Abstract
Prion protein (PrP) is expressed in many tissues and is required for susceptibility to scrapie and other prion diseases. To investigate the role of PrP expression in different cell types on pathology in retina and brain after scrapie infection, we examined transgenic mice expressing hamster PrP from the PrP promoter (tg7), the neuron-specific enolase promoter (tgNSE), or the astrocyte-specific glial fibrillary acidic protein promoter (tgGFAP). After intraocular inoculation with hamster scrapie, clinical disease developed in tg7 and tgNSE mice by 100 days and in tgGFAP mice by 350 days. Astrogliosis and scrapie-associated protease-resistant PrP (PrP-res) were detected in retina and brain before clinical onset. Retinal PrP-res was present in high amounts in both tg7 and tgNSE mice, however only tg7 mice developed retinal degeneration and extensive apoptosis. In contrast, in all three lines of mice high levels of brain PrP-res accompanied by neurodegeneration were observed. Thus, PrP expression on neurons or astrocytes was sufficient for development of scrapie-induced degeneration in brain but not in retina. The combined effects of PrP-res production in multiple cell types was required to produce retinal degeneration, whereas in brain PrP-res production by neurons or astrocytes alone was sufficient to cause neuronal damage via direct or indirect mechanisms.
Transmissible spongiform encephalopathies are a group of fatal degenerative brain diseases that occur naturally in primates and ruminants. They include scrapie in sheep, bovine spongiform encephalopathy in cattle, and several human diseases such as Creutzfeldt-Jakob disease, Kuru, and Gerstmann-Sträussler-Sheinker syndrome. An important feature of transmissible spongiform encephalopathies is the accumulation in the brain of a protease-resistant form of prion protein, PrPSc or PrP-res, which is formed by an incompletely understood posttranslational event from normal host-encoded protease-sensitive prion protein, termed PrPC or PrP-sen.1–3 Prion protein (PrP) is required for propagation of the scrapie agent and for development of clinical disease.4,5 PrP is attached to the cell surface by a glycolipid anchor and is normally expressed in a variety of cells and tissues, including neurons,6 astrocytes,7,8 lymphocytes,9 follicular dendritic cells,10 and tumor cell lines of various lineages.11
Studies of the pathology of scrapie have primarily focused on changes in the brain and spinal cord where the most characteristic lesion consists of spongy vacuolar degeneration of gray and sometimes white matter.12 As an extension of the central nervous system, the retina has increasingly featured in pathological studies because of its relatively simple architecture and well-described physiological properties. An early description of retinal degeneration in natural scrapie of sheep13 was followed by reports of retinal changes in laboratory rodents with experimental scrapie, and there is now a significant amount of information available about the targeting and infectivity of mouse scrapie in the rodent visual system.14,15 Retinopathy in mice after intraocular inoculation is dependent on the strain of scrapie and the genotype of mouse.16 Some scrapie strains (22C, ME7, 87A, and 87V) produce minimal or no retinal pathology, whereas other strains (79A and 139A) produce degeneration of the photoreceptor layer in several mouse genotypes. This pathology occurs in hamsters terminally sick with the 263K strain of scrapie,17,18 and retinal degeneration has also been observed in mice after infection with human Creutzfeldt-Jakob disease.19 Although neurons are clearly affected in the disease process, it is still unclear what role other cells have in scrapie pathogenesis.
The development of hamster PrP (HaPrP)-transgenic mice has made it possible to experimentally separate PrP-sen expression in neural or glial cell types in vivo. In the present study we took advantage of mice that differentially express hamster PrP (HaPrP) under the control of the neuron-specific enolase (NSE) promoter (tgNSE);20 the astrocyte-specific glial fibrillary acidic protein (GFAP) gene promoter (tgGFAP);21 or the endogenous PrP promoter (tg7), which drives the expression of HaPrP in many tissues.22 In these original studies intracerebral scrapie inoculation induced clinical disease in all three transgenic lines, although the incubation period for tgGFAP mice was found to be considerably longer than for the other two lines.
In this report, we have used the direct intraocular route for scrapie infection of these transgenic mouse lines to establish a localized infection for early histopathological analysis. Disease susceptibility as well as pathological effects on both retinal and brain tissue were analyzed. In these experiments all three lines of transgenic mice were susceptible to scrapie after intraocular inoculation, but the appearance and distribution of PrP-res in the retinas after infection varied in the different transgenic lines. Furthermore, these lines differed in the extent of retinal degeneration and apoptosis observed suggesting that the cell types expressing PrP-sen had a strong influence on the pathological processes in retina.
Materials and Methods
Infection of Mice
All mice were bred and raised at the Rocky Mountain Laboratories and were handled according to policies of the Rocky Mountain Laboratories Animal Care and Use Committee and all applicable federal guidelines. Adult (6 to 8 weeks old) animals were used for all experiments. The transgenic animals used in this study have been described.20–22 Mice were inoculated intraocularly, and for comparison were inoculated intracranially, intraperitoneally, and orally with hamster scrapie strain 263K. Before inoculation, mice were deeply anesthetized by intramuscular injection of a combination anesthetic cocktail containing ketamine, xylazine, and acepromazine. Approximately 2 × 105 intracranial 50% infectious doses (ID50) of hamster 263K scrapie infectivity in 1 μl of phosphate-buffered saline (pH 7.2) supplemented with 2% fetal bovine serum (Hyclone, Logan, UT) was injected unilaterally into the vitreous cavity using a 32-gauge needle attached to a 10-μl Hamilton syringe. After injection, the needle was left in the vitreal chamber for 1 minute to minimize leaking of the inoculum. Mice inoculated intracranially or intraperitoneally received 1 × 107 ID50 in a volume of 50 μl. Mice infected orally received 2 × 108 ID50 in 100 μl via a small-diameter flexible polypropylene catheter inserted over the base of the tongue ∼1 to 2 cm into the esophagus. All mice were observed several times each week for clinical signs of scrapie, which included weight loss, kyphosis, ataxia, and an exaggerated high-stepping gait most noticeable in the hind limbs.22 Mice exhibiting short incubation periods (less than 100 days) died within 1 to 4 days after the appearance of clinical symptoms, whereas mice exhibiting longer incubation periods had more prolonged clinical disease that lasted 10 to 14 days. Brain samples from mice in each group with clinical evidence of scrapie were analyzed for PrP-res by Western blotting to confirm the clinical diagnosis.20
Pathology and Immunohistochemistry
Mice used for histopathological analysis were lightly anesthetized, then trans-cardially perfused with 30 ml of phosphate-buffered saline. The eyes were removed and placed in Davidson’s fixative (3 parts of 100% ETOH, 2 parts of 37 to 40% formaldehyde, and 1 part glacial acetic acid) for 24 hours before dehydration and paraffin-embedding. Whole brains were removed and placed in 3.7% phosphate-buffered formalin for 3 to 5 days, then cut into 2- to 3-mm coronal sections before dehydration and paraffin-embedding. Serial 4-μm sections were cut using a standard Leica microtome, placed on positive-charged glass slides and dried overnight at 56°C. All histopathological procedures were performed on brain and eye sections from both mock-infected and scrapie-infected animals including appropriate immunohistochemical controls. Ocular sections of the retina were stained by standard hematoxylin and eosin (H&E) and analyzed for degeneration by measurement of retinal thickness and the loss of normal retinal architecture. Immunohistochemical analysis was performed using the Ventana automated Nexus stainer (Ventana, Tucson, AZ). Slides were deparaffinized, rehydrated to Tris-HCl buffer, pH 7.5 (GFAP staining), or 0.1 mmol/L citrate buffer, pH 6.0 (3F4 staining). Staining for GFAP used a standard avidin-biotin complex immunoperoxidase protocol using anti-GFAP at a dilution of 1:1000 (DAKO, Carpinteria, CA), biotinylated goat anti-rabbit IgG at a dilution of 1:250 (Vector Laboratories, Burlingame, CA), and amino-ethyl carbazole as substrate (Ventana). For the detection of PrP-res, it is necessary to perform antigen retrieval by treating the sections with formic acid, or subjecting them to autoclaving and/or proteinase K digestion.23,24 The sections in this study were rehydrated and autoclaved for 20 minutes at 122°C and 22 psi in 0.1 mmol/L citrate buffer (pH 6.0). PrP-res was then stained with mouse monoclonal antibody 3F4 cell culture supernatant (1:50).25 Detection was performed with biotinylated horse anti-mouse IgG diluted 1:250 (Vector Laboratories) followed by supersensitive streptavidin diluted 1:3 in Tris-HCl, pH 7.5, buffer (Biogenex, San Ramon, CA) and amino-ethyl carbazole. Staining for caspase-3 used rabbit anti-cleaved caspase-3 at a dilution of 1:100 (Cell Signaling, Beverly, MA), biotinylated goat anti-rabbit IgG at a dilution of 1:250 (Vector Laboratories), and amino-ethyl carbazole as a substrate. Terminal dUTP nick-end labeling (TUNEL) staining of retinas was performed manually using terminal deoxynucleotidyl transferase (TDT) enzyme from a NeuroTacs II in situ apoptosis detection kit (Trevigen, Gaithersburg, MD) according to the manufacturer’s instructions.
Real-Time Quantitative Reverse Transcriptase (RT)-Polymerase Chain Reaction (PCR)
Quantitative real-time RT-PCR was performed as previously described, with some modifications.26 Total RNA was isolated from whole eye and brain tissue of individual transgenic mice using an RNeasy mini kit (Qiagen, Valencia, CA), plus DNase treatment. Each RNA sample was reverse-transcribed and then PCR amplified in triplicate by using TaqMan one-step RT-PCR master mix (Applied Biosystems, Foster City, CA) in a total reaction volume of 10 μl. The relative quantity of the PrP target gene was normalized with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for each sample by amplifying GAPDH in separate triplicate reaction tubes. Normalized average relative PrP expression values for each sample were compared with the gene expression level of PrP from a normal hamster brain. Primer and probe sequences (Applied Biosystems) used for PrP RT-PCR were: forward, 5′-CTG GAG CAG GCC CAT GAT G; reverse, 5′-GCG GTA CAT GTT TTC ACG GTA GT; probe, 5′-CGG TCC TCC CAG TCG TTG CCA AA.
Immunoblot Analysis for PrP
Eye and brain tissues were analyzed for the presence of PrP-sen and PrP-res using Western blotting techniques as previously described20 with some modifications. For PrP-sen, 10% (w/v) homogenates of eye and brain tissue were made in modified RIPA buffer (50 mmol/L Tris-HCl, pH 7.4, 1% Triton X-100, 0.2% sodium deoxycholate, and 0.2% sodium dodecyl sulfate, with protease-inhibitor cocktail tablets added according to the manufacturers’ recommendation; Roche, Indianapolis, IN). To remove nucleic acids, the homogenates were treated with 20 U Benzoase nuclease (Novagen, Madison, WI) for 30 minutes at 37°C, and debris was removed by centrifugation at 12,000 × g for 10 minutes at 4°C. After normalization to total protein (BCA protein quantification assay; Pierce, Rockford, IL), homogenates were diluted in 2× sample buffer to a concentration of 1 mg of tissue equivalents or appropriate serial dilution per 10 μl, boiled for 5 minutes, and run on 16% polyacrylamide gels. Proteins were transferred to polyvinylidene difluoride membranes and probed with a 1:3000 dilution of monoclonal antibody 3F4 anti-hamster PrP for 2 hours at room temperature. Detection was with a 1:2500 dilution of goat anti-mouse IgG-alkaline phosphatase and standard enhanced chemifluorescence procedures (Amersham, Arlington Heights, IL). The blots were dried for 2 hours, scanned with a Storm phosphorimager, then analyzed with ImageQuaNT V5.2 software (Molecular Dynamics, Sunnyvale, CA).
PrP-res was isolated from eyes and brains of clinically sick animals as previously described,27 except proteinase K digestion was performed with 1 mg/ml of proteinase K for 30 minutes at 37°C, and the final pellets were resuspended by sonication in 100 μl of sample buffer. Five-mg tissue equivalents were run on 16% polyacrylamide gels and proteins were transferred to polyvinylidene difluoride membranes and probed with monoclonal antibody 3F4 as described above, except detection was with goat anti-mouse IgG-horseradish peroxidase (1:3000) and standard enhanced chemiluminescence procedures (Amersham).
Results
Susceptibility of Transgenic Mice to Hamster Scrapie
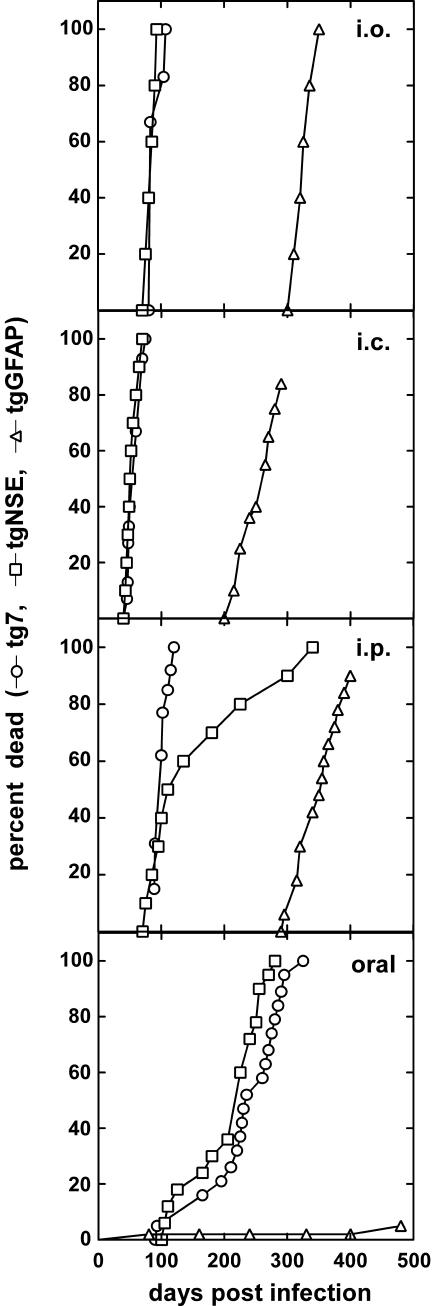
To study the effect of PrP expression in neurons or astrocytes on scrapie infection via the ocular route, tg7, tgNSE, and tgGFAP mice were inoculated intraocularly with 2 × 105 ID50 of hamster scrapie strain 263K. Tg7 mice and tgNSE mice had the most rapid course to clinical disease, and all mice died before 100 days after infection (Figure 1). In contrast, tgGFAP mice developed disease between 300 to 350 days after infection. These results demonstrated that expression of PrP in either neurons only or astrocytes only was sufficient to confer susceptibility to typical scrapie disease after intraocular scrapie inoculation.
Figure 1.
Kinetics of death of tg7, tgNSE, and tgGFAP mice after intraocular (i.o.), intracerebral (i.c.), intraperitoneal (i.p.), and oral inoculation of hamster scrapie strain 263K. The doses used for each type of inoculation are described in the Materials and Methods. The data are represented as percentage of animals dead versus days after infection, which were pooled from at least two independent experiments (tg7 mice: n = 6 intraocular, n = 15 intracranial, n = 13 intraperitoneal, and n = 19 oral; tgNSE mice: n = 5 intraocular, n = 10 intracranial, n = 10 intraperitoneal, and n = 12 oral; tgGFAP mice: n = 6 intraocular, n = 8 intracranial, n = 12 intraperitoneal, and n = 6 oral). The results of the intracranial inoculation were similar to what was published previously.20–22
In this experiment the ocular route was compared to intracranial, intraperitoneal, and oral routes in the three transgenic mouse lines. Intracranial inoculation of hamster scrapie strain 263K resulted in more rapid disease kinetics than was seen after intraocular inoculation (Figure 1). Tg7 and tgNSE mice became moribund and were sacrificed at 50 to 60 days after infection, and tgGFAP mice progressed more slowly to the endpoint clinical stage at 230 to 240 days after infection. This difference in the two routes was likely because of the 50-fold lower dose of inoculum administered intraocularly, because there was limited volume that can be given by this route.
Mice were also inoculated intraperitoneally and orally. By both routes in tg7 and tgNSE animals the interval to sacrifice was longer than with intracranial or intraocular routes (Figure 1); however, all mice eventually succumbed to scrapie by 300 days. The tgGFAP mice developed scrapie around 350 days after infection after intraperitoneal inoculation, and this time to disease was in the same range as that seen after intraocular infection. In contrast, only 5% of the tgGFAP mice developed disease after oral inoculation. The reduced susceptibility of tgGFAP mice to infection by the oral route suggested that PrP-expressing cells or mechanisms required for oral infection were limited in the tgGFAP mice.
HaPrP Is Expressed in the Ocular Tissue of Transgenic Mice
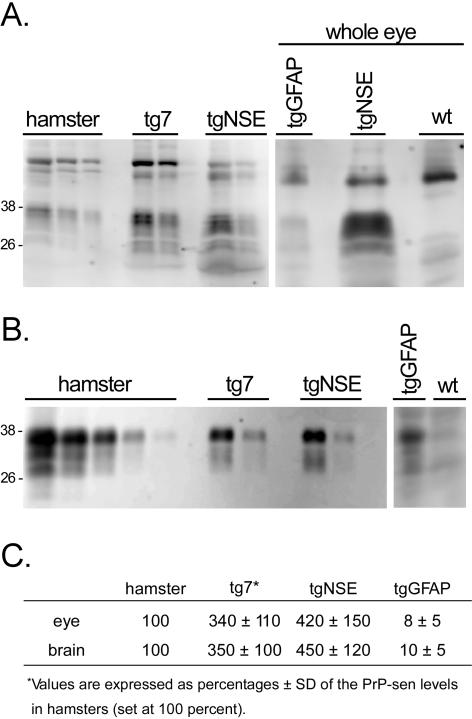
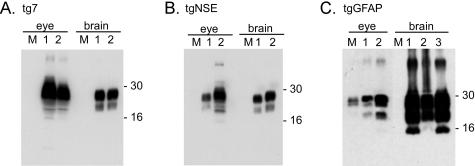
One possible explanation for the difference in disease tempo in intraocular-inoculated tg7 and tgNSE versus tgGFAP mice is that retinal or brain PrP-sen expression could differ in these mice. To determine the possible influence of PrP expression levels, we measured hamster PrP-sen in the eyes and brains of tg7, tgNSE, and tgGFAP mice by Western blotting with serial dilutions. By this method PrP-sen was readily detected from the ocular tissue of tg7 and tgNSE mice (Figure 2A). However, in the tgGFAP eye PrP-sen was detected only when the protein extract from the entire eye was analyzed in a single gel lane (Figure 2A, right). PrP-sen was detected at similar levels in the eyes of tg7 and tgNSE mice, and these levels were threefold to fourfold higher than in normal hamsters. In the eyes of tgGFAP mice PrP-sen levels were ∼10 times lower than hamster (Figure 2C).
Figure 2.
Western blot of PrP-sen from transgenic mice. Eye (A) and brain (B) tissue from hamster, tg7, tgNSE, tgGFAP, and nontransgenic (wt) mice were blotted and hamster PrP was detected with 3F4 monoclonal antibody and ECF as described in Materials and Methods. PrP-sen bands representing multiple glycosylated forms are indicated by the approximate molecular weights of 26 to 38 kd. A (right): Methanol-precipitated total protein from a whole tgGFAP eye to demonstrate detection of PrP-sen. tgNSE and wt mouse eyes are shown for comparison of positive and negative tissues, respectively. To determine amounts of PrP-sen each homogenate was adjusted to 1 mg of total protein/10 μl (tissue equivalents), then serial twofold dilutions were analyzed starting with (A) undiluted (hamster), 1:4 (tg); or (B) 1:2 (hamster), 1:32 (tg). C: At least three mice per group were analyzed for densitometric analysis. The relative amounts of PrP-sen from tg7, tgNSE, and tgGFAP were calculated by densitometric analysis after comparison with standard curves from hamster eye (undiluted to 1:16) or hamster brain (undiluted to 1:256). The values are expressed as percentage ± SD of PrP-sen in eye or brain compared to the level of PrP in hamster eye or brain, which was set at 100%.
Because PrP was difficult to detect from tgGFAP ocular tissue, analysis of PrP mRNA expression was performed to obtain additional comparative data. Real-time RT-PCR analysis showed that the level of PrP expression in tgGFAP eyes was 10-fold lower than in normal hamsters (data not shown); whereas the levels in tg7 and tgNSE eyes were threefold to fourfold higher than in normal hamsters. These results were in close agreement with the values obtained by Western blotting of proteins. In brain the relative PrP-sen levels in the three transgenic mouse lines were similar to what was seen in eye (Figure 2B), but as measured by RT-PCR each mouse line expressed approximately fourfold more PrP mRNA in brain than in eye (data not shown). In summary, the high levels of PrP expression in both the eye and brain of tg7 and tgNSE mice compared to tgGFAP mice could be contributing to the differences in disease tempo after intraocular inoculation.
PrP-res Accumulation in Transgenic Mouse Retinas
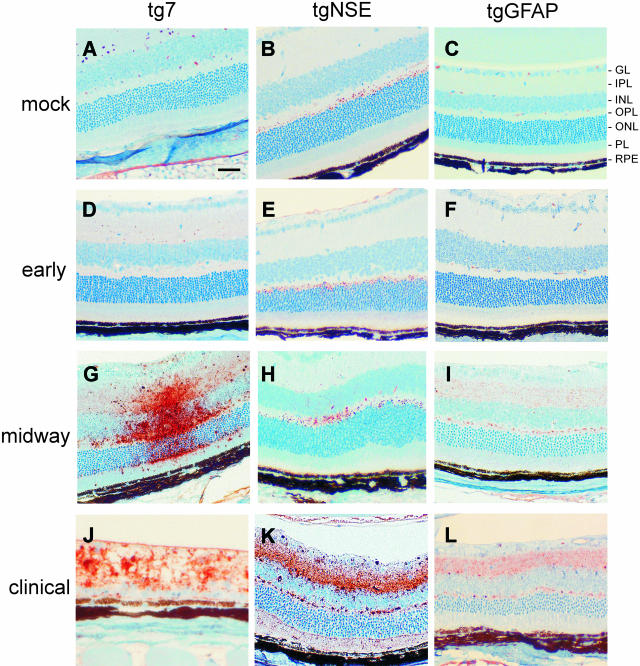
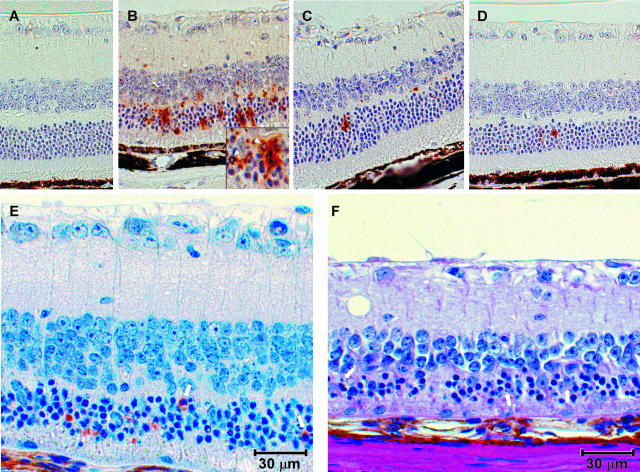
The timing and location of PrP-res deposition within the central nervous system are important features of the pathogenic process of transmissible spongiform encephalopathy diseases. To determine the extent of PrP-res accumulation in the eye after ocular infection, tg7, tgNSE, and tgGFAP retinas were examined throughout the course of disease for the appearance and accumulation of PrP-res by immunohistochemical staining using the 3F4 monoclonal antibody (Figure 3). In tg7 mice no PrP-res was observed early in infection (28 days after infection). In these mice PrP-res accumulation was first detected midway through the course of infection (45 days after infection, compare Figure 3, D and G). At this time point there was abundant diffuse and punctate PrP-res staining in all layers of the retina (Figure 3G). This finding was consistent in all animals evaluated (Table 1). By the time clinical symptoms were apparent (90 to 100 days after infection), the retinas had degenerated completely and residual PrP-res staining was still present (Figure 3J).
Figure 3.
Immunohistochemical analysis of PrP-res in hamster PrP transgenic mouse retinas. Shown are representative sections of mock-infected retinas age-matched from the midway or clinical time of disease (A–C), and retinas from early (28 days after infection) (D–F), midway (45 days after infection for tg7 and tgNSE; 140 days after infection for tgGFAP) (G–I), and clinical (90 to 100 days after infection for tg7 and tgNSE; 350 to 355 days after infection for tgGFAP) (J–L) time points of disease after intraocular infection of tg7, tgNSE, and tgGFAP mice with hamster scrapie. A–F: No specific PrP-res immunoreactivity was detected in mock-infected retinas or retinas from early time points of infection. The small amount of staining seen in the OPL of tgNSE mice at the early time point was also seen in mock-infected mice and was therefore not PrP-res-specific. Specific PrP-res immunoreactivity was detected midway in infection in the plexiform layers of all three transgenic retinas tested (G–I) and accumulated primarily in the inner retinal layers of the retinas of tgNSE and tgGFAP mice (J and K, IPL and OPL). J: In contrast with the tgNSE and tgGFAP retinas, the retinas from tg7 mice were completely degenerated by the time of clinical disease. GL, ganglion cell layer; IPL, inner plexiform layer; INL, inner nuclear layer; PL, photoreceptor layer; RPE, retinal pigment epithelium. Scale bar, 20 μm. Original magnifications: ×20 (A, I, K–L); ×40 (J).
Table 1.
Summary of Retinal Pathology after Intraocular Infection of Transgenic Mice
| Time* | Mice† | Retinal degeneration‡ | GFAP§ | PrP-res¶ | Western blot∥ |
|---|---|---|---|---|---|
| Early | tg7 | 0/7 | 0/7 | 0/7 | 0/2 |
| tgNSE | 0/4 | 0/4 | 0/4 | 0/2 | |
| tgGFAP | 0/4 | 0/4 | 0/4 | 0/2 | |
| Midway | tg7 | 6/7 | 7/7 | 7/7 | 2/2 |
| tgNSE | 0/3 | 3/3 | 3/3 | 0/2 | |
| tgGFAP | 0/3 | 0/3 | 3/3 | 0/2 | |
| Clinical | tg7 | 5/5 | 5/5 | 5/5 | 2/2 |
| tgNSE | 0/3 | 3/3 | 3/3 | 2/2 | |
| tgGFAP | 0/3 | 3/3 | 3/3 | 2/2 |
Three time points were analyzed for each strain of tg mouse. Early, 28 dpi for all three strains; midway, 42 dpi for tg7 and tgNSE, and 140 dpi for tgGFAP; clinical, 90 to 100 dpi for tg7 and tgNSE, and 320 to 350 dpi for tgGFAP.
One mock-infected animal from each strain was included for analysis at each time point, and always scored negative for disease-specific pathology.
Retinal degeneration was characterized by atrophy, cell loss, and disorganization of layers as shown in Figures 3J, 4G, and 4J. Number of positive mice/total.
Staining of up-regulated GFAP. Number of mice with staining exceeding controls/total mice studied. See Figure 4.
Number of mice with PrP-res staining/total studied. See Figure 3.
Western blots for PrP-res were performed on 5-mg tissue equivalents of age-matched ocular tissues. Number of positive mice/total tested.
In tgNSE mice, the retinas had a background level of immunoreactivity characterized by small, uniform punctate staining in the outer plexiform layer (OPL) (Figure 3B). Western blot analysis of eye homogenates after proteinase K digestion confirmed this background staining was not PrP-res (data not shown). The same time points were evaluated for tgNSE mice as tg7 mice, and early in infection there was no detectable increase in PrP-res staining (Figure 3E). By the midway point, PrP-res was detectable in the OPL and inner nuclear layer, and was characterized by both diffuse and small punctate staining (Figure 3H). At the time of clinical disease (90 to 100 days after infection) both the intensity and distribution of PrP-res staining were amplified in all layers of the retina, especially in the inner retinal layers (inner and outer plexiforms and nerve fiber layers) (Figure 3K, IPL and OPL), which exhibited the largest amount of PrP-res accumulation.
The retinas from tgGFAP mice were also examined at several times from early to the clinical time of disease (28 to 355 days after infection). Because the disease course was much slower in tgGFAP mice, the midway and clinical time points were much later than in the other two transgenic lines. Similar to tg7 and tgNSE mice, PrP-res was first detected in these mice midway through the course of disease (140 days after infection, Figure 3I). At this time there was a uniform, diffuse PrP-res staining pattern in the plexiform layers. The amount of PrP-res staining in the plexiform layers increased as the disease progressed to the clinical stage (355 days after infection) (Figure 3, compare I and L). More PrP-res accumulated as small plaque-like deposits in the OPL compared to the inner plexiform layer, and this was consistent in all mice that were evaluated (Table 1). There was very little or no PrP-res staining observed in the outer nuclear layer or photoreceptor layer at any time during the course of infection (Figure 3; F, I, and L). Interestingly, in both tgNSE and tgGFAP mice the appearance of PrP-res in the retinas after intraocular infection was primarily in the plexiform layers; with lesser involvement of the nuclear layers even late in disease.
Histopathology and Müller Cell Astrocytosis after Intraocular Inoculation of Hamster Scrapie
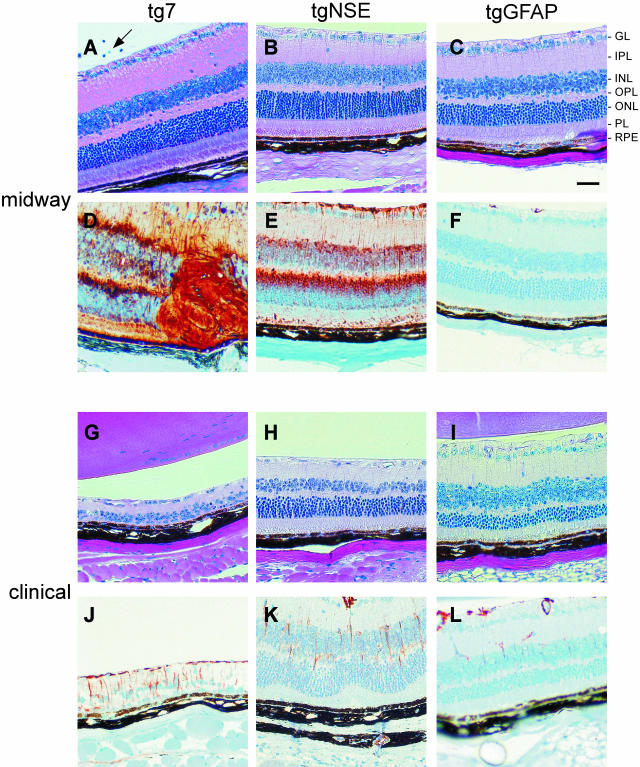
The up-regulation of GFAP in glial astrocytes of the brain is a prominent feature of transmissible spongiform encephalopathy diseases. The retina contains a specialized type of astroglia, the Müller cell, which under normal conditions express minimal levels of GFAP.28 However, the levels of GFAP are markedly increased in Müller cells after retinal degeneration, detachment, or disease. To determine the extent of retinal damage after scrapie infection, retinas of transgenic mice were examined for pathology by H&E and for reactive astrocytosis by anti-GFAP immunostaining (Figure 4). By midway through the infection, some tg7 retinas had more degeneration or atrophy of the outer nuclear layer (Table 1) as well as some vitritis (Figure 4A, arrow) and retinal vasculitis. This was in contrast to the tgNSE and tgGFAP retinas (Figure 4, B and C), which had no ocular inflammation (vitritis or vasculitis), and thinning of the outer nuclear layer was observed in only a few retinas at the midway point. By the clinical stage of disease, the tg7 retinas had progressed to loss of photoreceptor cells (Figure 4G). In contrast, the overall architecture of the tgNSE and tgGFAP retinas remained intact, but there was mild thinning of the outer nuclear layer and some degeneration of the inner and outer segments of the photoreceptor layer (PL) when compared to retinas earlier in the course of disease (Figure 4, compare H with B and I with C).
Figure 4.
Histopathological analysis and Müller cell astrocytosis in PrP-transgenic mouse retinas. Shown are representative retinal sections midway (A–F) and at the clinical time (G–L) of disease (see Figure 3 legend for precise times). Retinal sections were stained with H&E for histopathological analysis (A–C and G–I) and with anti-GFAP antibody for astrocytosis (D–F and J–L). A (arrow): Vitritis in tg7 mouse at the midway point of infection. Scale bar, 20 μm. Original magnifications, ×20.
Retinas from all three transgenic mice were examined early (28 days after infection) for GFAP up-regulation, and none demonstrated any anti-GFAP staining above the levels seen in mock-infected retinas (Table 1). In tg7 and tgNSE mice, by the midway point of disease there was a marked increase in the number and intensity of GFAP-expressing Müller cells (Figure 4, D and E). The Müller cell nuclei are located in the INL and their cell processes extend across from external to internal limiting membrane. Hence, the staining was observed in every layer of the retina. However, this expression was transient because there was much less GFAP staining by the clinical time point (Figure 4, J and K). In contrast, in the tgGFAP mice GFAP staining was not seen at the midpoint of infection (Figure 4F), and was only slightly detectable in these retinas by the clinical stage of disease (Figure 4L). Notably, the relationship of GFAP staining to PrP-res staining varied in the three transgenic strains studied. In the tgNSE mice abundant GFAP expression preceded the appearance of PrP-res (compare Figure 4E with Figure 3H); in tg7 mice GFAP and PrP-res expression were synchronous (compare Figure 4D with Figure 3G); and in tgGFAP mice GFAP expression was delayed compared to PrP-res (compare Figure 4F with Figure 3I).
Retinal Apoptosis in Scrapie-Infected Transgenic Mice
Because there was a marked difference in the levels of PrP-res accumulation, astrocytosis, and ultimate retinal destruction in the transgenic mice after intraocular scrapie infection, we next studied the retinas for indications of apoptotic cell death. Serial sections of the same ocular tissue used for PrP-res and GFAP immunohistochemical analysis were stained for TUNEL at midway and clinical time points (Figure 5). Midway through disease, tg7 mice showed abundant TUNEL staining primarily in the outer nuclear layer (Figure 5B), while retinas from tgNSE and tgGFAP mice did not show any detectable staining (data not shown). However, at the clinical time of disease retinas from tgNSE and tgGFAP mice did show occasional positive TUNEL staining, which was mostly in the outer nuclear layer (Figure 5, C and D). In tg7 mice, but not in tgNSE or tgGFAP mice, there was positive immunostaining for anti-active caspase-3 (Figure 5E). Furthermore, in these same mice in areas with more advanced retinal degeneration there was no detectable caspase staining. Instead, there was ample morphological evidence of apoptosis with degenerative nuclear changes and apoptotic bodies, particularly in the outer nuclear layer (Figure 5F). Thus, the retinal degeneration observed in the retinas from tg7 mice was associated with apoptotic cell death.
Figure 5.
Apoptosis in retinas of scrapie-infected transgenic mice. Shown are representative retinal sections stained by TUNEL of mock-infected tg7 mice (A) and scrapie-infected tg7 (B), tgNSE (C), and tgGFAP (D) mice. tgGFAP and tgNSE retinas were stained at the clinical time of disease, and tg7 retinas were stained midway through disease (see Figure 3 legend for precise times). B: Characteristic pyknotic nuclei from apoptotic cells in tg7 retinas are indicated by white arrowheads (inset). E: Staining with anti-cleaved caspase-3 at a higher magnification shows typical cytoplasmic staining (white arrows). F: In a different mouse with more severe retinal degeneration, H&E staining shows retinal atrophy and cell loss in outer nuclear layer with typical pyknotic nuclei and apoptotic bodies (white arrows). Original magnifications, ×40.
PrP-res in Transgenic Eye and Brain Tissue
Because detection of PrP-res by immunohistochemistry is not a quantitative method PrP-res was also measured by Western blotting of retinal extracts from scrapie-infected transgenic mice at the time of clinical disease. In the eye, tg7 mice accumulated the highest levels of PrP-res (Figure 6A). The levels in tgNSE mice were slightly lower than in tg7, whereas in tgGFAP mice the levels were markedly lower than both tg7 and tgNSE mice (Figure 6, B and C). Therefore, in all three transgenic mice the relative amounts of PrP-res that accumulated in the retinal tissues were consistent with the retinal PrP-sen expression levels (Figure 2).
Figure 6.
Western blot of PrP-res from clinically sick transgenic mice. Shown are representative blots from tg7 (A), tgNSE (B), and tgGFAP (C) eye and brain tissue at the clinical time of infection after intraocular inoculation with hamster scrapie. Each 10% (w/v) homogenate was treated with proteinase K as described in the Materials and Methods, and PrP-res bands representing multiple glycosylation forms are indicated by approximate molecular weight markers (16 to 30 kd). M (mock), uninfected brain or eye homogenate. Lanes 1 to 3 represent protein from up to three individual animals. All blots were loaded with an equivalent amount of protein (5 mg/10 μl tissue equivalents). All exposure times were equivalent except for tgGFAP eye, which was 100-fold longer because of low levels of detectable PrP-res. This very long exposure also resulted in visualization of a band in the lane with mock-infected eye. It is unclear whether this is a non-PrP protein or actual PrP-res from the adjacent lane.
Because intraocular inoculation resulted in brain disease for all three strains of transgenic mice, we also evaluated the PrP-res from brains of clinically sick animals. Large amounts of PrP-res were detected from the brains of all three strains of mice and the glycoform patterns were typical of those seen after infection with strain 263K (Figure 6). The tgGFAP mice accumulated even more brain PrP-res compared to tgNSE and tg7 mice (Figure 6C). This was surprising because PrP-sen expression in both brain and eye of tgGFAP mice was ∼10% of the level seen in hamsters and ∼3% of that seen in tg7 and tgNSE (Figure 2C).
Pathology and PrP-res Distribution in the Brains of Clinically Ill Transgenic Mice
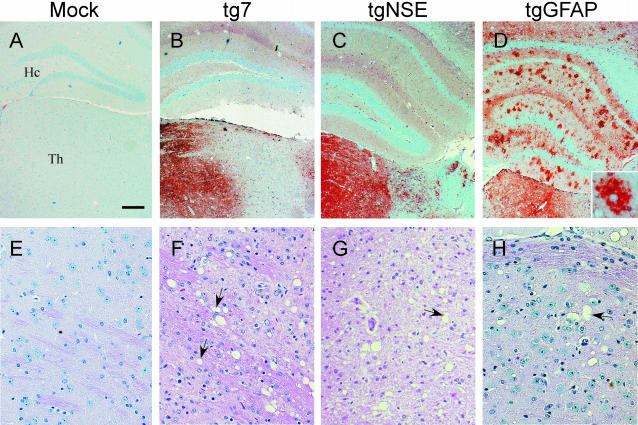
Because there was a large accumulation of PrP-res in the brains of tgGFAP mice after intraocular inoculation, we investigated the distribution of PrP-res in brain tissue of the transgenic mice at the clinical time of disease (Figure 7). The regional distribution of PrP-res in the brains of both tg7 (Figure 7B) and tgNSE (Figure 7C) mice infected with hamster scrapie intraocularly was similar to the distribution seen previously after intracranial inoculation of tgNSE mice.20 In the tg7 mice, there was abundant diffuse PrP-res found in the thalamus (Figure 7B). We also observed diffuse staining in the superior colliculus and olfactory bulb, and there was some focal, plaque-like staining in the cerebral cortex (not shown). There was only rarely detectable PrP-res staining in the basal ganglia, hippocampus, or cerebellar cortex. In tgNSE mice, the majority of PrP-res immunostaining in the thalamus had a diffuse staining pattern similar to tg7 mice (Figure 7C). However, in the hypothalamic region the PrP-res was located in or adjacent to neuronal perikarya and axons and was characterized by intense focal, punctate staining. In tgGFAP mice there was also abundant PrP-res staining in thalamus, but these mice differed from tg7 and tgNSE mice because they had unusual focal, spherical, or ovoid PrP-res staining in the hippocampus (Figure 7D). There was also abundant, more diffuse staining in the cerebral cortex (Figure 7D, top), as well as intense staining in the cerebellar granule cell layer (data not shown).
Figure 7.
Immunohistochemical analysis of PrP-res and spongiosis in hamster PrP transgenic mouse brains. Shown are representative sections of brains from clinical time points of disease after intraocular infection with hamster scrapie of aged-matched mock-infected tg7 mice (A and E) and scrapie-infected tg7 (B and F), tgNSE (C and G), and tgGFAP (D and H) mice. A–D: Thalamus (Th) and hippocampal (Hc) region immunohistochemically stained with monoclonal antibody 3F4, showing diffuse and punctate staining for PrP-res. D, inset: The hippocampal region of the tgGFAP mice. E–H: Representative sections of thalamus stained with H&E to show characteristic spongy lesions (arrows) at the time of clinical disease. Scale bar, 20 μm. Original magnifications: ×4 (A–D); ×20 (E–H).
In contrast to the observations in the retinas, in the brain the accumulations of PrP-res correlated with the appearance of spongiosis in all three lines of transgenic mice. The most obvious spongiosis was observed in the thalamic region of the brain (Figure 7; F, G, and H) where the most intense diffuse staining for PrP-res was observed for mice at the clinical stage of disease. However, there was neurodegeneration with spongiosis and astrocytosis in all brain areas with detectable PrP-res. Brain tissue from mock-infected animals did not have any detectable immunostaining or show any evidence of spongiosis (Figure 7, A and E).
Similar to what has been observed previously after intraocular inoculations of scrapie,15,29 the appearance of PrP-res in both tg7 and tgNSE mice appeared sequentially in ascending visual target regions of the brain throughout the course of infection; firstly in the retina, next in the contralateral lateral geniculate nucleus of the thalamus, then in the superior colliculus, and finally in the visual projection of the cerebral cortex (data not shown). In contrast, the accumulation of PrP-res in the tgGFAP mice did not follow a clear progression through the visual system throughout the course of disease (Table 2). PrP-res was first observed in both the retina and lateral geniculate nucleus at the midpoint of infection (140 days after infection), however, subsequently PrP-res did not appear in the superior colliculus, but instead was detected in multiple brain regions outside the visual system including the cortex, hippocampus, and granule cell layer of the cerebellum at 200 days after infection. This widespread distribution was maintained through the final disease progression to the clinical stage at 355 days after infection. Therefore, in tgGFAP mice progression of PrP-res detection did not appear to follow the neurons of the visual system as it did in tg7 and tgNSE mice. Instead, PrP-res accumulations appeared to diverge into various adjacent regions after initial establishment in the retina and lateral geniculate nucleus.
Table 2.
PrP-res Distribution in the Brain of tgGFAP Mice
| Days after infection | 84 | 112 | 140 | 200 | 252 | 355 |
|---|---|---|---|---|---|---|
| Visual projection regions | ||||||
| Retina | − | − | +* | + | ++ | +++ |
| Lateral geniculate nucleus | − | − | + | + | + | ++ |
| Superior colliculus | − | − | − | − | − | − |
| Visual cortex (occipital) | − | − | − | − | − | + |
| Other brain regions | ||||||
| Neocortex (frontal/parietal) | − | − | − | + | ++ | +++ |
| Hippocampus | − | − | − | + | ++ | +++ |
| Cerebellum (deep cerebellar nuclei) | − | − | − | − | − | − |
| Cerebellum (granule cell layer) | − | − | − | + | ++ | +++ |
Indicates the intensity of PrP-res staining in that region.
Discussion
In the present experiments intraocular inoculation of hamster scrapie was used to infect three lines of transgenic mice expressing PrP in differing cell types. Although all three mouse lines succumbed to scrapie disease and showed scrapie-induced spongiosis and neurodegeneration in brain, there were marked differences among these mice in the extent of retinal damage observed. Tg7 mice had evidence of vitritis, retinal vasculitis, apoptosis, and atrophy, whereas tgNSE and tgGFAP mice showed minimal signs of retinal apoptosis and no retinal atrophy (Figures 3, 4, and 5). In contrast to tgGFAP mice, tg7 mice and tgNSE expressed the transgene and protein at similar high levels (Figure 2). Both had high levels of PrP-res accumulation in retina, but differed in retinal degeneration (Figure 3 and 6). One possible explanation for the differences in retinal damage might be that slightly higher ocular PrP-res levels in the tg7 mice compared to the tgNSE mice (Figure 6) are sufficient to cause extensive retinal degeneration. However, individual tgNSE mice appeared to overlap with tg7 mice in their PrP-res levels (Figure 6), so quantitative differences in PrP-res accumulation are not likely to explain the consistent differences seen in retinal degeneration in these two transgenic lines. An alternative possibility is that PrP-res accumulation in different retinal cell types could contribute to degeneration in an additive or complementary manner. Perhaps in the tgNSE mice, PrP-res generation exclusively in neurons is not sufficient to induce the severe retinal pathology, whereas in tg7 mice PrP-res generation in neurons plus Müller cells, microglia, and/or other glial cells might induce more severe damage by triggering multiple pathogenic pathways.
Retinal Müller cells are likely to be involved in the retinal pathology observed in this system. Numerous GFAP-positive Müller cells were detected at 42 days after infection in both the tg7 and the tgNSE mice; however, these cells were not prominent at 90 to 100 days after infection, ie, at the time of clinical scrapie (Figure 4). In tg7 mice PrP-res may be generated directly by GFAP-positive Müller cells, and injury to these cells could contribute to the pathogenesis by interrupting one of their normal neuron support roles, such as maintaining normal glutamate levels.30 It is also possible that PrP-res-positive Müller cells could release neurotoxic products such as chemokines, cytokines, or oxidative molecules.31,32 In the tgNSE mice Müller cells appeared to be activated transiently in response to the neuronal injury caused by the neuronal PrP-res generation, but this process did not proceed to neuronal death. We cannot exclude the possibility that the observation of transient GFAP-positive staining was because of death of the Müller cell by the clinical stage of disease, however this is unlikely because the TUNEL staining of tgNSE retinas was minimal throughout the course of disease (Figure 5 and data not shown). An alternative possibility could be that GFAP-positive staining disappeared in the tgNSE mice because the neuronal injury incurred was itself minor and transient.
Microglia might also play a role in the retinal degeneration seen in tg7 mice. In typical scrapie brain pathogenesis microglial activation is a prominent feature33 and in recent experiments ocular inoculation using scrapie-infected cell homogenates resulted in rapid recruitment of microglia into the retina.34 In these same experiments neurons and astrocytes exposed to scrapie in vitro were able to secrete soluble factors capable of attracting microglia. It is possible microglia can participate both as a primary cause of injury and as a component of the host response to the injury.35 In tg7 mice microglia could express PrP-sen, so they would also generate PrP-res, which could contribute to the retinal damage. Whether or not activated microglia are simply a consequence of the neurodegeneration or if they contribute to disease progression in chronic neurodegenerative diseases is not known,36 and further experiments will be required to determine the possible contributions of both microglia and Müller cells to the retinal degeneration seen in tg7 mice.
In all three transgenic mouse lines studied by intraocular inoculation in the present experiments generation of PrP-res was detected in the brain and in the contralateral retina (data not shown). These data were in agreement with previous results showing that scrapie infectivity spreads from the eye to the brain via the optic nerve and follows the neuroanatomy of the visual system in the brain14 and that spread also proceeds down the optic nerve from the brain to the eye.37 Similarly, in the present study PrP-res could be detected sequentially in the visual system in retina, lateral geniculate nucleus, and finally in the superior colliculus of tg7 and tgNSE mice. In contrast, in tgGFAP mice the pathway of spread of PrP-res from eye to brain was different. PrP-res was first detected in retina at 140 days after infection and appeared simultaneously in the lateral geniculate nucleus (Table 2). At the next time point tested (200 days after infection) PrP-res was detected in several regions not associated with the visual system including cerebral cortex, hippocampus, hypothalamus, and cerebellum. Thus, our data suggest that in tgGFAP mice PrP-res did not spread via the neurons of the visual system, but more likely spread by another mechanism, possibly by sequential conversion in adjacent glial cells.38,39
In the present experiments all three transgenic mouse lines developed typical scrapie central nervous system signs of disease including ataxia, kyphosis, weakness, tremor, paralysis, and somnolence. Brain pathology and neurodegeneration was also typical of scrapie with PrP-res deposition, astrogliosis, and vacuolation, even in the two mouse lines (tgGFAP and tgNSE) that had no obvious evidence of retinal degeneration. The time to death was much shorter in tg7 and tgNSE mice (100 days) compared to tgGFAP mice (335 days), and these differences appeared to correlate with the levels of hamster PrP transgene expression in brain in these mouse lines. In contrast to what was seen in the retina, the amount of PrP-res deposition in brain was far higher in the tgGFAP mice than in tg7 and tgNSE (Figure 6). Our previous electron microscopic studies of the tgGFAP mice indicated that PrP-res was deposited exclusively around astrocytes,40 and in these earlier studies there was evidence of severe damage to adjacent neuronal processes, suggesting that the PrP-res generated by astrocytes had an indirect pathological effect on PrP-negative neurons. Similar conclusions were drawn from in vitro studies using cells from tgGFAP mice after stimulation with a toxic PrP peptide.41 Possibly the indirect neurotoxic effects of astrocytic PrP-res are less toxic than the direct effects of neuronal PrP-res, thus requiring accumulation of astrocytic PrP-res to higher levels to achieve a significant clinical effect. This would explain the longer incubation times to clinical disease and the higher PrP-res levels in brain of tgGFAP mice at the time of sacrifice. An alternative hypothesis is that the early sites of PrP-res accumulation in tgGFAP mice are less important clinically than those involved early in the tg7 and tgNSE mice. If so, the tgGFAP mice would survive longer until the more clinically important regions were damaged, and during this time higher levels of PrP-res would accumulate as we observed. We consider this latter hypothesis less likely because in all three transgenic lines the earliest site of PrP-res appears to be the thalamus; and in tgNSE mice this thalamic accumulation appears to be sufficient to cause clinical signs leading to death. Whereas in tgGFAP mice thalamic PrP-res is found midway through the disease course, but higher levels in thalamus and elsewhere appear to be required for clinical disease and death.
Acknowledgments
We thank S. Priola, G. Baron, J. Portis, and S. Best for critical reading of the manuscript and helpful suggestions; and K. Meade-White and A. Raines for excellent technical assistance.
Footnotes
Address reprint requests to Dr. Bruce Chesebro, Laboratory of Persistent Viral Diseases, Rocky Mountain Laboratories, National Institutes of Allergy and Infectious Diseases, 903 S. 4th St., Hamilton, MT 59840. E-mail: bchesebro@niaid.nih.gov.
References
- Chesebro B, Race R, Wehrly K, Nishio J, Bloom M, Lechner D, Bergstrom S, Robbins K, Mayer L, Keith JM. Identification of scrapie prion protein-specific mRNA in scrapie-infected and uninfected brain. Nature. 1985;315:331–333. doi: 10.1038/315331a0. [DOI] [PubMed] [Google Scholar]
- Oesch B, Westaway D, Walchli M, McKinley MP, Kent SB, Aebersold R, Barry RA, Tempst P, Teplow DB, Hood LE. A cellular gene encodes scrapie PrP 27-30 protein. Cell. 1985;40:735–746. doi: 10.1016/0092-8674(85)90333-2. [DOI] [PubMed] [Google Scholar]
- Basler K, Oesch B, Scott M, Westaway D, Walchli M, Groth DF, McKinley MP, Prusiner SB, Weissmann C. Scrapie and cellular PrP isoforms are encoded by the same chromosomal gene. Cell. 1986;46:417–428. doi: 10.1016/0092-8674(86)90662-8. [DOI] [PubMed] [Google Scholar]
- Bueler H, Aguzzi A, Sailer A, Greiner RA, Autenried P, Aguet M, Weissmann C. Mice devoid of PrP are resistant to scrapie. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- Brandner S, Isenmann S, Raeber A, Fischer M, Sailer A, Kobayashi Y, Marino S, Weissmann C, Aguzzi A. Normal host prion protein necessary for scrapie-induced neurotoxicity. Nature. 1996;379:339–343. doi: 10.1038/379339a0. [DOI] [PubMed] [Google Scholar]
- Kretzschmar HA, Prusiner SB, Stowring LE, DeArmond SJ. Scrapie prion proteins are synthesized in neurons. Am J Pathol. 1986;122:1–5. [PMC free article] [PubMed] [Google Scholar]
- Moser M, Colello RJ, Pott U, Oesch B. Developmental expression of the prion protein gene in glial cells. Neuron. 1995;14:509–517. doi: 10.1016/0896-6273(95)90307-0. [DOI] [PubMed] [Google Scholar]
- van Keulen LJ, Schreuder BE, Meloen RH, Poelen-van den Berg M, Mooij-Harkes G, Vromans ME, Langeveld JP. Immunohistochemical detection and localization of prion protein in brain tissue of sheep with natural scrapie. Vet Pathol. 1995;32:299–308. doi: 10.1177/030098589503200312. [DOI] [PubMed] [Google Scholar]
- Cashman NR, Loertscher R, Nalbantoglu J, Shaw I, Kascsak RJ, Bolton DC, Bendheim PE. Cellular isoform of the scrapie agent protein participates in lymphocyte activation. Cell. 1990;61:185–192. doi: 10.1016/0092-8674(90)90225-4. [DOI] [PubMed] [Google Scholar]
- McBride PA, Eikelenboom P, Kraal G, Fraser H, Bruce ME. PrP protein is associated with follicular dendritic cells of spleens and lymph nodes in uninfected and scrapie-infected mice. J Pathol. 1992;168:413–418. doi: 10.1002/path.1711680412. [DOI] [PubMed] [Google Scholar]
- Caughey B, Race RE, Chesebro B. Detection of prion protein mRNA in normal and scrapie-infected tissues and cell lines. J Gen Virol. 1988;69:711–716. doi: 10.1099/0022-1317-69-3-711. [DOI] [PubMed] [Google Scholar]
- Fraser H. The pathology of a natural and experimental scrapie. Front Biol. 1976;44:267–305. [PubMed] [Google Scholar]
- Barnett KC, Palmer AC. Retinopathy in sheep affected with natural scrapie. Res Vet Sci. 1971;12:383–385. [PubMed] [Google Scholar]
- Fraser H, Dickinson AG. Targeting of scrapie lesions and spread of agent via the retino-tectal projection. Brain Res. 1985;346:32–41. doi: 10.1016/0006-8993(85)91091-1. [DOI] [PubMed] [Google Scholar]
- Scott JR, Fraser H. Transport and targeting of scrapie infectivity and pathology in the optic nerve projections following intraocular infection. Prog Clin Biol Res. 1989;317:645–652. [PubMed] [Google Scholar]
- Foster JD, Fraser H, Bruce ME. Retinopathy in mice with experimental scrapie. Neuropathol Appl Neurobiol. 1986;12:185–196. doi: 10.1111/j.1365-2990.1986.tb00049.x. [DOI] [PubMed] [Google Scholar]
- Hogan RN, Baringer JR, Prusiner SB. Progressive retinal degeneration in scrapie-infected hamsters: a light and electron microscopic analysis. Lab Invest. 1981;44:34–42. [PubMed] [Google Scholar]
- Buyukmihci N, Goehring-Harmon F, Marsh RF. Retinal degeneration during clinical scrapie encephalopathy in hamsters. J Comp Neurol. 1982;205:153–160. doi: 10.1002/cne.902050206. [DOI] [PubMed] [Google Scholar]
- Hogan RN, Kingsbury DT, Baringer JR, Prusiner SB. Retinal degeneration in experimental Creutzfeldt-Jakob disease. Lab Invest. 1983;49:708–715. [PubMed] [Google Scholar]
- Race RE, Priola SA, Bessen RA, Ernst D, Dockter J, Rall GF, Mucke L, Chesebro B, Oldstone MB. Neuron-specific expression of a hamster prion protein minigene in transgenic mice induces susceptibility to hamster scrapie agent. Neuron. 1995;15:1183–1191. doi: 10.1016/0896-6273(95)90105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeber AJ, Race RE, Brandner S, Priola SA, Sailer A, Bessen RA, Mucke L, Manson J, Aguzzi A, Oldstone MB, Weissmann C, Chesebro B. Astrocyte-specific expression of hamster prion protein (PrP) renders PrP knockout mice susceptible to hamster scrapie. EMBO J. 1997;16:6057–6065. doi: 10.1093/emboj/16.20.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race R, Oldstone M, Chesebro B. Entry versus blockade of brain infection following oral or intraperitoneal scrapie administration: role of prion protein expression in peripheral nerves and spleen. J Virol. 2000;74:828–833. doi: 10.1128/jvi.74.2.828-833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamoto T, Tateishi J, Tashima T, Takeshita I, Barry RA, DeArmond SJ, Prusiner SB. Amyloid plaques in Creutzfeldt-Jakob disease stain with prion protein antibodies. Ann Neurol. 1986;20:204–208. doi: 10.1002/ana.410200205. [DOI] [PubMed] [Google Scholar]
- Hardt M, Baron T, Groschup MH. A comparative study of immunohistochemical methods for detecting abnormal prion protein with monoclonal and polyclonal antibodies. J Comp Pathol. 2000;122:43–53. doi: 10.1053/jcpa.1999.0343. [DOI] [PubMed] [Google Scholar]
- Kascsak RJ, Rubenstein R, Merz PA, Tonna-DeMasi M, Fersko R, Carp RI, Wisniewski HM, Diringer H. Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J Virol. 1987;61:3688–3693. doi: 10.1128/jvi.61.12.3688-3693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimcheff DE, Askovic S, Baker AH, Johnson-Fowler C, Portis JL. Endoplasmic reticulum stress is a determinant of retrovirus-induced spongiform neurodegeneration. J Virol. 2003;77:12617–12629. doi: 10.1128/JVI.77.23.12617-12629.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Race R, Ernst D, Jenny A, Taylor W, Sutton D, Caughey B. Diagnostic implications of detection of proteinase K-resistant protein in spleen, lymph nodes, and brain of sheep. Am J Vet Res. 1992;53:883–889. [PubMed] [Google Scholar]
- Sarthy PV, Fu M, Huang J. Developmental expression of the glial fibrillary acidic protein (GFAP) gene in the mouse retina. Cell Mol Neurobiol. 1991;11:623–637. doi: 10.1007/BF00741450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser H. Neuronal spread of scrapie agent and targeting of lesions within the retino-tectal pathway. Nature. 1982;295:149–150. doi: 10.1038/295149a0. [DOI] [PubMed] [Google Scholar]
- Garcia M, Vecino E. Role of Muller glia in neuroprotection and regeneration in the retina. Histol Histopathol. 2003;18:1205–1218. doi: 10.14670/HH-18.1205. [DOI] [PubMed] [Google Scholar]
- Peterson KE, Errett JS, Wei T, Dimcheff DE, Ransohoff R, Kuziel WA, Evans L, Chesebro B. MCP-1 and CCR2 contribute to non-lymphocyte-mediated brain disease induced by Fr98 polytropic retrovirus infection in mice; role for astrocytes in retroviral pathogenesis. J Virol. 2004;78:6449–6458. doi: 10.1128/JVI.78.12.6449-6458.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagar A, Shapshak P, Fujimura R, Ownby R, Heyes M, Eisdorfer C. The role of macrophage/microglia and astrocytes in the pathogenesis of three neurologic disorders: HIV-associated dementia, Alzheimer disease, and multiple sclerosis. J Neurol Sci. 2002;202:13–23. doi: 10.1016/s0022-510x(02)00207-1. [DOI] [PubMed] [Google Scholar]
- Rezaie P, Lantos PL. Microglia and the pathogenesis of spongiform encephalopathies. Brain Res Brain Res Rev. 2001;35:55–72. doi: 10.1016/s0165-0173(01)00042-x. [DOI] [PubMed] [Google Scholar]
- Marella M, Chabry J. Neurons and astrocytes respond to prion infection by inducing microglia recruitment. J Neurosci. 2004;24:620–627. doi: 10.1523/JNEUROSCI.4303-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DR. Microglia and prion disease. Microsc Res Tech. 2001;54:71–80. doi: 10.1002/jemt.1122. [DOI] [PubMed] [Google Scholar]
- Perry VH, Newman TA, Cunningham C. The impact of systemic infection on the progression of neurodegenerative disease. Nat Rev Neurosci. 2003;4:103–112. doi: 10.1038/nrn1032. [DOI] [PubMed] [Google Scholar]
- Hogan RN, Bowman KA, Baringer JR, Prusiner SB. Replication of scrapie prions in hamster eyes precedes retinal degeneration. Ophthalmic Res. 1986;18:230–235. doi: 10.1159/000265439. [DOI] [PubMed] [Google Scholar]
- Baron GS, Wehrly K, Dorward DW, Chesebro B, Caughey B. Conversion of raft associated prion protein to the protease-resistant state requires insertion of PrP-res (PrP(Sc)) into contiguous membranes. EMBO J. 2002;21:1031–1040. doi: 10.1093/emboj/21.5.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diedrich JF, Bendheim PE, Kim YS, Carp RI, Haase AT. Scrapie-associated prion protein accumulates in astrocytes during scrapie infection. Proc Natl Acad Sci USA. 1991;88:375–379. doi: 10.1073/pnas.88.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey M, Goodsir CM, Race RE, Chesebro B. Scrapie-specific ultrastructural neuronal lesions occur in transgenic mice expressing prion protein only in astrocytes. Ann Neurol. 2004;55:781–792. doi: 10.1002/ana.20093. [DOI] [PubMed] [Google Scholar]
- Brown DR. Prion protein peptide neurotoxicity can be mediated by astrocytes. J Neurochem. 1999;73:1105–1113. doi: 10.1046/j.1471-4159.1999.0731105.x. [DOI] [PubMed] [Google Scholar]