCA1 antibody was used in SIRIUS-ABCA1/AlbTet-Cre transfected cells and the results are shown in Figure 2c. This clearly demonstrates that ABCA1 was progressively reduced based on the time period (48, 72, and 96 hours) after infection. The ABCA1 quantity dropped to approximately 30% of the original level 96 hours after the transfection.
Figure 2.
Generation of reSIRIUS-ABCA1 and knockdown of ABCA1 via recombination in HepG2 cells after doxycycline (2 μg/ml) treatment. a: PCR analysis of recombined SIRIUS-ABCA1 after treatment (for 2 hours at 37°C) with purified Cre recombinase using specific primers (p1581 and p4214 primers, see Materials and Methods). b: Upper panel, fluorescence pictures of HepG2 cells transfected either with pLEGFP-C1 alone or SIRIUS-ABCA1 alone, or co-transfected with pLEGFP-C1 and AlbTet-Cre or SIRIUS-ABCA1 and AlbTet-Cre, respectively. Photograph displays significant down-regulation of GFP expression in SIRIUS-ABCA1/AlbTet-Cre transfected cells. Lower panel, Western blot of Cre recombinase genomic-PCR analyses to demonstrate the recombination process. Cellular lysates (1: pLEGFP-C1 alone, 2: pLEGFP-C1/AlbTet-Cre, 3: SIRIUS-ABCA1 alone, and 4: SIRIUS-ABCA1/AlbTet-Cre) were prepared 72 hours after transfection with different combinations of vectors. Lower left panel, Western blot analysis of Cre recombinase. There was significant expression of the Cre recombinase in pLEGFP-C1/AlbTet-Cre and SIRIUS-ABCA1/AlbTet-Cre transfected HepG2 cells after doxycycline treatment. Cre polyclonal antibody (Novagen) was used in this study. The commercial Cre recombinase was incorporated as a positive control (con). PCNA was used as an internal control. Lower right panel, genomic-PCR analysis showed that the recombination occurred in HepG2 cells when transfected with SIRIUS-ABCA1/AlbTet-Cre after doxycycline treatment. c: Expression of ABCA1 protein was detected by Western blot in SIRIUS-ABCA1/AlbTet-Cre transfected cells. Result indicates that the ABCA1 expression in SIRIUS-ABCA1/AlbTet-Cre co-transfected cells was progressively decreased according to the time (48, 72, and 96 hours) after transfection. The β-actin was used as an internal control.
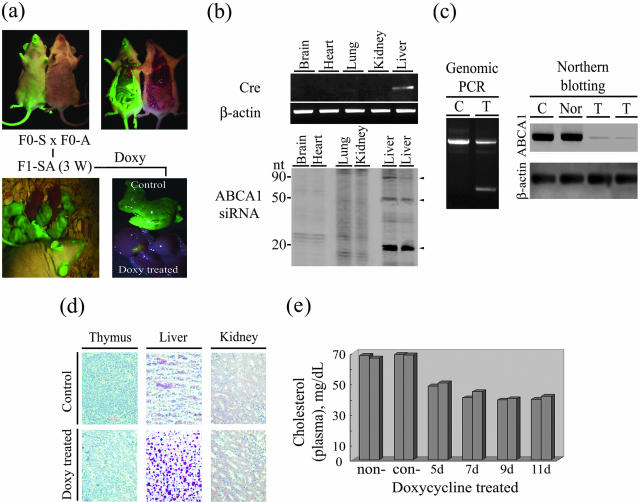
Having proved in HepG2 cells that AlbTet-Cre does produce Cre recombinase, which in turn recombines the two sites of loxP in SIRIUS-ABCA1 and expresses functional ABCA1 siRNA, the SIRIUS-ABCA1 and AlbTet-Cre plasmids were prepared by CsCl gradient and injected into mouse fertilized eggs to generate transgenic mice. Blood samples from the tail were collected from the newly born transgenic mice and the genomic DNA was extracted for transgene analysis by genomic-PCR with specific primers (see Materials and Methods). These mice were named F0-S and F0-A for SIRIUS-ABCA1 and AlbTet-Cre incorporation, respectively. Among 50 newborn mice injected with the exogenous DNA fragments, four animals (14%, 4 of 28) exhibited the integration of SIRIUS-ABCA1 (F0-S) and 27% (6 of 22) exhibited the integration of AlbTet-Cre (F0-A). GFP expression in the F0-S animals and their organs can all be visualized under a fluorescent anatomy-microscopy as shown in Figure 3a. F1 offspring of the F0-S and F0-A were produced through spontaneous mating soon after the F0 animals reached puberty (named F1-SA, Figure 3a, bottom). It was shown that the F1-SA animals carry intact SIRUS-ABCA1 and AlbTet-Cre genes as proved by GFP expression under a fluorescent anatomy-microscopy as described earlier for the initial F0-S (Figure 3a). Southern blotting was also conducted to confirm the integration of exogenous DNA into the mouse genomic DNA (data not shown). Doxycycline treatment of F1-SA, however, abolished GFP expression through the reaction elicited after Cre expression. Tissue-specific expression of Cre is active via AlbTet promoter control, as shown by the differential expression of fluorescence between the liver and gall bladder (Figure 3a, lower right panel). To demonstrate the controlled expression of foreign genes, both Cre and ABCA1 siRNA were detected by RT-PCR and Northern blotting assays, respectively. To increase the electrophoretic separation and resolution of RNA with a length less than 100 nt, we used 15% polyacrylamide gels in the presence of 7 mol/L urea instead of agarose gel. As shown in Figure 3b, Cre and ABCA1 siRNA only express in liver from a F1-SA transgenic animal after doxycycline treatment. Notably, there are no signals detectable in the tissues of brain, heart, lung, or kidney.
Figure 3.
The phenotype and symptoms of ABCA1 gene knockdown mice. a: The two separate lines of transgenic animals carrying AlbTet-Cre (F0-A) and SIRIUS-ABCA1 (F0-S) plasmid, respectively. The two separate lines of transgenic mice were mated to generate offspring (F1-SA) that carry both transgenes in a single mouse. F1-SA animals were examined under a fluorescent anatomy-microscopy to visualize the expression pattern of green fluorescent, like in F0-S founders. F1-SA mice at 3 weeks old were supplied with or without doxycycline by drinking and injection as described in Materials and Methods. Liver tissues from F1-SA mice were sectioned and visualized at 96 hours after doxycycline treatment. The fluorescent anatomy-microscopy showed that a reduction in EGFP occurred in F1-SA when treated with doxycycline. b: Cre recombinase and ABCA1-siRNA in various tissues of a F1-SA animal. Upper panel, RT-PCR analysis of Cre-recombinase expression in brain, heat, lung, kidney, and liver tissues. Cre-recombinase was only detected in the liver of a F1-SA mouse. β-Actin was used as an internal control. Lower panel, Northern blot analysis of ABCA1 siRNA in brain, heart, lung, kidney and liver tissues of F1-SA mouse. ABCA1 siRNA was only detected in the liver of two doxycycline-treated F1-SA mice. The different fragments of ABCA1 siRNA are indicated by arrowheads. c: Genomic-PCR analysis of the recombination of SIRIUS-ABCA1 in the livers of doxycycline-treated (treated, T) and -untreated mice (control, C). Left panel, doxycycline-treated F1-SA mice showed the presence of a less than 0.5-kb PCR product same as was shown in the HepG2 cells in Figure 2a. Right panel, Northern blot analysis of ABCA1 in doxycycline-treated and control F1-SA mice. β-actin was used as an internal control. d: Microscopic slides of Oil Red O staining of lipid in the thymus, livers, and kidney derived from non-induced and doxycycline-induced F1-SA mice (Magnification, ×200). Liver specimens exhibited significantly higher levels of cholesteryl ester staining after doxycycline treatment. e: Plasma cholesterol distribution in mice on different days after doxycycline treatment. Fifty μl of plasma were obtained from each of the 12 transgenic animals after doxycycline-treated (5 days to 11 days), together with non-treated (non-) and doxycycline-treated normal control (con-) mice. All three groups were subjected to cholesterol measurement as described in Materials and Methods.
Besides the fluorescence visualization, Cre-induced loxP recombination can also be proved by a genomic PCR study of SIRUS-ABCA1 using specific primers. Genomic DNAs from the liver were isolated from F1-SA mice treated with doxycycline. The 2.6-kb segment of DNA was reduced to less than 0.5-kb after doxycycline treatment (Figure 3c, left panel) and the result was the same as in the Cre treatment studies of HepG2 cells described earlier in Figure 2, a and b. The actual reduction of ABCA1 protein in doxycycline-treated F1-SA mice can be seen by Northern blot analysis (Figure 3c, right panel). ABCA1 is reduced to an almost non-detectable level 96 hours after doxycycline treatment whereas untreated F1-SA (C: non-induced) as well as normal mice (Nor) exhibited a high level of ABCA1. The ultimate proof of a successful transgenic model in this case, of course, is the deposition of cholesterol in liver due to the inhibition of ABCA1 in F1-SA transgenic mice. Cholesterol was detected using the Oil Red O staining method and its presence in tissue specimens was compared between transgenic animals with doxycycline treatment and untreated control mice. Lipid staining in the liver of a doxycycline-treated F1-SA mouse shows an overwhelming red color whereas the color is almost non-detectable in the liver of the untreated control ones (Figure 3d). Cholesterol accumulation was not observed in thymus and kidney. To reconfirm the phenomenon of ABCA1 inhibition in F1-SA mice, plasma cholesterol levels were also measured after doxycycline treatment. Untreated (non-) F1-SA and doxycycline treated (con-) normal mice were incorporated as control groups. In this experiment, two F1-SA mice were sacrificed for plasma collection at each time point. As shown in Figure 3e, a significant 35 to 40% reduction in the plasma cholesterol level was observed on the fifth day of doxycycline treatment in drinking water. The distribution of cholesterol is clearly shown following doxycycline inducement and demonstrates a gene-related decrease in plasma cholesterol of F1-SA mice. This is similar to the clinical phenotype of Tangier disease patients, with the knockdown of the ABCA1 gene in mice resulting in a marked decrease in the total plasma cholesterol. Such a cholesterol decrease in blood and an accumulation in liver are the major defects associated with ABCA-1-related Tangier disease.11 Other symptoms are not discussed in this paper.
These results clearly show that this approach has created a stable, inheritable, and inducible transgenic-animal model with the hallmark of high cholesterol content in liver using this new transgenic system for mimicking genetic diseases.
Discussion
Transgenic models using specific promoters have provided a wealth of information about the function of specific genes.14,15 However, the limitations of promoter-controlled transgene expression have become clear. The constitutive system has no control over the timing of the expression, which depends entirely on the properties of the promoters used. The promoters in this setting are active constitutively, many starting early in the embryonic stage. If the transgene product happens to be toxic to the organism, it can be highly detrimental to the developing embryo. Also, if a defect of the target gene is important to embryonic development, it may have a lethal impact during embryogenesis.16,17 Therefore, a constitutive overexpression transgenic system is not suitable for answering questions concerning specific genetic defects and their relationship to particular specific proteins. To address the limitations of the constitutive overexpression system, many laboratories have invested tremendous amounts of time and effort to establish conditional or inducible transgenic modeling systems. Several different inducible systems are currently in use including Mfp, tetracycline, and Cre-loxP systems.14,17–19 In order that an inducible transgenic system is able to work properly, three criteria are necessary: a regulatory unit, a responsive element linked to a target gene, and an induction agent. Furthermore, an ideally regulated transgene should have the following features: construction of transgene should be easy and have a low cost; transgene expression can be initiated or terminated rapidly and reversibly by a simple external inducer; together with a specific promoter, the inducible system should be able to provide temporal and spatial control over transgene expression; and the transgene should have no basal level expression in the silent state, whereas high-level expression should be achieved when the system is activated. When judged by these criteria, none of the currently available systems are even close to ideal and without apparent disadvantages. For example, not only is the knockout system of Cre-loxP difficult to construct and expensive to maintain in stem cells but it also has a high frequency of embryonic fatality and a low germline transmission efficiency.20
During the development of specific gene-knockout techniques, one of the most exciting findings in recent years has been the discovery of RNAi. In this study, we have combined the RNAi, Cre-loxP, and tetracycline-on systems to form a cluster target for specific gene knockout and under spatiotemporal control. We have demonstrated here that our SIRIUS-Cre system is heritable for the case of SIRIUS-ABCA1 transgenic mice. An inducible and significant syndrome involving a defect in ABCA1 was observed in these transgenic mice after doxycycline induction. This system fulfills the criteria of an ideally regulated transgene described above very well.
However, if the defect in the target gene is important to life, it may have a lethal impact after the treatment of doxycycline. To further improve pathological and pharmacological studies, a recoverable gene-knockdown system is necessary. Therefore, it should be possible to modify the current system and develop a recoverable SIRIUS-Cre system to rescue such transgenic animals from death. Such an approach for the new SIRIUS-Cre system presented here is described as follows. The SIRIUS-ABCA1 vector is modified by insertion of a TRE (tetracycline responsible element) fragment in the U6 promoter and under control of rtTA, such as in AlbTet-Cre vector. When doxycycline is removed, the rtTA silencer will bind to albumin and U6 promoters, and thus cause the blocking of siRNA expression. The blocking of siRNA expression will allow the target gene to re-express, thus avoiding transgenic animal death.
The relationship between Tangier disease, a congenital absence of serum α-lipoprotein resulting in the accumulation of cholesteryl esters in tissues, and the ABCA1 gene has been suggested.21 An animal model for the disease has been established using traditional techniques after lengthy trials and many fetal deaths.11 Our SIRIUS-Cre system is easy to construct and the F1 transgene mice were obtained at the first mating. Most important of all, the ABCA1 knockout can be achieved anytime after the animal is born through chemical induction. When compared with the Christiansen-Weber’s model, cholesteryl ester accumulation was only observed in the liver in our animal due to the tissue-specific promoter that we used for the tetracycline-on system. In addition, plasma cholesterol was about 40% less in doxycycline-treated F1-SA when compared with non-treated mice in our study.
In this study, we have demonstrated that an improved siRNA method with spatiotemporal and conditional knockout control was established and this system is much more powerful than those previously used to create animal models of genetic diseases.
Acknowledgments
We thank Dr. Ralph Kirby, Department of Life Science, National Yang-Ming University, for his criticisms and suggestions on the manuscript.
Footnotes
Address reprint requests to Winston C.Y. Yu, Ph.D., National Health Research Institutes, 3F, 109, Min-Chuan East Road, Sec 6, Taipei 114, Taiwan, R.O.C. E-mail: winston@nhri.org.tw.
Supported by a grant from National Health Research Institutes, Taiwan.
References
- McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nat Rev Genet. 2002;3:737–747. doi: 10.1038/nrg908. [DOI] [PubMed] [Google Scholar]
- Scherr M, Morgan MA, Eder M. Gene silencing mediated by small interfering RNAs in mammalian cells. Curr Med Chem. 2003;10:245–256. doi: 10.2174/0929867033368493. [DOI] [PubMed] [Google Scholar]
- Manche L, Green SR, Schmedt C, Mathews MB. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol Cell Biol. 1992;12:5238–5248. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- Paddison PJ, Caudy AA, Hannon GJ. Stable suppression of gene expression by RNAi in mammalian cells. Proc Natl Acad Sci USA. 2002;99:1443–1448. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasuwa H, Kaseda K, Einarsdottir T, Okabe M. Small interfering RNA and gene silencing in transgenic mice and rats. FEBS Lett. 2002;532:227–230. doi: 10.1016/s0014-5793(02)03680-3. [DOI] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Tiscornia G, Singer O, Ikawa M, Verma IM. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc Natl Acad Sci USA. 2003;100:1844–1848. doi: 10.1073/pnas.0437912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Ihrig MM, McManus MT, Gertler FB, Scott ML, Van Parijs L. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells, and transgenic mice by RNA interference. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- Shinagawa T, Ishii S. Generation of Ski-knockdown mice by expressing a long double-strand RNA from an RNA polymerase II promoter. Genes Dev. 2003;17:1340–1345. doi: 10.1101/gad.1073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen-Weber TA, Voland JR, Wu Y, Ngo K, Roland BL, Nguyen S, Peterson PA, Fung-Leung WP. Functional loss of ABCA1 in mice causes severe placental malformation, aberrant lipid distribution, and kidney glomerulonephritis as well as high-density lipoprotein cholesterol deficiency. Am J Pathol. 2000;157:1017–1029. doi: 10.1016/S0002-9440(10)64614-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly S, Gardner JM, McClelland A, Kaleko M. High-level tissue-specific expression of functional human factor VIII in mice. Hum Gene Ther. 1996;7:183–195. doi: 10.1089/hum.1996.7.2-183. [DOI] [PubMed] [Google Scholar]
- Choo KB, Chen HH, Cheng WT, Chang HS, Wang M. In silico mining of EST databases for novel pre-implantation embryo-specific zinc finger protein genes. Mol Reprod Dev. 2001;59:249–255. doi: 10.1002/mrd.1029. [DOI] [PubMed] [Google Scholar]
- Stricklett PK, Nelson RD, Kohan DE. The Cre/loxP system and gene targeting in the kidney. Am J Physiol. 1999;276:F651–F657. doi: 10.1152/ajprenal.1999.276.5.F651. [DOI] [PubMed] [Google Scholar]
- Udvadia AJ, Linney E. Windows into development: historic, current, and future perspectives on transgenic zebrafish. Dev Biol. 2003;256:1–17. doi: 10.1016/s0012-1606(02)00083-0. [DOI] [PubMed] [Google Scholar]
- Ranger AM, Malynn BA, Korsmeyer SJ. Mouse models of cell death. Nat Genet. 2001;28:113–118. doi: 10.1038/88815. [DOI] [PubMed] [Google Scholar]
- Zuo J. Transgenic and gene targeting studies of hair cell function in mouse inner ear. J Neurobiol. 2002;53:286–305. doi: 10.1002/neu.10128. [DOI] [PubMed] [Google Scholar]
- Acres B, Apostolopoulos V, Balloul JM, Wreschner D, Xing PX, Ali-Hadji D, Bizouarne N, Kieny MP, McKenzie IF. MUC1-specific immune responses in human MUC1 transgenic mice immunized with various human MUC1 vaccines. Cancer Immunol Immunother. 2000;48:588–594. doi: 10.1007/PL00006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryding AD, Sharp MG, Mullins JJ. Conditional transgenic technologies. J Endocrinol. 2001;171:1–14. doi: 10.1677/joe.0.1710001. [DOI] [PubMed] [Google Scholar]
- Lewandoski M. Conditional control of gene expression in the mouse. Nat Rev Genet. 2001;2:743–755. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- Schmitz G, Langmann T. Structure, function, and regulation of the ABC1 gene product. Curr Opin Lipidol. 2001;12:129–140. doi: 10.1097/00041433-200104000-00006. [DOI] [PubMed] [Google Scholar]