Abstract
An environmental isolate of Serratia fonticola resistant to carbapenems was shown to contain a genetic determinant encoding a metallo-β-lactamase of the subclass B2. The Sfh-I enzyme exhibits some divergence from the previously characterized enzymes of this subclass. This is the first example of a naturally occurring metallo-β-lactamase in Enterobacteriaceae.
Metallo-β-lactamases (MBLs) are enzymes that show carbapenemase activity (3) and require a zinc cofactor (16). The environmental microbiota is presumed to constitute an important reservoir of these harmful genetic determinants. A survey study concerning the bacterial populations of untreated drinking waters in the region of Trás-os-Montes (northeastern Portugal) revealed the presence of bacterial isolates resistant to carbapenems. One isolate had a high level of resistance to imipenem, with a MIC of 32 μg/ml. For identification at the species level, the 16S rRNA gene was amplified with universal primers (Table 1) and sequenced. The sequence clearly assigns the strain to the species Serratia fonticola. This result agrees with the identification obtained with the API-32GN System (BioMérieux, Marcy l'Etoile, France). The strain was designated S. fonticola UTAD54. Determination of the MICs of antibiotics with E-test gradient strips (AB Biodisk, Solna, Sweden) indicated resistance to penicillins, cephalothin, and cefuroxime, antibiotics to which other strains of the same species are susceptible (Table 2).
TABLE 1.
Oligonucleotide primers used in this study
| PCR target and primer | Sequence 5′-3′ | Reference |
|---|---|---|
| 16S rRNA gene | ||
| rD1 | AAG GAG GTG ATC CAG CC | 17 |
| fD1 | AGA GTT TGA TCC TGG CTC AG | 17 |
| BSF517/17 | GCC AGC AGC CGC GGT AA | 18 |
| Class I integrons | ||
| Int 1F | GGC ATC CAA GCA GCA AG | 7 |
| Int 1B | AAG CAG ACT TGA CCT GA | 7 |
| fonA-like genes | ||
| FON_f | GAT CGA TAC CGC CGA TAA TTC GC | This study |
| FON_r | ACG GCG ATA TCG TTA GTG GTA CC | This study |
| Sfh-I-encoding gene | ||
| SFH-I_f | GAT CTC GAG ATG GCT TCT GAA AAA AAC TTA ACG | This study |
| SFH-I_r | GAT GAA TTC TTA CTT AGG CGC CTT CTC AAG CAG | This study |
TABLE 2.
MICs of antibiotics for S. fonticola LMG7887 (type strain), S. fonticola CIP103580, S. fonticola UTAD54, E. coli XL2 Blue(pSF14), and E. coli XL2 Blue (reference strain)
| Antibiotic(s)a | MIC (μg/ml)
|
||||
|---|---|---|---|---|---|
| S. fonticola LMG7882T | S. fonticola CIP103580 | S. fonticola UTAD54 | E. coli XL2 Blue(pSF14) | E. coli XL2 Blue | |
| Amoxicillin | 12 | 64 | >256 | 4 | 4 |
| Amoxicillin-CLA | 0.75 | 1.5 | 12 | 3 | 3 |
| Piperacillin | 1.0 | 2.0 | 32 | 0.75 | 0.75 |
| Piperacillin-TZB | 0.19 | 1.0 | 0.38 | 0.5 | 0.5 |
| Ticarcillin-CLA | 1.9 | 1.5 | 1.5 | 1.5 | 0.75 |
| Mecillinam | NDb | ND | >256 | 0.75 | 0.75 |
| Cephalothin | 24 | ND | >256 | 24 | 6 |
| Cefoxitin | 6 | 6 | 6 | 4 | 4 |
| Cefuroxime | 16 | 48 | >256 | 2 | 2 |
| Cefotaxime | 0.125 | 0.50 | 2 | 0.064 | 0.064 |
| Ceftazidime | 0.023 | 0.125 | 0.064 | 0.19 | 0.19 |
| Cefepime | 0.016 | 0.023 | 0.032 | 0.032 | 0.032 |
| Cefpirome | 0.016 | ND | 0.032 | 0.016 | 0.016 |
| Imipenem | 0.25 | 0.38 | >32 | >32 | 0.125 |
| Meropenem | 0.016 | 0.047 | >32 | >32 | 0.008 |
| Aztreonam | 0.094 | 0.25 | 1.5 | 0.19 | 0.19 |
CLA, clavulanic acid; TZB, tazobactam.
ND, not done.
A shotgun cloning approach was used to assess the genetic determinant of imipenem resistance. For that purpose, the total DNA of S. fonticola UTAD54 was partially digested with Sau3AI and the resulting fragments were ligated to the Escherichia coli vector pBGS19+ (13) linearized with BamHI. After transformation on E. coli XL2 Blue (Stratagene, La Jolla, Calif.), the resulting genomic library was amplified by cultivation in the presence of kanamycin and stored as several aliquots. Small volumes of the genomic library were plated on LA medium supplemented with imipenem at a concentration of 3 μg/ml. One clone containing a recombinant plasmid carrying an insert of 5 kb was chosen for further characterization. The recombinant plasmid was named pSF14. The susceptibility of the recombinant clone to several antibiotics was tested (Table 2). In contrast to untransformed E. coli, the recombinant strain showed resistance to carbapenems. E. coli XL2 Blue(pSF14) exhibited decreased in vitro susceptibility to several β-lactams in comparison with XL2 Blue. The relative decrease in susceptibility was greater with penicillins and carbapenems than with cephalosporins. Susceptibility to aztreonam was not affected. S. fonticola UTAD54 exhibited low overall susceptibility to β-lactams (Table 2).
With a primer-walking strategy, the complete nucleotide sequence of both strands of the insert contained in pSF14 was determined. Analysis of the nucleotide sequence revealed the presence of an open reading frame contained in a short region of approximately 1 kb, with a moles percent G+C content of 42.5%. This value is outside the range of the moles percent G+C content attributed to the genome of S. fonticola (48.8 to 52.5%) (6). The open reading frame encodes a polypeptide of 253 amino acids. A BLAST (1) search showed that the deduced amino acid sequence displays similarity to β-lactamases belonging to molecular class B (2). The highest similarity score was obtained with the CphA enzyme of A. hydrophila (8). This new MBL was named Sfh-I, and its genetic determinant was named blaSfh-I.
The deduced amino-terminal sequence of the protein exhibits features typical of prokaryotic signal peptides targeting protein secretion into the periplasmic space via the general secretory pathway. According to known patterns (10), the most likely cleavage site is located between the amino acids at positions 21 and 22, which are alanine and serine, respectively. The theoretical molecular masses of the complete and mature forms of the Sfh-I protein are 28.50 and 26.13 kDa, respectively.
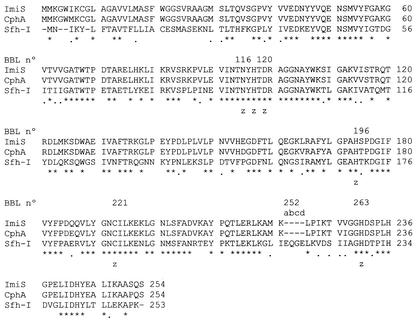
As shown in Fig. 1, the amino acid sequence of Sfh-I could be aligned with the sequences of the subclass B2 enzymes without introducing major gaps. The exception is a short insertion sequence of four amino acids (Glu-Gln-Gly-Glu) located at position 252 according to the BBL numbering scheme (5). The Sfh-I sequence retains four invariant residues shared by the other class B β-lactamases (His-118, Asp-120, His-196, and His-263 [BBL numbering]). The motif His-X-His-X-Asp (positions 116 to 120 [BBL consensus]), present in the MBLs belonging to subclasses B1 and B3, is changed to the related sequence Asn-Tyr-His-Thr-Asp, a distinctive feature of subclass 2 MBLs. This motif is presumed to be involved in the coordination of the two zinc ions found in the active site of these enzymes. The amino acids Asp-120, Cys-221, and His-263 (consensus BBL), which are presumably involved in the binding of the second zinc ion in the enzymes of subclass B2, are conserved in Sfh-I.
FIG. 1.
Alignment of the amino acid sequences of MBLs included in subclass B2. Residues conserved in the three enzymes are indicated by asterisks; dots indicate functionally homologous residues. The BBL numbering scheme is used for residues conserved in MBLs. Amino acids presumptively involved in binding to zinc are indicated by the letter z.
Pairwise alignment (14) of the sequence of Sfh-I with those of other MBLs confirmed the closest relationship of the new enzyme to the subclass B2 enzymes. In fact, Sfh-I displays 53.8% identity of residues with CphA from A. hydrophila and 52.2% identity with ImiS from Aeromonas veronii (Table 3). Identity to subclass 1 enzymes is between 17.9% (IMP-1) and 23.4% (IND-1). Comparison to the newly characterized MBL SPM-1 from Pseudomonas aeruginosa (15) revealed an identity of 15.4%. Lower identity percentages are found when comparisons are made with subclass B3 enzymes: the range is 11.5% (THIN-B) to 14.6% (CAU-1). These results are consistent with the classification of Sfh-I as a member of subclass B2.
TABLE 3.
Pairwise percent amino acid identities between Sfh-I and other class B β-lactamases
| β-Lactamase (accession no.) | % Identity between amino acid sequencesa
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IND-1b | BlaB | CcrA | Bc-II | VIM-1 | IMP-1 | SPM-1 | ImiS | CphA | Sfh-I | L1 | CAU-1 | |
| BlaB (X96858) | 43.1 | |||||||||||
| CcrA (M63556) | 28.5 | 25.3 | ||||||||||
| Bc-II (M11189) | 33.5 | 32.9 | 29.7 | |||||||||
| VIM-1 (Y18050) | 25.5 | 23.3 | 27.3 | 33.1 | ||||||||
| IMP-1 (S71932) | 28.0 | 26.9 | 28.9 | 27.2 | 22.6 | |||||||
| SPM-1 (CAD37801) | 25.1 | 23.7 | 18.5 | 22.2 | 20.3 | 28.5 | ||||||
| ImiS (Y10415) | 21.8 | 21.3 | 18.9 | 21.0 | 20.3 | 18.3 | 22.8 | |||||
| CphA (X57102) | 22.2 | 21.3 | 20.5 | 21.8 | 20.7 | 18.7 | 23.6 | 96.5 | ||||
| Sfh-I | 23.4 | 20.5 | 19.3 | 18.3 | 16.9 | 17.9 | 15.4 | 52.0 | 53.5 | |||
| L1 (X75074) | 10.9 | 11.2 | 10.8 | 12.5 | 14.7 | 12.2 | 10.9 | 11.0 | 12.2 | 12.6 | ||
| CAU-1 (AJ308331) | 14.2 | 11.6 | 12.4 | 13.2 | 13.5 | 13.4 | 12.7 | 14.6 | 15.0 | 14.6 | 28.3 | |
| THIN-B (AJ250876) | 29.1 | 10.8 | 14.1 | 13.6 | 15.0 | 13.8 | 10.5 | 13.4 | 13.8 | 11.5 | 32.1 | 29.4 |
Identity values are expressed as percentages calculated with the complete amino acid sequences, including the signal peptides. The values were derived by dividing the number of perfect matches by the length of the shorter sequence, excluding gaps, by a multiple-alignment method.
The accession number of IND-1 is AF099139.
Specific oligonucleotide primers were designed that amplify the blaSfh-I gene of strain UTAD54 (Table 1). With DNA from S. fonticola strains LMG 7882T, DSM 9663, and CIP 103850, no amplification products were obtained. The amplicon from S. fonticola UTAD54 labeled with digoxigenin (Roche Molecular Biochemicals, Indianapolis, Ind.) was used to probe Southern blots of digested and undigested DNAs isolated from the same strains. This approach did not detect blaSfh-I-like sequences in strains LMG 7882T, DSM 9663, and CIP 103850 (data not shown). The probe clearly hybridizes to the band representing the undigested chromosomal DNA of UTAD54. With different extraction methods, no DNA of plasmid origin was detectable on ethidium bromide-stained agarose gels after electrophoresis, suggesting that blaSfh-I is a chromosomal gene. The presence of integrons was also checked with a primer set designed to anneal to the 5′ and 3′ conserved sequences of flanking regions containing integrated gene cassettes (Table 1). Amplifications by PCR did not produce DNA fragments (data not shown).
The high MICs of antibiotics to which the recombinant E. coli XL2 Blue(pSF14) is susceptible (e.g., amoxicillin and narrow- and expanded-spectrum cephalosporins) indicated the existence of additional β-lactam resistance mechanisms in UTAD54. The presence of class A β-lactamases in a clinical isolate of S. fonticola was previously reported (11); later, a gene encoding a β-lactamase 96% homologous to that one (SFO-1; GenBank accession number AB003148) was detected in Enterobacter cloacae (9). Also, six nucleotide sequences have been deposited in the GenBank database (accession numbers AJ251239 to -44) that represent six variants of the fonA gene for a class A β-lactamase present in different S. fonticola strains. The fonA1 variant is from the type strain. Specific primers based on the nucleotide sequences of those β-lactamases were designed (Table 1). Amplification by PCR resulted in a DNA fragment with the expected size (approximately 550 bp) in all of the S. fonticola strains studied, including UTAD54. The MBL-producing bacteria are known to possess other β-lactamase-encoding determinants, and that is also the case with S. fonticola UTAD54.
The environmental microbiota can constitute an important reservoir of genetic determinants of antibiotic resistance. Recently, MBLs were detected in bacteria that are not of clinical origin, as is the case with THIN-B from Janthinobacterium lividum (12) and CAU-1 from Caulobacter crescentus (4). In this work, we have identified a putative MBL-encoding gene in an environmental isolate that is a member of the family Enterobacteriaceae.
Nucleotide sequence accession numbers.
The nucleotide sequence of the 16S rRNA gene of S. fonticola UTAD54 was deposited in the GenBank database and assigned accession number AY236502. The blaSfh-I nucleotide sequence was deposited in the GenBank database and assigned accession number AF197943.
Acknowledgments
This work was financed by the Fundação para a Ciência e a Tecnologia through Ph.D. grants to Artur Alves (SFRH/BD/10389/2002) and Isabel Henriques (SFRH/BD/5275/2001).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ambler, R. P. 1980. The structure of β-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci. 289:321-331. [DOI] [PubMed] [Google Scholar]
- 3.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Docquier, J.-D., F. Pantanella, F. Giuliani, M. C. Thaller, G. Amicosante, M. Galleni, J. M. Frère, K. Bush, and G. M. Rossolini. 2002. CAU-1, a subclass B3 metallo-β-lactamase of low substrate affinity encoded by an ortholog present in the Caulobacter crescentus chromosome. Antimicrob. Agents Chemother. 46:1823-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galleni, M., J. Lamotte-Brasseur, G. M. Rossolini, J. Spencer, O. Dideberg, J. M. Frère, and The Metallo-β-lactamase Working Group. 2001. Standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 45:660-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimont, P. A. D., and F. Grimont. 1984. Genus Serratia, p. 477-484. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. The Williams & Wilkins Co., Baltimore, Md.
- 7.Lévesque, C., L. Pyché, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Massidda, O., G. M. Rossolini, and G. Satta. 1991. The Aeromonas hydrophila cphA gene: molecular heterogeneity among metallo-β-lactamases. J. Bacteriol. 173:4611-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsumoto, Y., and M. Inoue. 1999. Characterization of SFO-1, a plasmid-mediated inducible class A β-lactamase from Enterobacter cloacae. Antimicrob. Agents Chemother. 43:307-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nielsen, H., J. Engelbrecht, S. Brunak, and G. V. Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 11.Péduzzi, J., S. Farzaneh, A. Reynaud, M. Barthelemy, and R. Labia. 1997. Characterization and amino acid sequence analysis of a new oxyimino cephalosporin-hydrolyzing class A beta-lactamase from Serratia fonticola CUV. Biochim. Biophys. Acta 1341:58-70. [DOI] [PubMed] [Google Scholar]
- 12.Rossolini, G. M., M. A. Condemi, F. Pantanella, J. D. Docquier, G. Amicosante, and M. C. Thaller. 2001. Metallo-β-lactamase producers in environmental microbiota: new molecular class B enzyme in Janthinobacterium lividum. Antimicrob. Agents Chemother. 45:837-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spratt, B. G., P. J. Hedge, S. Teheesen, A. Edelman, and J. K. Broome-Smith. 1986. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene 41:337-342. [DOI] [PubMed] [Google Scholar]
- 14.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Toleman, M. A., A. M. Simm, T. A. Murphy, A. C. Gales, D. J. Biedenbach, R. N. Jones, T. R. Walsh. 2002. Molecular characterization of SPM-1, a novel metallo-beta-lactamase isolated in Latin America: report from the SENTRY antimicrobial surveillance programme. J. Antimicrob. Chemother. 50:673-679. [DOI] [PubMed] [Google Scholar]
- 16.Wang, Z. W., W. Fast, A. M. Valentine, and S. J. Benkovic. 1999. Metallo-β-lactamases: structure and mechanism. Curr. Opin. Chem. Biol. 3:614-622. [DOI] [PubMed] [Google Scholar]
- 17.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and J. R. Lupski. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilmotte, A., G. Van der Auwera, and R. De Wachter. 1993. Structure of the 16S ribosomal RNA of the thermophilic cyanobacterium Chlorogloeopsis HTF (“Mastigocladus laminosus HTF”) strain PCC7518, and phylogenetic analysis. FEBS Lett. 317:96-100. [DOI] [PubMed] [Google Scholar]