Abstract
We describe an allele-specific PCR assay to detect mutations in three codons of the rpoB gene (516, 526, and 531) in Mycobacterium tuberculosis strains; mutations in these codons are reported to account for majority of M. tuberculosis clinical isolates resistant to rifampin (RIF), a marker of multidrug-resistant tuberculosis (MDR-TB). Three different allele-specific PCRs are carried out either directly with purified DNA (single-step multiplex allele-specific PCR), or with preamplified rpoB fragment (nested allele-specific PCR [NAS-PCR]). The method was optimized and validated following analysis of 36 strains with known rpoB sequence. A retrospective analysis of the 287 DNA preparations from epidemiologically unlinked RIF-resistant clinical strains from Russia, collected from 1996 to 2002, revealed that 247 (86.1%) of them harbored a mutation in one of the targeted rpoB codons. A prospective study of microscopy-positive consecutive sputum samples from new and chronic TB patients validated the method for direct analysis of DNA extracted from sputum smears. The potential of the NAS-PCR to control for false-negative results due to lack of amplification was proven especially useful in the study of these samples. The developed rpoB-PCR assay can be used in clinical laboratories to detect RIF-resistant and hence MDR M. tuberculosis in the regions with high burdens of the MDR-TB.
The spread of multidrug-resistant tuberculosis (MDR-TB) has increased worldwide and reached epidemic proportions in many countries. MDR Mycobacterium tuberculosis strains are considered those resistant to at least rifampin (RIF) and isoniazid. RIF is a key component of the World Health Organization DOTS (directly observed therapy, short course) regimen: because RIF monoresistance is extremely rare and development of isoniazid resistance usually precedes that to RIF, resistance to the latter is considered to be the MDR marker (8). It has been shown in many studies that RIF resistance in up to 95 to 98% of resistant strains is caused by mutations in the rpoB gene encoding the RNA polymerase β-subunit (20, 21). These mutations are generally described in the short 81-bp region in rpoB (RIF resistance-determining region: codons 507 to 533 [21]). In addition, RIF resistance may be caused by mutations in other parts of rpoB outside the hot spot region, such as codon 176 (146 [21]) in the N-terminal part (9) and codons 541 and 553 (18). Methods used so far to detect rpoB mutations associated with RIF-resistance were direct DNA sequencing, PCR-single-strand conformation polymorphism, heteroduplex mobility, dot spot, RNA-RNA mismatch, and some other assays (reviewed in references 4 and 25). Previously, we have described a multiplex allele-specific PCR (MAS-PCR) assay based on standard PCR and agarose gel electrophoresis to detect mutations in katG315 (15) and embB306 (14) in M. tuberculosis strains. A similar assay to detect rpsL43, rpoB531, and katG315 mutations was very recently published by Victor et al. (24). The MAS-PCR method in our design (14, 15) uses two outer primers that flank a region under study and invariably anneal on the conserved DNA targets, plus a wild-type-allele-specific inner primer that stops in its 3′ end at the targeted codon and amplifies a wild-type-allele specific fragment. An alteration of the base that corresponds to the 3′-end of the specific primer causes the primer-template mismatch that prevents polymerase to extend the primer and results in nonamplification of the indicative fragment.
Notably, rpoB mutations in three codons (516, 526, or 531) account for the majority of RIF-resistant strains (70 to 95%), especially in the areas with high incidence of MDR-TB (3, 20). Previously, it has been shown on small samples of strains from the St. Petersburg area of Russia (12, 13) and other Russian regions (7, 22) that mutations in these three codons in rpoB were on average found in 93.6% of RIF-resistant strains; that is, mutational analysis of the remaining rpoB hot spot codons only insignificantly contributed for prediction of RIF resistance in Russian isolates. In the present study we report a simple, rapid, and inexpensive assay based on the allele-specific PCR methodology targeting rpoB mutations to detect RIF-resistant M. tuberculosis. We evaluated this method with a collection of clinical strains from northwestern Russia, collected from 1996 to 2002, and with DNA samples from the microscopy-positive sputum smears.
MATERIALS AND METHODS
Description of study samples.
The subject of the study consisted of three sets of samples that corresponded to the specific objectives.
(i) Set one.
Thirty-six M. tuberculosis strains with known rpoB sequences served for optimization of the PCR assay conditions. These strains included H37Rv, 17 strains from St. Petersburg (12, 16) and 18 DNA samples kindly provided by J. D. A. van Embden, The National Institute of Public Health and the Environment (Bilthoven, The Netherlands), within the framework of the coordinated research project E1.50.15 (International Atomic Energy Agency, Vienna, Austria) and had the following distribution of the rpoB alleles: wild type, 12 strains; 531TTG, 7 strains; 531TGG, 2 strains; 516GTC, 4 strains; 516TAC, 3 strains; 516GGC, 1 strain; 522CAG, 1 strain; 526TAC, 2 strains; 526GAC, 3 strains; 526CTC, 1 strain.
(ii) Set two.
A total of 114 auramine or Ziehl-Neelsen-stained sputum smears served to evaluate the developed rpoB-PCR assay for rapid direct analysis of human samples. These sputum samples from epidemiologically unlinked patients (new and chronic TB) were selected from specimens consecutively analyzed in the bacteriology laboratory and identified as microscopy positive (1+, 2+, or 3+ cell count [27]). They were further subjected to simultaneous analysis by culture-based susceptibility testing and the rpoB-PCR assay, the nested version (see below).
(iii) Set three.
The 287 M. tuberculosis isolates recovered from 287 adult patients with newly and previously diagnosed pulmonary TB served to validate the developed rpoB-PCR assay in our setting (northwestern Russia). These patients were admitted to the hospitals of St. Petersburg Research Institute of Phthisiopulmonology and City Anti-Tuberculosis Dispensary of St. Petersburg (1996 to 2002) and proven unlinked by standard epidemiological investigation. For each patient, only the first available isolate was included in the study. Löwenstein-Jensen medium was used for cultivation of isolates. Susceptibility testing for anti-TB drugs was done by the method of absolute concentration as previously described (26); in particular, an isolate was considered RIF-resistant when bacterial growth occurred at a concentration of 20 μg of the drug per ml (26). Strain H37Rv was included as a control in each susceptibility test. The used method of absolute concentration was previously shown in our setting to give concordant results with those generated by the proportion method in a comparative study conducted with the National Mycobacterial Reference Laboratory in Turku, Finland (26).
DNA isolation and fingerprinting and quality control.
DNA from cultured cells was extracted as described by van Embden et al. (23). Strain differentiation was performed by spoligotyping (10) and IS6110 restriction fragment length polymorphism typing (23) as described previously.
DNA from Ziehl-Neelsen-stained sputum smears was obtained as described previously for auramine stained smears (14). The lysates were further subjected to phenol-chloroform-isoamyl alcohol (25:24:1) extraction, followed by isopropanol overnight precipitation (−20°C) of supernatant and dissolving in 0.3× Tris-EDTA buffer.
Control for contamination during microbiological and genetic experiments was performed as recommended in references 28 and 5, respectively. In particular, a negative control sputum slide was processed along with the test slides. A control of possible contamination with previously amplified amplicons was performed by including a negative control sample (distilled water) in each PCR run; no contamination was detected.
MAS-PCR assay.
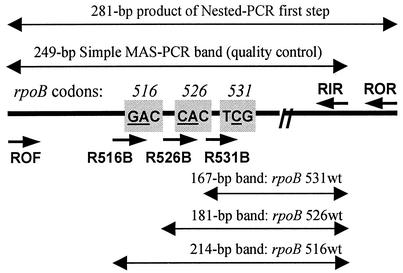
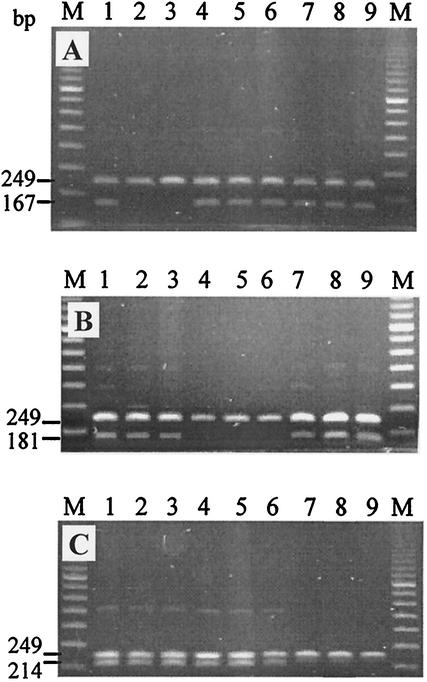
A simple single-step MAS-PCR assay was used for analysis of purified DNA from cultured cells and consisted of three independent allele-specific PCRs targeting three rpoB codons (codons 516, 526, and 531). The inner forward primers (R516B, R526B, and R531B) are positioned so that their 3′-OH ends pair with the second bases of the respective codons in the case of wild-type allele (Fig. 1). Consequently, in the absence of mutation in these positions in rpoB531/526/516, wild-type allele-specific fragments (167 or 181 or 214 bp, respectively) are amplified by the reverse primer RIR and an inner forward primer (Fig. 2A to C, respectively). If a mutation occurs, this results in mismatch at the 3′-end of the “wild type” inner primer and, under appropriate stringent PCR conditions, in the absence of the allele-specific PCR product (Fig. 2). Two outer primers ROF and RIR flank the entire rpoB region under study (rpoB positions 1252 to 1500 in strain H37Rv [http://genolist.pasteur.fr/TubercuList]) and amplify a 249-bp fragment in all strains (Fig. 2). The quality of the MAS-PCR is controlled by invariable amplification of this 249-bp fragment in all alleles (Fig. 1 and 2). The following primers were used for three MAS-PCRs targeting three different codons of the rpoB gene: two outer primers, forward ROF (5′-GTCGCCGCGATCAAGGA) and reverse RIR (5′-TGACCCGCGCGTACAC), and inner primers R516B (5′-GCTGAGCCAATTCATGGA), R526B (5′-GTCGGGGTTGACCCA), or R531B (5′-ACAAGCGCCGACTGTC). Purified DNA sample (0.1 to 0.5 μl) was added to PCR mixture (final volume of 20 μl) that contained MgCl2 (3 mM for rpoB526- and rpoB531-PCR or 1.5 mM for rpoB516-PCR), 1 U of recombinant Taq DNA polymerase (MBI Fermentas), 200 μM concentrations of each of the deoxynucleoside triphosphates (dNTPs), outer primers ROF (1 pmol) and RIR (20 pmol for rpoB526- and rpoB531-PCR, or 10 pmol for rpoB516-PCR) and one of allele-specific inner primers R531B (35 pmol) or R526B (30 pmol), or R516B (15 pmol). The reactions of rpoB526-PCR and rpoB531-PCR were performed in a PTC-100 thermal controller (MJ Research, Inc.) under the following conditions: initial denaturation at 96°C for 3 min; 5 cycles of 95°C for 45 s, 74°C for 1 min, and 72°C for 20 s; 5 cycles of 95°C for 40 s, 73°C for 50 s, and 72°C for 20 s; 22 cycles 94°C for 50 s, 70°C for 40 s, and 70°C for 20 s; and final elongation at 72°C for 3 min. The conditions of rpoB516-PCR were as follows: 96°C for 3 min; 30 cycles of 95°C for 50 s, 65°C for 40 s, and 72°C for 20 s; and 72°C for 3 min. The amplified fragments (10 μl) were electrophoresed in 1.5% standard agarose gels (Quantum Bioprobe) and visualized under UV light.
FIG. 1.
Schematic view of the rpoB gene fragment targeted by the allele-specific PCR assays. Short arrows depict the primers, long double-headed arrows represent PCR fragments, either invariable (249-bp) or allele-specific (167, 181, or 214 bp); lengths not to scale. The targeted rpoB codons are in shaded boxes; the mutated bases are underlined.
FIG. 2.
Profiles generated by single-step MAS-PCR assay with purified DNA preparations from clinical M. tuberculosis strains targeting three rpoB codons: codon 531 (A), codon 526 (B), and codon 516 (C). Lanes: 1, H37Rv strain; 2 and 3, strains with rpoB531 mutant alleles (TCG and TTG); 4 to 6, strains with rpoB526 mutant alleles (GAC, TAC, and CTC); 7 to 9, strains with rpoB516 mutant alleles (GTC, TAC, and GGC); M, 100-bp DNA ladder (Amersham Bioscience).
Nested allele-specific PCR (NAS-PCR) assay.
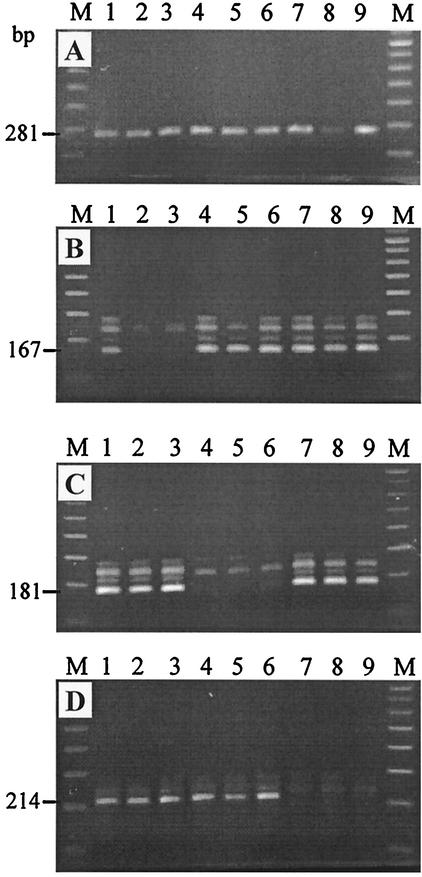
The assay includes preliminary amplification of the larger portion of rpoB with outer primers ROF and ROR (5′-GGTACGGCGTTTCGATGAAC) (Fig. 3A). This 281-bp fragment serves as a template for three subsequent allele-specific PCRs targeting three rpoB codons and performed at the same cycling conditions (Fig. 3B to D). The first PCR step of the NAS-PCR assay was performed under the following conditions: 96°C for 3 min; 30 cycles (38 for sputum slide preparations) of 95°C for 50 s, 62°C for 40 s, and 72°C for 30 s; and 72°C for 3 min. Purified DNA sample (0.1 μl) or sputum slide preparation (7 to 10 μl) were added to the PCR mixture (final volume of 30 μl) that contained 5 pmol of each primer ROF and ROR, 1.5 mM MgCl2, 1 U of recombinant Taq DNA polymerase, and 200 μM concentrations of each of dNTPs. The amplified 281-bp fragment (5 μl) was electrophoresed in 1.5% agarose gels and visually evaluated under UV light. Second step of the NAS-PCR assay comprised three specific PCRs performed simultaneously at the same cycling conditions: 96°C for 3 min; 5 cycles of 95°C for 45 s, 74°C for 30 s, and 72°C for 20 s; 5 cycles of 95°C for 40 s, 73°C for 40 s, and 72°C for 20 s; 12 cycles of 95°C for 40 s, 70°C for 40 s, and 72°C for 20 s; and 72°C for 3 min. The product of the first PCR (5 to 1 μl depending on its concentration; e.g., see Fig. 3A, lane 8 versus other lanes) was added to the PCR mixture (final volume of 20 μl) that contained 3 mM MgCl2, 1 U recombinant Taq DNA polymerase, 200 μM concentrations of each of dNTPs, reverse consensus primer RIR (20 pmol for rpoB531- and rpoB526-PCR, and 10 pmol for rpoB516-PCR) and one of allele-specific primers R531B (20 pmol), or R526B (20 pmol), or R516B (10 pmol). The amplified fragments (10 μl) were electrophoresed in 1.5% agarose gels and visualized under UV light.
FIG. 3.
Profiles generated by two-step nested allele-specific PCR assays with sputum slides DNA preparations. (A) First-step PCR with outer rpoB derived primers. (B to D) Analysis of three rpoB codons, codons 531, 526, and 516, respectively, by allele-specific PCR assays. Lanes: 1, H37Rv strain; 2 and 3, strains with rpoB531 mutant alleles (TCG and TTG); 4 to 6, strains with rpoB526 mutant alleles (GAC, TAC, and CTC), 7 to 9, strains with rpoB516 mutant alleles (GTC, TAC, and GGC); M, 100-bp DNA ladder (Amersham Bioscience).
RESULTS AND DISCUSSION
The MAS-PCR assay (both variants, simple and nested) was optimized to detect rpoB alterations in the 36 strains for which the rpoB sequence data were available; profiles for different alleles are shown in Fig. 2 and 3. Mutations in codons 516, 526, and 531 were successfully detected by the respective specific assays. After the stringency of PCR had been stably adjusted, the assay was repeated three times on these test strains to assess reproducibility. The MAS-PCR method successfully identified rpoB531, rpoB526, and rpoB516 changes in all isolates that harbored such mutations in all three reactions per strain. Furthermore, we have demonstrated that R526B and R516B primers detected not only mutations in the second bases of the codons 526 and 516 (that exactly corresponded to 3′ ends of the primers) but also mutations in their first bases that were in the −1 position with respect to the 3′ end of a primer. One possible explanation of this finding is that the melting temperature of the primer can be more crucial factor than DNA-DNA mismatch at the 3′-OH end of the primer, and hence wild-type-allele specific primers simply did not anneal on mutated template under selected stringent annealing temperatures. On the other hand, a mutation in rpoB522 was not detected by MAS-PCR although the R526B primer spans this codon.
Following validation and optimization performed on DNA samples with known rpoB sequence, these assays were used: (i) NAS-PCR, for analysis of sputum slide DNA samples from different patients, and (ii) MAS-PCR, for analysis of purified DNA preparations from 287 RIF-resistant isolates with different IS6110 restriction fragment length polymorphism profiles and recovered from unlinked patients, collected from 1996 to 2002.
A prospective study evaluated NAS-PCR for the direct analysis of microscopy-positive sputum smears. Analysis of DNA preparations from sputum slides generally yielded good amplification of the targeted rpoB fragment which was similar to that from purified DNA from cultured cells (e.g., see Fig. 3A). The rpoB 281-bp fragment was amplified in 110 of 114 samples (nonamplification could be due to polymerase inhibition or DNA target degradation). An rpoB mutation was detected in 72 of the 110 PCR-positive samples. Phenotypic susceptibility testing revealed that 87 isolates were RIF-resistant (83 [95.4%] were MDR) including two samples for which rpoB-PCR product was not amplified. Thus, NAS-PCR analysis detected a rpoB mutation in 72 out of 87 (82.8%) RIF-resistant samples. The high proportion of RIF-resistant isolates from the sputum-positive samples (87 [76.3%] of 114) is remarkable; however, this is a reflection of the real situation with MDR-TB in Russia. The MDR-TB rate in the St. Petersburg area of Russia in 2002 was high both for primary and acquired resistance: 36.3 and 75.2%, respectively. Similarly, the MDR-TB rate in the Samara region in central Russia was recently reported to vary from 43 to 65% depending on setting (6). This such situation with MDR-TB in Russia may in part be explained by poor adherence to treatment protocols and, more generally, by social economic instability in Russia while the transition period in nineties when the drug supply was frequently cut and the therapy was interrupted and/or inadequate (11, 17). Consequently, active transmission of MDR strains accounted for high MDR-TB rate in newly diagnosed patients in different Russian settings, including St. Petersburg (6, 16, 22).
The two-step design of the NAS-PCR assay provides the multiple quality assurance to control for false-negative results due to lack of amplification. This is especially useful for direct analysis of human samples, as we have demonstrated with sputum slides. The quality of DNA from such clinical samples may be poor and the first PCR step is needed to generate a quality template for subsequent allele-specific reactions. The output of the first-step PCR is monitored by short-run gel-electrophoresis. If amplification is weak (e.g., Fig. 3A, lane 8), than more amount of the 281-bp PCR product is used as a substrate for the second step allele-specific PCR (e.g., 5 μl instead of 1 μl). Furthermore, for easier and unambiguous interpretation of the PCR profiles of the test strains, each run of allele-specific PCR should include: (i) a wild-type strain (H37Rv), as a positive control of amplification of the allele-specific fragment, (ii) and a strain with known mutation in the targeted codon, as a negative control of nonamplification due to mutation. Thus, a test strain is compared in agarose gel versus positive and negative control lanes and the absence of wild-type-allele specific fragment is considered as a presence of mutation and hence indicates a drug-resistant phenotype.
A retrospective study of the collection of DNA samples from strains isolated in northwestern Russia in 1996 to 2002 revealed mutations in one of the analyzed rpoB codons in 247 out of 287 (86.1%) RIF-resistant strains. This is similar to the prevalence of these mutations in RIF-resistant strains worldwide (average of 85% [20]). This rate was previously reported for the specific Russian settings to vary from 86.7 to 100% (7, 12, 13, 22), and such a variation in percentage may be due to the fact that strains were isolated in different regions and periods of time. A possibility of limited and biased sampling should also be kept in mind.
The rpoB codons 516, 526, and 531 are the most frequently mutated worldwide though a variation in relative frequencies of mutations in these codons has been described in M. tuberculosis isolates from different geographic locations (reviewed in references 3 and 20). Nevertheless, simultaneous analysis of these three codons uncovers the majority of RIF-resistant isolates, especially in the regions with high rate of MDR-TB. The inherent sensitivity limit of both the assay described here (82.8 to 86.1% in our setting) and other genotypic methods targeting the rpoB RIF resistance-determining region is explained by the fact that a certain number of isolates (up to 10 to 13% in some studies [2, 19, 22]) harbor mutations in other parts of rpoB outside the hot spot region or may have other molecular mechanisms of resistance (e.g., a permeability barrier as in Neisseria [1]). Therefore the molecular methods cannot completely substitute culture-based phenotypic tests but may successfully complement them.
To conclude, the described rpoB-PCR assays can be used to detect the considerable proportion of RIF-resistant (and hence MDR) M. tuberculosis isolates in Russia and other middle- or low-income countries that correspond to the regions with highest levels of MDR-TB. The NAS-PCR assay was shown applicable for rapid, direct analysis of sputum slides though a further study on a larger and more representative selection of human samples from other settings (laboratories) is required to validate our findings. The proposed assays are rapid and easy to perform, and the results are easy to interpret. Furthermore, the procedure is inexpensive and requires only standard PCR and electrophoresis equipment, and its implementation would be useful for timely delivery of adequate antituberculous therapy.
Acknowledgments
We thank Elena Limeschenko and Anna Vyazovaya for excellent technical assistance, J. D. A. van Embden for kindly providing us with some DNA samples, Maxim Filipenko for rpoB sequencing of some strains, and Lidia Steklova and Vyacheslav Zhuravlev for providing us with some of the clinical isolates and sputum slides. We are grateful to Marie-Françoise Saron for careful reading of the manuscript and language corrections. We also acknowledge two anonymous reviewers for their valuable comments and suggestions.
This study was partly supported by the Délégation Générale au Réseau International des Instituts Pasteur et Instituts Associés, Institut Pasteur, Paris, and the International Atomic Energy Agency (research contract 9924).
REFERENCES
- 1.Abadi, F. J. R., P. E. Carter, P. Cash, and T. Hugh Pennington. 1996. Rifampin resistance in Neisseria meningitidis due to alterations in membrane permeability. Antimicrob. Agents Chemother. 40:646-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartfai, Z., A. Somoskovi, C. Kodmon, N. Szabo, E. Puskas, L. Kosztolanyi, E. Farago, J. Mester, L. M. Parsons, and M. Salfinger. 2001. Molecular characterization of rifampin-resistant isolates of Mycobacterium tuberculosis from Hungary by DNA sequencing and the line probe assay. J. Clin. Microbiol. 39:3736-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavusoglu, C., S. Hilmioglu, S. Guneri, and A. Bilgic. 2002. Characterization of rpoB mutations in rifampin-resistant clinical isolates of Mycobacterium tuberculosis from Turkey by DNA sequencing and line probe assay. J. Clin. Microbiol. 40:4435-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cockerill, F. R., III. 1999. Genetic methods for assessing antimicrobial resistance. Antimicrob. Agents Chemother. 43:199-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dragon, E. A., J. P. Spadoro, and R. Madej. 1993. Quality control of polymerase chain reaction, p. 160-168. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology. American Society for Microbiology, Washington, D.C.
- 6.Drobniewski, F., Y. Balabanova, M. Ruddy, L. Weldon, K. Jeltkova, T. Brown, N. Malomanova, E. Elizarova, A. Melentyev, E. Mutovkin, S. Zhakharova, and I. Fedorin. 2002. Rifampin- and multidrug-resistant Russian civilians and prison inmates: dominance of the Beijing strain family. Emerg. Infect. Dis. 8:1320-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Generozov, E. V., T. A. Akopian, V. M. Govorun, L. N. Chernoussova, E. E. Larionova, S. N. Savinkova, T. G. Smirnova, V. I. Golyshevskaia, and A. G. Khomenko. 2000. Molecular characteristics of multiresistant clinical strains of Mycobacterium tuberculosis isolated in Russia. Mol. Genet. Mikrobiol. Virusol. N1:11-17. (In Russian.) [PubMed]
- 8.Gillespie, S. H. 2002. Evolution of drug resistance in Mycobacterium tuberculosis: clinical and molecular perspective. Antimicrob. Agents Chemother. 46:267-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heep, M., B. Brandstätter, U. Rieger, N. Lehn, E. Richter, S. Rusch-Gerdes, and S. Niemann. 2001. Frequency of rpoB mutations inside and outside the cluster I region in rifampin-resistant clinical Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 39:107-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. D. A. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimerling, M. E. 2000. The Russian equation: an evolving paradigm in tuberculosis control. Int. J. Tuberc. Lung Dis. 4(Suppl. 2):S160-S167. [PubMed] [Google Scholar]
- 12.Marttila, H. J., H. Soini, E. Eerola, E. Vyshnevskaya, B. I. Vyshnevskiy, T. F. Otten, A. V. Vasilyef, and M. K. Viljanen. 1998. A Ser315Thr substitution in KatG is predominant in genetically heterogeneous multidrug-resistant Mycobacterium tuberculosis isolates originating from the St. Petersburg area in Russia. Antimicrob. Agents Chemother. 42:2443-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mokrousov, I., I. Filliol, E. Legrand, C. Sola, T. Otten, E. Vyshnevskaya, E. Limeschenko, B. Vyshnevskiy, O. Narvskaya, and N. Rastogi. 2002. Molecular characterization of multiple-drug-resistant Mycobacterium tuberculosis isolates from North-Western Russia and analysis of rifampin resistance using RNA/RNA mismatch analysis as compared to the line probe assay and sequencing of the rpoB gene. Res. Microbiol. 153:213-219. [DOI] [PubMed] [Google Scholar]
- 14.Mokrousov, I., O. Narvskaya, E. Limeschenko, T. Otten, and B. Vyshnevskiy. 2002. Detection of ethambutol-resistant Mycobacterium tuberculosis strains by multiplex allele-specific PCR assay targeting embB306 mutations. J. Clin. Microbiol. 40:1617-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mokrousov, I., T. Otten, M. Filipenko, A. Vyazovaya, E. Chrapov, E. Limeschenko, L. Steklova, B. Vyshnevskiy, and O. Narvskaya. 2002. Detection of isoniazid-resistant Mycobacterium tuberculosis strains by multiplex allele-specific PCR assay targeting katG codon 315 variation. J. Clin. Microbiol. 40:2509-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Narvskaya, O., T. Otten, E. Limeschenko, N. Sapozhnikova, O. Graschenkova, L. Steklova, A. Nikonova, M. L. Filipenko, I. Mokrousov, and B. Vyshnevskiy. 2002. Nosocomial outbreak of multidrug-resistant tuberculosis caused by a strain of Mycobacterium tuberculosis W-Beijing family in St. Petersburg, Russia. Eur. J. Clin. Microbiol. Infect. Dis. 21:596-602. [DOI] [PubMed] [Google Scholar]
- 17.Perelman, M. I. 2000. Tuberculosis in Russia. Int. J. Tuberc. Lung Dis. 4:1097-1103. [PubMed] [Google Scholar]
- 18.Pozzi, G., M. Meloni, E. Iona, G. Orrù, O. F. Thoresen, M. L. Ricci, M. R. Oggioni, L. Fattorini, and G. Orefici. 1999. rpoB mutations in multidrug-resistant strains of Mycobacterium tuberculosis isolated in Italy. J. Clin. Microbiol. 37:1197-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian, L., C. Abe, T. P. Lin, M. C. Yu, S. N. Cho, S. Wang, and J. T. Douglas. 2002. rpoB genotypes of Mycobacterium tuberculosis Beijing family isolates from East Asian countries. J. Clin. Microbiol. 40:1091-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramaswamy, S. V., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuberc. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 21.Telenti, A., P. Imboden, F. Marchesi, D. Lowrie, S. Cole, M. J. Colston, L. Matter, K. Schopfer, and T. Bodmer. 1993. Detection of rifampicin-resistant mutations in Mycobacterium tuberculosis. Lancet 341:647-650. [DOI] [PubMed] [Google Scholar]
- 22.Toungoussova, O., P. Sandven, A. Mariandyshev, N. Nizovtseva, G. Bjune, and D. A. Caugant. 2002. Spread of drug-resistant Mycobacterium tuberculosis strains of the Beijing genotype in the Archangel Oblast, Russia. J. Clin. Microbiol. 40:1930-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Embden, J. D. A., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnik, and P. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Victor, T. C., H. Lee, S. N. Cho, A. M. Jordaan, G. van der Spuy, P. D. van Helden, and R. Warren. 2002. Molecular detection of early appearance of drug resistance during Mycobacterium tuberculosis infection. Clin. Chem. Lab. Med. 40:876-881. [DOI] [PubMed] [Google Scholar]
- 25.Victor, T. C., P. D. van Helden, and R. Warren. 2002. Prediction of drug resistance in M. tuberculosis: molecular mechanisms, tools and applications. IUBMB Life 53:231-237. [DOI] [PubMed] [Google Scholar]
- 26.Viljanen, M. K., B. I. Vyshnevskiy, T. F. Otten, E. Vyshnevskaya, M. Marijamäki, H. Soini, P. J. Laippala, and A. V. Vasilyef. 1998. Survey of drug-resistant tuberculosis in northwestern Russia from 1984 through 1994. Eur. J. Clin. Microbiol. Infect. Dis. 17:177-183. [DOI] [PubMed] [Google Scholar]
- 27.World Health Organization. 1998. Laboratory services in tuberculosis control. Part II. Microscopy, p. 43. World Health Organization, Geneva, Switzerland.
- 28.World Health Organization. 1998. Laboratory services in tuberculosis control. Part III. Culture, p. 77. World Health Organization, Geneva, Switzerland.