Abstract
The influenza virus neuraminidase (NA) inhibitors zanamivir and oseltamivir were introduced into clinical practice in various parts of the world between 1999 and 2002. In order to monitor the potential development of resistance, the Neuraminidase Inhibitor Susceptibility Network was established to coordinate testing of clinical isolates collected through the World Health Organization influenza surveillance network from different regions of the world (M. Zambon and F. G. Hayden, Antivir. Res. 49:147-156, 2001). The present study establishes the baseline susceptibilities prior to and shortly after the introduction of the NA inhibitors. Over 1,000 clinical influenza isolates recovered from 1996 to 1999 were tested. Susceptibilities were determined by enzyme inhibition assays with chemiluminescent or fluorescent substrates with known NA inhibitor-resistant viruses as controls. The 50% inhibitory concentrations (IC50s) depended upon the assay method, the drug tested, and the influenza virus subtype. By both assays, the mean zanamivir IC50s were 0.76, 1.82, and 2.28 nM for the subtype H1N1 (N1), H3N2 (N2), and B NAs, respectively, and the oseltamivir IC50s were 1.2, 0.5, and 8.8 nM for the N1, N2, and B NAs, respectively. The drug susceptibilities of known zanamivir- and oseltamivir-resistant viruses with the NA mutations E119V, R292K, H274Y, and R152K fell well outside the 95% confidence limits of the IC50s for all natural isolates. Sequence analysis of the NAs of viruses for which the IC50s were above the 95% confidence limits and several control isolates for which the IC50s were in the normal range revealed variations in some previously conserved residues, including D151, A203, T225, and E375 (N2 numbering). Known resistance mutations are both influenza virus subtype and drug specific, but there was no evidence of naturally occurring resistance to either drug in any of the isolates.
Influenza is an infection of the upper respiratory tract which causes significant morbidity and mortality, and the impact of influenza virus infections is estimated to run into billions of dollars worldwide. Vaccination plays a major role in the prevention of influenza and the associated complications. However, due to limited use of the influenza vaccine, constant antigenic drift of the viruses, and the ever-present potential for antigenic shift and an associated pandemic, there is an important role for antiviral therapy in the management of influenza. Amantadine has been available in some countries for more than 25 years, but it is effective only against influenza virus A subtypes and can have adverse effects on the intestinal and central nervous systems. Furthermore, resistant viruses are readily generated in up to 30% of patients and mutant viruses are still virulent and readily transmitted (5, 11). Resistant viruses are also circulating in the community and can be isolated from patients who have never been exposed to amantadine (14, 19, 30).
In a new approach to drug development, zanamivir was designed on the basis of knowledge of the three-dimensional structure of the influenza virus neuraminidase (NA) and its interaction with the cell surface receptors (27). The drug is based on the structure of the transition-state analogue 2,3-dehyro-2-deoxy-N-acetylneuraminic acid, with a single substitution of a guanidinium group on the 4′ position on the sugar ring. Zanamivir targets highly conserved residues in the influenza virus NA and was the first specific inhibitor of both influenza A and B viruses. Zanamivir is administered by oral inhalation, which deliver high concentrations of the drug to the respiratory tract, where the virus is replicating. On the basis of the antiviral properties of zanamivir, oseltamivir was subsequently developed as an orally active NA inhibitor (17). The structure of oseltamivir is based on a cyclohexene ring rather than a sugar ring and has a substitution of an amino group at the 4′ position on the ring, while a hydrophobic pentyl ether group replaces the glycerol side chain at the 6′ position. It is taken orally as the ethyl ester prodrug oseltamivir phosphate, which is converted to the active oseltamivir carboxylate by hepatic esterases.
Zanamivir and oseltamivir were introduced into clinical practice in various parts of the world between 1999 and 2002. The drugs act by inhibiting NA activity, which prevents the release and spread of progeny virions from infected cells. Resistant viruses can be generated in vitro, but only after several passages in the presence of drug. Altered susceptibility can arise due to mutations in either the hemagglutinin (HA) or the NA (20). HA is responsible for attachment of the virus to cellular sialic acid receptors, and the role of NA is to cleave the sialic acids from receptors and virus proteins so that the progeny virions can spread. HA mutations, which are the predominant mutation in cell culture, appear to lower the affinity of the HA for receptors, so that there is less dependence on the NA activity for elution of progeny virions. However, the lower affinity of HA may be restricted to interactions with receptors on the cells in which the virus was passaged. Hence, in a different cell there may be no altered susceptibility to the drugs. At this stage, as there is no suitable cell line which bears receptors comparable to those seen in the human respiratory tract, there is no reliable assay for the screening of clinical isolates for HA mutations. Sequencing of the HA gene for potential mutations in residues involved in receptor binding is the only method available for evaluation of potential HA resistance in clinical isolates.
Variants with mutations in their NAs have been isolated only after extensive passage in cell culture in the presence of either zanamivir or oseltamivir (2, 9, 22, 25; J. Carr, J. Ives, N. Roberts, L. Kelly, R. Lambkin, J. Oxford, C. Y. Tai, D. Mendel, and F. Hayden, Antivir. Res. 46:A59 [abstr. 79], 2000; Z. M. Wang, C. Y. Tai, and D. B. Mendel, Antivir. Res. 46:A60 [abstr. 81], 2000). Due to the differences in the chemical structures of the inhibitors, there are mutations in the NA that specifically affect the binding to each of the drugs. No resistance has been detected in previously healthy patients treated with zanamivir (3, 12). One isolate with reduced sensitivity and an R152K NA mutation (and an HA mutation) was obtained from an immunocompromised child after a prolonged infection with an influenza B virus (8).
Mutations in the NA causing resistance to oseltamivir have arisen in both challenge studies and in patients with naturally acquired infections (7, 16). The rates of resistance are estimated to vary from 0.4 to 1% in the adult population and from 4 to 8% in the pediatric population (16, 24), which may be a reflection of the longer duration of virus shedding in children or a higher virus load, perhaps as a result of replication in the presence of limited preexisting immunity. There appear to be NA subtype-specific mutations, with R292K in a conserved residue in the NA active site being the predominant mutation in the H3N2 (N2) subtype. An E119V NA mutation has also been described in an N2 clinical isolate, and an H274Y NA mutation has also been described in an H1N1 (N1) clinical isolate. Unlike amantadine-resistant viruses, all of the NA mutants have been compromised in their enzyme activities, stabilities, or infectivities in vitro and in animal models (9, 13, 15, 21, 22).
Because NA inhibitors are a new class of anti-influenza drugs with limited use in the clinical setting to date, there are public health concerns about the potential for the emergence and spread of drug-resistant viruses. The global Neuraminidase Inhibitor Susceptibility Network was established to coordinate testing of clinical isolates in order to monitor both the potential development of resistance in individual isolates and a general decrease in the susceptibilities of isolates to the inhibitors (29). The purposes of this study were to establish the baseline NA susceptibilities of clinical isolates prior to the introduction of the NA inhibitors and to determine whether there were any naturally occurring resistant isolates. The experimental design involved testing of natural clinical influenza virus isolates collected from humans worldwide through the World Health Organization global influenza program. The isolates had undergone limited passage in tissue culture. Isolates were tested by enzyme inhibition assays, the 50% inhibitory concentrations (IC50s) and 95% confidence intervals were determined, and the distributions of the IC50s over time and by geography and influenza virus subtype were compared. Sequence analysis of the isolates for which the inhibitory concentrations were the highest, along with a selection of isolates for which the IC50s were near the mean values for each subtype, was also carried out to determine whether there was any correlation with the sequences of their NAs and those of known resistant isolates.
MATERIALS AND METHODS
Viruses and cells.
Clinical isolates representing the circulating influenza A virus subtypes N1 and N2 and influenza B viruses were obtained from the regional collaborating World Health Organization influenza reference and research laboratories in Australia, Japan, the United Kingdom, and the United States. Samples were obtained from 1996 to 1999 prior to the introduction of the NA inhibitors to generate the baseline data for determination of the susceptibilities of the different NA subtypes. Paired clinical isolates consisting of documented NA inhibitor-resistant viruses and the sensitive parent isolates were provided by Margaret Tisdale (GlaxoSmithKline, Stevenage, United Kingdom) and Noel Roberts (Roche Products Ltd., Welwyn Garden City, United Kingdom). A B/Memphis/20/96 isolate with an R152K NA mutation was selected during treatment of an immunocompromised child with zanamivir (8). An A/Texas/36/9 H1N1 isolate with an H274Y NA mutation, an A/Wuhan/359/95-like N2 isolate with an E119V NA mutation, and an A/Sydney/5/97 A/H3N2 isolate with an R292K NA mutation were selected during oseltamivir treatment (16). MDCK cells from one of the authors were infected with all viruses to provide sufficient stock volumes to evaluate sensitivities by both the fluorometric and the chemiluminescent assays. The virus stocks were stored at −70°C.
Since the presence of phenol red interferes with the chemiluminescent assay, all cultures were grown in phenol red-free Eagle minimum essential medium (EMEM; Gibco, Grand Island, N.Y.) containing 10% fetal bovine serum (Summit Biotechnology, Fort Collins, Colo.) as well as 1 mM l-glutamine, 1% HEPES, and 1% penicillin-streptomycin (all from Gibco). Medium for growth of viruses was EMEM supplemented with 0.14% bovine serum albumin fraction V and 2.5 μg of tosylsulfonyl phenylalanyl chloromethyl ketone-trypsin (Worthington Biochemical Co., Lakewood, N.J.) per ml.
Inhibitors.
Zanamivir was provided by Margaret Tisdale (GlaxoSmithKline). Oseltamivir carboxylate (GS4071), the active compound of the ethyl ester prodrug oseltamivir phosphate (GS4104), was supplied by Noel Roberts (Roche Products Ltd.).
NA assays.
Two different assays were used to evaluate the sensitivity of NA to zanamivir and oseltamivir. All samples were assayed in duplicate in each assay. The fluorescent assay used methyl umbelliferone N-acetyl neuraminic acid (MUNANA) as the substrate, based on the method of Potier et al. (23). Virus samples were initially titrated to obtain a dilution of virus in the linear portion of the enzyme activity curve. A signal-to-noise ratio of >2:1 is considered optimal for use in the inhibition assay. For inhibition assays 10 μl of drug and 10 μl of diluted virus were mixed and preincubated for 30 min at 37°C in black Optiplates (Packard, Meriden, Conn.). The final drug concentrations in the assay ranged from 0.0038 to 1,000 nM in serial fourfold dilutions. A total of 30 μl of 100 μM MUNANA was then added in 32.5 mM 2-(N-morpholino)ethanesulfonic acid (MES; pH 6.5; Sigma Chemical Company) buffer containing 4 mM CaCl2. After 1 h at 37°C, the reaction was stopped by addition of 150 μl of freshly prepared 0.14 M NaOH in 83% ethanol. Fluorometric determinations were quantified with a Packard FluoroCount fluorimeter by using an excitation wavelength of 365 nm and an emission wavelength of 450 nm.
The second assay, a chemiluminescent assay, used the NA-Star (Tropix, Bedford, Mass.) 1,2-dioxetane derivative of 100 μM sialic acid as the substrate (4). The virus was initially serially diluted in 32.5 mM MES (pH 6.0)-4 mM CaCl2 containing NA-Star in white Optiplates (Packard). The reaction mixture was then incubated at 37°C for 15 min with shaking. For the inhibition assays, 40 μl of diluted NA with a signal-to-noise ratio of 40:1 was preincubated with 10 μl of drug for 30 min at room temperature. Final drug concentrations ranged from 0.028 to 550 nM. The reactions were started by the addition of 5 μl of a 1:15 dilution of NA-Star, and the reaction mixtures were incubated for 15 min at 37°C. Chemiluminescent light emission was triggered by the addition of 55 μl of Light Emission Accelerator II (Sapphire II enhancer in 0.1 M diethanolamine [pH 10]; Tropix) to each well. As the half-life of the mixture is 5 min, all assays were immediately read on an Applied Biosystems (Foster City, Calif.) NORTHSTAR Luminometer.
Statistical analysis.
IC50s were calculated by using the Robosage Microsoft Excel software add-in for curve fitting and calculation of the IC50s. The distribution of the log10 IC50s is displayed by using box plots.
To show the correspondence between the two assays, the log10 IC50s obtained by the fluorescent assay were plotted against the log10 IC50s obtained by the chemiluminescent assays for influenza A virus subtypes N1 and N2 and influenza B virus.
Sequencing.
The NAs of the 24 isolates for which the greatest deviation above the mean IC50 of each drug was detected in each of the assays were selected for sequencing. All sequencing was carried out at Professional Genetics Laboratories, Uppsala, Sweden (now Capio Diagnostics AB, Eskilstuna, Sweden). The NAs of control isolates of each type and subtype for which IC50s were near the mean were also selected for sequencing.
RESULTS
NA assays. (i) Susceptibilities of isolates
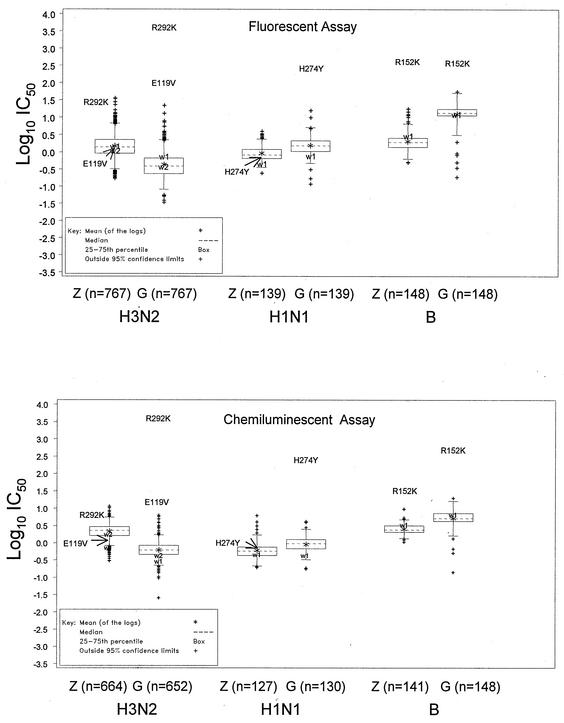
For screening of the NA susceptibilities of viruses isolated prior to the use of drugs, 1,054 influenza viruses collected from 1996 to 1999 from different geographical regions were expanded in MDCK cells and the susceptibilities of their NAs were determined. Viruses selected for screening were representative of the major strains circulating worldwide and were in proportion to the virus subtypes circulating in different regions of the world during that period: 139 influenza A virus subtype N1 isolates, 767 influenza A virus subtype N2 isolates, and 148 influenza B virus isolates. Susceptibilities to zanamivir and oseltamivir carboxylate were evaluated by both chemiluminescent and fluorescent assays, and these results were compared for each type or subtype. Box plots showing the distribution of the log10 IC50s are shown in Fig. 1. The N2 viruses were generally more sensitive to oseltamivir in both assays, with mean IC50s of 0.62 and 0.43 nM by the chemiluminescent and fluorescent assays, respectively, whereas the mean IC50s of zanamivir were 2.17 and 1.48 nM, respectively. The N1 viruses were slightly more sensitive to zanamivir, with mean zanamivir IC50s of 0.61 and 0.92 nM by the chemiluminescent and fluorescent assays, respectively, whereas the mean oseltamivir IC50s were 0.92 and 1.54 nM, respectively. For influenza B viruses the mean IC50s of zanamivir were approximately twofold lower than those of oseltamivir carboxylate by the chemiluminescent assay (2.57 and 5.21 nM, respectively) and nearly sixfold lower than those of oseltamivir carboxylate by the fluorescent assay (2.02 and 12.46 nM, respectively). Thus, the influenza B viruses had lower levels of sensitivity to oseltamivir by the fluorescent assay than by the chemiluminescent assay but by both assays had reduced susceptibilities to both drugs compared to the susceptibilities of the N1 and N2 viruses.
FIG. 1.
Box plots of the log10 of the mean IC50s of each of the inhibitors for each virus type by the fluorescent and chemiluminescent assays. The ends of the solid lines extending on either side of the box represent the approximate 95% confidence limits. Extreme values are those that lie outside the 95% confidence limits. The position of the text label for each mutant corresponds to the IC50. w1 and w2, values for paired wild-type controls for mutants.
The mean of the log10 IC50s of each drug for each subtype determined by each assay are listed in Table 1. Comparison of the sensitivities of the isolates obtained from the different geographic regions over the 4-year sampling period showed no significant differences in the mean IC50 for each type or subtype. This confirms that there is a reproducible difference in the sensitivities of the different types or subtypes of influenza viruses to the two drugs by the two different assays.
TABLE 1.
Mean zanamivir and oseltamivir carboxylate IC50s for clinical influenza isolates collected worldwide from 1996 to 1999 determined by fluorescent and chemiluminescent assays
| Time or location of sample collection and drug | Geometric mean IC50 (nM)a
|
|||||
|---|---|---|---|---|---|---|
| N2
|
N1
|
B
|
||||
| Chemi | Fluoro | Chemi | Fluoro | Chemi | Fluoro | |
| All samples | ||||||
| Zanamivir | 2.17 | 1.48 | 0.61 | 0.92 | 2.57 | 2.02 |
| Oseltamivir | 0.62 | 0.43 | 0.92 | 1.54 | 5.21 | 12.46 |
| 1996 | ||||||
| Zanamivir | 2.06 | 1.49 | 0.69 | 0.8 | 2.83 | 2.29 |
| Oseltamivir | 0.6 | 0.46 | 1.03 | 1.59 | 3.9 | 13.41 |
| 1997 | ||||||
| Zanamivir | 2.36 | 1.68 | 0.61 | 0.96 | 2.73 | 2.29 |
| Oseltamivir | 0.65 | 0.49 | 0.9 | 1.59 | 4.85 | 12.53 |
| 1998 | ||||||
| Zanamivir | 2.11 | 1.44 | 0.55 | 0.95 | 2.62 | 2.02 |
| Oseltamivir | 0.61 | 0.36 | 0.87 | 1.43 | 4.77 | 10.89 |
| 1999 | ||||||
| Zanamivir | 2.09 | 1.04 | 0.39 | 0.66 | 2.31 | 1.76 |
| Oseltamivir | 0.63 | 0.36 | 0.56 | 1.32 | 6.53 | 14.78 |
| 1996-1999 | ||||||
| North America | ||||||
| Zanamivir | 2.54 | 1.73 | 0.51 | 0.8 | 2.38 | 2.03 |
| Oseltamivir | 0.66 | 0.46 | 0.68 | 1.29 | 5.47 | 12.49 |
| Europe | ||||||
| Zanamivir | 2.23 | 1.38 | 1.6 | 0.81 | 2.31 | 1.76 |
| Oseltamivir | 0.69 | 0.5 | 0.98 | 1.42 | 5.42 | 12.91 |
| Asia | ||||||
| Zanamivir | 1.99 | 1.33 | 0.65 | 1.03 | 2.93 | 2.31 |
| Oseltamivir | 0.59 | 0.39 | 0.94 | 1.73 | 5.22 | 12.9 |
| Africa | ||||||
| Zanamivir | 1.87 | 1.47 | 0.58 | 0.73 | 1.98 | 1.78 |
| Oseltamivir | 0.53 | 0.39 | 0.88 | 1.3 | 5.36 | 10.83 |
| South and Latin America | ||||||
| Zanamivir | 2 | 1.28 | 0.5 | 1.06 | 3.21 | 2.73 |
| Oseltamivir | 0.58 | 0.37 | 0.83 | 1.56 | 5.24 | 13.37 |
| Oceania | ||||||
| Zanamivir | 2.06 | 2.44 | 0.66 | 0.75 | 2.53 | 1.75 |
| Oseltamivir | 0.6 | 0.4 | 0.8 | 1.29 | 3.47 | 8.57 |
Chemi, chemiluminescent assay; Fluoro, fluorescent assay.
(ii) Comparison of fluorescent and chemiluminescent assays.
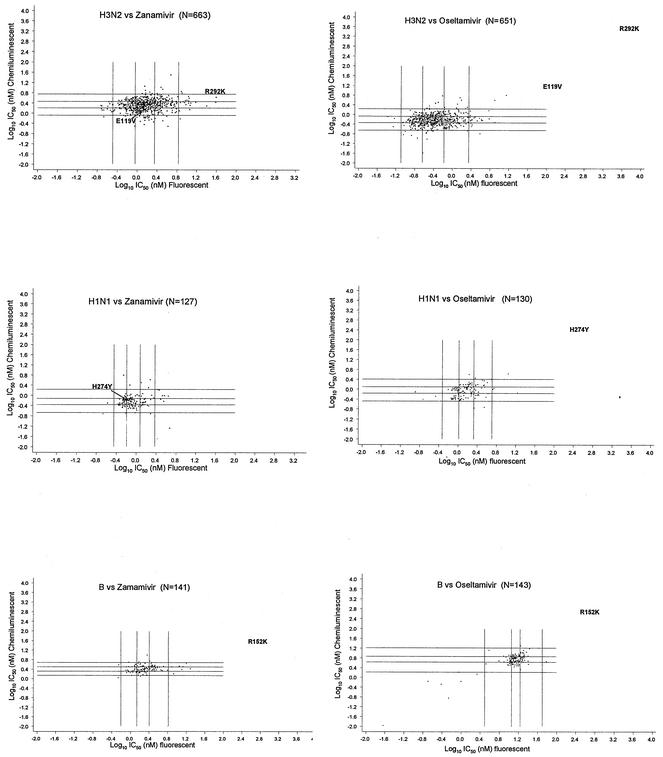
The grid plots in Fig. 2 show the correspondence of the results of the two assays. For all subtypes of influenza viruses and for both drugs, IC50s in the upper extremes by one assay generally fell within the normal to upper extremes by the other assay, with very few exceptions. Similarly, IC50s for isolates within the lower extremes by one assay fell within the lower or middle extremes by the other assay. There appeared to be very few discordant pairings, with the IC50s for only two isolates being in the upper extreme by the fluorescent assay and the lower extreme by the chemiluminescent assay, and no pairing opposite this was obtained for any of the isolates.
FIG. 2.
Grid plots showing the correspondence between inhibition by the chemiluminescent and fluorescent assays for individual samples. Each sample was assayed in duplicate. The four vertical and horizontal lines refer to the upper and lower 95% confidence limits and the 25th and 75th percentiles. Values lying outside the upper and lower 95% confidence limits and the four corners of the plots show where an extreme value by one assay corresponds to an extreme value by the other assay for a particular isolate. The IC50s of at least one of the drugs fall in the upper extreme values for mutant NAs. The position of the text label for each mutant corresponds to its IC50s by both assays. w1 and w2, values for paired wild-type controls for mutants.
A panel of resistant clinical isolates and the corresponding parental wild-type viruses was initially used to evaluate the fluorescent and chemiluminescent assays (28). These mutants were selected with different drug treatments and had different resistance profiles. The influenza B virus R152K mutant demonstrated cross-resistance to both drugs. However, it was more resistant to zanamivir than to oseltamivir carboxylate by the fluorescent assay (150- and 30-fold, respectively, compared to the susceptibility of the wild-type parent); in contrast, it was resistant to oseltamivir carboxylate rather than to zanamivir by the chemiluminescent assay (80- and 10-fold, respectively). The N1 virus with H274Y and the N2 virus with E119V were more resistant only to oseltamivir carboxylate by either assay (300- to 600-fold and 50- to 100-fold, respectively, compared to the susceptibilities of the wild-type parents). The N2 virus with R292K demonstrated >9,000-fold increased resistance to oseltamivir carboxylate and 4- to 25-fold increased resistance to zanamivir compared to the susceptibility of the wild-type parent. Although their relative resistance was found to depend upon the assay, by both assays the IC50s of at least one of the drugs for all the mutants fell outside the 95% confidence limits and were clearly distinguishable from the values for the parents (Fig. 1 and 2) (28).
Sequence analysis.
The top 24 isolates with outlier IC50s, that is, those for which IC50s were above the 95% confidence limits, by each assay with each drug were selected for sequencing, along with subtype controls for which IC50s were near the means. Sequence analysis of the NAs revealed variations in some residues previously considered to be conserved (26). All sequence numbering is based on the numbering for N2 (26). The most common variation found in all three subtypes was at D151 to G, V, N, or E. One of 10 N1 isolates, 1 of 42 influenza B virus isolates, and 7 of 38 N2 isolates had a D151 variation. One influenza B virus isolate had a variation of the previously conserved residue, T225I (T223 in influenza B virus). The apparent coevolution of two amino acids was also observed in several of the influenza B viruses. E375 (E377 in influenza B virus) was previously considered to be conserved in all influenza virus isolates, and all influenza B virus isolates recovered since 1959 had an A203I variation (I201 in influenza B virus). Several influenza B virus isolates had an E375G variation, and they all had a concomitant coevolution of the I203L variation (Table 2).
TABLE 2.
IC50 for inhibition of NA enzyme activity by zanamivir and oseltamivir carboxylate in chemiluminescent and fluorescent assays for outliers with variations in previously conserved residues in influenza B virus N1 and N2 isolates
| Virus strain, subtype | Mean IC50 (nM)a
|
Variation | |||
|---|---|---|---|---|---|
| Zanamivir
|
Oseltamivir
|
||||
| Chemi | Fluoro | Chemi | Fluoro | ||
| B/Beijing/09/97, B | 3.32 | 5.13b | 4.09 | 10.91 | D151Nc 1203/E375 |
| B/Chile/3045/97, B | 4.55b | 2.08 | 4.79 | 16.48 | T225I L203/G375 |
| B/Hawaii/04/97, B | 3.39 | 2.93 | 13.41b | 3.87 | L203/G375 |
| B/New Jersey/11/97, B | 2.30 | 6.34b | 3.66 | 3.36 | L203/G375 |
| B/Japan/697/97, B | 4.09b | 1.62 | 5.13 | 14.45 | L203/G375 |
| Controls | |||||
| B/Sydney/13/97, B | 2.07 | 1.39 | 4.66 | 10.46 | L203/G375 |
| B/Switz/6940/98, B | 2.74 | 2.04 | 4.75 | 13.93 | L203/G375 |
| A/Auckland/6/96, N1 | 2.01 | 1.76 | 2.84b | 2.28 | D151N |
| Controls | |||||
| A/Taiwan/1348/97, N1 | 0.73 | 0.85 | 0.91 | 0.99 | |
| A/Neimenggu/36/98, N1 | 0.20 | 1.95 | 1.49 | 3.50 | |
| A/Durb/113/97, N1 | 0.61 | 0.76 | 0.69 | 1.45 | |
| A/Brazil/207/96, N2 | 2.06 | 0.73 | 3.23b | 0.50 | D151G |
| A/Minnesota/06/96, N2 | 2.54 | 12.03b | 1.48 | 6.01b | D151E |
| A/Delaware/01/97, N2 | 6.25b | 8.01 | 1.16 | 0.88 | D151N |
| A/Den/22511/98, N2 | 1.96 | 25.24b | 0.50 | 0.28 | D151V |
| A/Stock/1/99, N2 | 6.73b | 1.79 | 1.65 | 0.33 | D151N |
| A/Victoria/54/96, N2 | 13.91b | 0.66 | D151N | ||
| A/Fukushima/114/96, N2 | 2.969 | 3.834 | 0.632 | 0.206 | D151G |
| Controls | |||||
| A/Guangdong/57/98, N2 | 0.87 | 0.74 | 0.03 | 0.24 | |
| A/Athens/23/97, N2 | 2.03 | 0.19 | 0.37 | 0.96 | |
| A/Sydney/16/97, N2 | 2.28 | 1.93 | 0.69 | 0.31 | |
| A/Thess/23/98, N2 | 1.53 | 0.19 | 0.45 | 0.93 | |
Chemi, chemiluminescence assay; Fluoro, fluorescence assay.
Selected as an outlier, with the IC50 of the inhibitor being above the 95% confidence intervals for this subtype; IC50 for controls were near the mean for that subtype.
Variations in residues previously thought to be conserved in all influenza viruses are boldfaced.
Despite the variations in residues previously considered to be conserved, there was no correlation between any variation and a consistent pattern of altered sensitivity to either inhibitor by either assay.
Since residue D151 was thought to be involved as a proton donor in the catalytic reaction (26), variation at this site would be expected to affect enzyme activity, as was observed by Ghate and Air (6), who generated a D151E mutant NA which had less than 10% of the activity of the wild-type enzyme. With the available data we investigated the relative activities of the NAs of the various D151 variants in our panel. As no NA-specific sera were available for quantification of the amount of NA protein to determine the relative specific activity (2), we compared the dilutions of the variants and wild-type viruses used for the enzyme assays as an indication of relative enzyme activity. If the D151 residue is critical for the catalytic activity of the enzyme, then mutation at this site would lead to a loss or a decrease in enzyme activity or replication of the virus would be compromised due to the loss of NA activity. Either would mean that significantly more virus would be needed for the same amount of NA activity. However, comparable dilutions of virus were used to achieve similar levels of enzyme activity (Table 3). Hence, there was no significant decrease in either the enzyme activity or the yields of the D151 variant viruses.
TABLE 3.
Comparative dilutions used for samples with wild-type viruses and D151 variants for enzyme inhibition assays and D151 codon usage
| Virus strain, subtype | Variation | Codon | Dilution fora:
|
|
|---|---|---|---|---|
| Chemi | Fluoro | |||
| B/Bangkok/4030/98, B | D151 (wtb) | GAC | 1:2 | Neat |
| B/Madrid/g-818/98, B | D151 (wt) | GAC | 1:64 | 1:4 |
| B/Switz/9359/99, B | D151 (wt) | GAC | 1:80 | 1:6 |
| B/Beijing/09/97, B | I203 | AUC | 1:32 | Neat |
| D151 (wt) | GAC (wt) | |||
| B/Beijing/22/97, B | I203, D151Nc | AUC AAC | 1:16 | 1:2 |
| B/Chile/3045/97, B | E150D, D151 (wt) | GAG | 1:80 | 1:4 |
| GAC (wt) | ||||
| T225I | AUA | |||
| A/Nanchang/04/98, N1 | D151 (wt) | GAC (wt) | 1:6 | Neat |
| A/Auckland/6/96, N1 | D151N | AAC | 1:8 | 1:2 |
| A/Dakar/5/97, N2 | D151 (wt) | GAU | 1:2 | 1:1.5 |
| A/Delaware/04/97, N2 | D151 (wt) | GAU (wt) | 1:6 | 1:1.5 |
| A/Eng/23/96, N2 | D151 (wt) | GAU | Neat | Neat |
| A/Stock/8/97, N2 | D151 (wt) | GAU | 1:12 | 1:3 |
| A/Victoria/69/97, N2 | D151 (wt) | GAU | 1:2 | 1:2 |
| A/Brazil/207/96, N2 | D151G | GGU | 1:1.5 | 1:1.5 |
| A/Fukushima/114/96, N2 | D151G | GGU | 1:2 | 1:15 |
| A/Minnesota/06/96, N2 | D151E | GAG | 1:3 | 1:2 |
| A/Delaware/01/97, N2 | D151N | AAU | 1:6 | 1:2 |
| A/Stock/1/99, N2 | D151N | AAU | 1:8 | 1:2 |
| A/Victoria/54/96, N2 | D151N | AAU | 1:2 | 1:2 |
| A/Den/22511/98, N2 | D151V | GUU | 1:3 | 1:1.5 |
Chemi, chemiluminescence assay; Fluoro, fluorescence assay.
wt, wild type.
Variations in residues previously thought to be conserved in all influenza viruses are boldfaced.
DISCUSSION
Our results demonstrate that recently circulating human influenza A and B viruses, collected before introduction of the NA inhibitors into clinical use, were all susceptible to both zanamivir and oseltamivir carboxylate and that, in contrast to the M2 inhibitors (amantadine and rimantadine), there was no evidence for naturally occurring resistance.
With the approval and use of the NA inhibitors for the treatment of influenza, there was concern over the possible emergence and spread of potentially resistant viruses and a potential gradual shift in the susceptibilities of clinical isolates to the inhibitors. The Neuraminidase Inhibitor Susceptibility Network was established to monitor the susceptibilities of influenza viruses to these inhibitors over a period of several years (29) to determine whether there was any evidence of the spread of resistant virus within the community. More than 1,000 isolates of influenza A and B virus subtypes were obtained over a period of 3 years, from 1996 to 1999, from all parts of the globe in order to establish the baseline susceptibilities of their NAs prior to introduction of the NA inhibitors between 1999 and 2002. Susceptibilities were evaluated by two different assays. One assay uses a fluorescent substrate, MUNANA (23), and the second uses a chemiluminescent substrate, NA-Star (4). We observed differences in the range of susceptibilities of the influenza A and B viruses greater than those previously determined by assays comparing smaller numbers of clinical isolates (1, 10). This variation in susceptibility indicates that caution must be exercised in the interpretation of phenotypic data obtained in future testing of clinical isolates for IC50s after the rate of NA inhibitor use increases. The results indicated that the two assays provided generally comparable values. Comparisons of the sensitivities of the enzyme to the two inhibitors showed that isolates of the N1 and B subtypes were slightly more sensitive to zanamivir, whereas the isolates of the N2 subtype were slightly more sensitive to oseltamivir. Isolates known to be resistant were clearly distinguishable by all assays, with the IC50s for the isolates lying above the 95% upper confidence limits. It should be noted that due to the drug-specific nature of the resistance seen to date, it is essential that isolates be screened against both inhibitors, since isolates may be resistant to only one of the inhibitors.
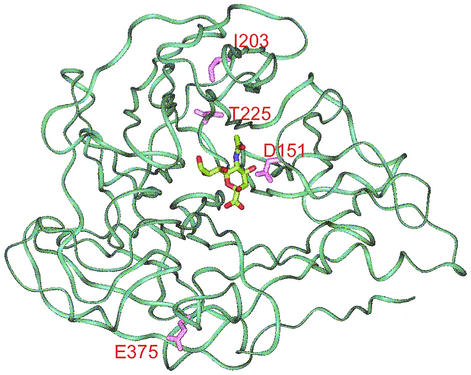
Sequence analysis of the NAs from 100 isolates for which the IC50s were outliers by one of the assays with one of the two drugs, plus the NAs from normal subtype controls, revealed variations at several amino acids which were previously considered (26) conserved: D151, T225, and E375 (D148, T223, and E377, respectively, in influenza B virus). Residue T225 is a neighbor to active-site residue R224, but as the sample was diluted 1:80 for the chemiluminescent enzyme assay, the variation had no obvious effect on enzyme activity and there were no significant differences in drug sensitivities between isolates with and without this variation. While residues A203 and E375 are both conserved in all influenza A virus N1 and N2 isolates, the sequences of earlier influenza B virus isolates in the influenza virus sequence database (18) revealed that the I203 variation (I201 in influenza B virus) was present in most influenza B virus isolates as far back as 1959, with the L203 variation detected after 1996. The E375G variation was also detected concomitantly in isolates with the I203L variation. Among the sequences of all influenza B virus isolates in the influenza virus sequence database (18), the sequence of only one isolate had each of the single variations; the sequences of all others had neither or both. To determine whether these could be compensatory variations, the residues were mapped on the X-ray structure of the influenza B virus NA (Fig. 3). In fact, these residues are on opposite sides of the head of the NA monomer and are quite distant even between the dimers; hence, it is unlikely that they are compensating variations, and although the residues are conserved in the N1 and N2 subtypes, neither of these residues plays a critical role in either enzyme activity or enzyme structure. Neither of these variations correlated with any altered sensitivity to NA inhibitors.
FIG. 3.
X-ray crystallographic model of influenza virus B/Beijing/87 NA (Protein Data Bank reference 1NSC). The amino acids at positions 203 and 375, which have coevolved, as well as residues D151 and T225 (positions 201, 377, 148, and 223, respectively, in the numbering for influenza B virus), were mapped. Since positions 203 and 375 fall on opposite sides of the head, they are clearly not compensatory variations.
Surprisingly, the conserved residue showing the greatest number of variations was D151. This residue was thought to be highly conserved, acting as a proton donor in the catalytic reaction (26). However, there were a variety of different amino acids in the B, N1, and N2 subtypes, including 151E, 151N, 151G, and 151V. As residues 151N, 151G, and 151V could not act as proton donors, D151 does not play a critical role in catalysis, which supports the earlier findings of Ghate and Air (6), although they saw a 10-fold reduction in kcat, suggesting some role for D151 in catalysis. In contrast, there was no clear indication of reduced enzyme activity for any of the NAs with variations seen in this study, including the same D151E variation generated by Ghate and Air (6). Furthermore, since all of the viruses in the present study were from viable clinical isolates, they were not compromised in their growth either in vivo or in vitro. Although the D151 variation was conserved across N1, N2, and B viruses, analysis of the available sequences of subtype N1 virus isolates recovered since 1933 and subtype B virus isolates recovered since 1940 revealed that they all used the same GAC codon for D151; in contrast, all N2 virus isolates used the GAU codon. Analysis of the codons used at position 151 in each of the variants reveals that the amino acid in each variant was coded for by a single base change. However, as there was no clear evidence of decreased enzyme activity and no apparent compensatory variations, the role of D151 remains unclear.
In summary, we have obtained baseline data for the sensitivities of contemporary strains of influenza viruses to the NA inhibitors zanamivir and oseltamivir prior to introduction of these drugs into clinical practice. Importantly, there were no major seasonal or geographical variations in the IC50s, and no naturally occurring resistant variants were isolated anywhere in the world. These data can now act as a valuable reference set for future screening for potential resistant variants and for determination of any future shift in the baseline sensitivities to the inhibitors.
In addition, the extensive sequencing of almost 100 NAs revealed variations in residues previously considered conserved. However, none of the variations was correlated with altered susceptibility to either drug by either assay, and no clinical isolates were found to have mutations known to confer NA resistance, even when the IC50 lay outside the 95% confidence intervals of the mean value for the subtype.
Acknowledgments
We thank N. Wetherall, C. Hodges-Savola, and J. Zeller, ViroMed Laboratories, Inc., Minnetonka, Minn., for performance of NA susceptibility assays and Brian Smith for the molecular modeling of the influenza B virus NA.
A. Hampson, A. Hay, F. G. Hayden, A. Klimov, J. McKimm-Breschkin, M. Tashiro, and M. Zambon are members of the Neuraminidase Inhibitor Susceptibility Network. The other members are M. Aymard, Universite Claude Bernard, Lyon, France; C. Macken, Los Alamos National Laboratory, Los Alamos, N.M.; R. Webster, St. Jude Children's Research Hospital, Memphis, Tenn.; and A. Monto, University of Michigan School of Public Health, Ann Arbor.
REFERENCES
- 1.Bantia, S., C. D. Parker, S. L. Ananth, L. L. Horn, K. Andries, P. Chand, P. L. Kotian, A. Dehghani, Y. El-Kattan, T. Lin, T. L. Hutchison, J. A. Montgomery, D. L. Kellog, and Y. S. Babu. 2001. Comparison of the anti-influenza virus activity of RWJ-270201 with those of oseltamivir and zanamivir. Antimicrob. Agents Chemother. 45:1162-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blick, T. J., T. Tiong, A. Sahasrabudhe, J. N. Varghese, P. M. Colman, G. J. Hart, R. C. Bethell, and J. L. McKimm-Breschkin. 1995. Generation and characterization of an influenza virus neuraminidase variant with decreased sensitivity to the neuraminidase-specific inhibitor 4-guanidino-Neu5Ac2en. Virology 214:475-484. [DOI] [PubMed] [Google Scholar]
- 3.Boivin, G., N. Goyette, I. Hardy, F. Aoki, A. Wagner, and S. Trottier. 2000. Rapid antiviral effect of inhaled zanamivir in the treatment of naturally occurring influenza in otherwise healthy adults. J. Infect. Dis. 181:1471-1474. [DOI] [PubMed] [Google Scholar]
- 4.Buxton, R. C., B. Edwards, R. R. Juo, J. C. Voyta, M. Tisdale, and R. C. Bethell. 2000. Development of a sensitive chemiluminescent neuraminidase assay for the determination of influenza virus susceptibility to zanamivir. Anal. Biochem. 280:291-300. [DOI] [PubMed] [Google Scholar]
- 5.Englund, J. A., R. E. Champlin, P. R. Wyde, H. Kantarjian, R. L. Atmar, J. Tarrand, H. Yousuf, H. Regnery, A. I. Klimov, N. J. Cox, and E. Whimbey. 1998. Common emergence of amantadine- and rimantadine-resistant influenza A viruses in symptomatic immunocompromised adults. Clin. Infect. Dis. 26:1418-1424. [DOI] [PubMed] [Google Scholar]
- 6.Ghate, A. A., and G. M. Air. 1998. Site-directed mutagenesis of catalytic residues of influenza virus neuraminidase as an aid to drug design. Eur. J. Biochem. 258:320-331. [DOI] [PubMed] [Google Scholar]
- 7.Gubareva, L. V., L. Kaiser, M. N. Matrosovich, Y. Soo-Hoo, and F. G. Hayden. 2001. Selection of influenza virus mutants in experimentally infected volunteers treated with oseltamivir. J. Infect. Dis. 183:523-531. [DOI] [PubMed] [Google Scholar]
- 8.Gubareva, L. V., M. N. Matrosovich, M. K. Brenner, R. C. Bethell, and R. G. Webster. 1998. Evidence for zanamivir resistance in an immunocompromised child infected with influenza B virus. J. Infect. Dis. 178:1257-1262. [DOI] [PubMed] [Google Scholar]
- 9.Gubareva, L. V., M. J. Robinson, R. C. Bethell, and R. G. Webster. 1997. Catalytic and framework mutations in the neuraminidase active site of influenza viruses that are resistant to 4-guanidino-Neu5Ac2en. J. Virol. 71:3385-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gubareva, L. V., R. G. Webster, and F. G. Hayden. 2001. Comparison of the activities of zanamivir, oseltamivir, and RWJ-270201 against clinical isolates of influenza virus and neuraminidase inhibitor-resistant variants. Antimicrob. Agents Chemother. 45:3403-3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayden, F. G., R. B. Belshe, R. D. Clover, A. J. Hay, M. G. Oakes, and W. Soo. 1989. Emergence and apparent transmission of rimantadine-resistant influenza A virus in families. N. Engl. J. Med. 321:1696-1702. [DOI] [PubMed] [Google Scholar]
- 12.Hayden, F. G., L. V. Gubareva, A. S. Monto, T. C. Klein, M. J. Elliot, J. M. Hammond, S. J. Sharp, M. J. Ossi, et al. 2000. Inhaled zanamivir for the prevention of influenza in families. N. Engl. J. Med. 343:1282-1289. [DOI] [PubMed] [Google Scholar]
- 13.Herlocher, M. L., J. Carr, J. Ives, S. Elias, R. Truscon, N. Roberts, and A. S. Monto. 2002. Influenza virus carrying an R292K mutation in the neuraminidase gene is not transmitted in ferrets. Antivir. Res. 54:99-111. [DOI] [PubMed] [Google Scholar]
- 14.Houck, P., M. Hemphill, S. LaCroix, D. Hirsh, and N. Cox. 1995. Amantadine-resistant influenza A in nursing homes. Identification of a resistant virus prior to drug use. Arch. Intern. Med. 155:533-537. [PubMed] [Google Scholar]
- 15.Ives, J. A., J. A. Carr, D. B. Mendel, C. Y. Tai, R. Lambkin, L. Kelly, J. S. Oxford, F. G. Hayden, and N. A. Roberts. 2002. The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antivir. Res. 55:307-317. [DOI] [PubMed] [Google Scholar]
- 16.Jackson, H. C., N. Roberts, Z. M. Wange, and R. Belshe. 2000. Management of influenza. Use of new antivirals and resistance in perspective. Clin. Drug Investig. 20:447-454. [Google Scholar]
- 17.Kim, C. U., W. Lew, M. A. Williams, H. Liu, L. Zhang, S. Swaminathan, N. Bischofberger, M. S. Chen, D. B. Mendel, C. Y. Tai, W. G. Laver, and R. C. Stevens. 1997. Influenza neuraminidase inhibitors possessing a novel hydrophobic interaction in the enzyme active site: design, synthesis, and structural analysis of carbocyclic sialic acid analogues with potent anti-influenza activity. J. Am. Chem. Soc. 119:681-690. [DOI] [PubMed] [Google Scholar]
- 18.Macken, C., H. Lu, J. Goodman, and L. Boykin. 2001. The value of a database in surveillance and vaccine selection, p. 103-106. In A. Osterhaus, N. Cox, and A. Hampson (ed.), Options for the control of influenza IV. Elsevier Science, Amsterdam, The Netherlands.
- 19.Masuda, H., H. Suzuki, H. Oshitani, R. Saito, S. Kawasaki, M. Nishikawa, and H. Satoh. 2000. Incidence of amantadine-resistant influenza A viruses in sentinel surveillance sites and nursing homes in Niigata, Japan. Microbiol. Immunol. 44:833-839. [DOI] [PubMed] [Google Scholar]
- 20.McKimm-Breschkin, J. L. 2000. Resistance of influenza viruses to neuraminidase inhibitors—a review. Antivir. Res. 47:1-17. [DOI] [PubMed] [Google Scholar]
- 21.McKimm-Breschkin, J. L., M. McDonald, T. J. Blick, and P. M. Colman. 1996. Mutation in the influenza virus neuraminidase gene resulting in decreased sensitivity to the neuraminidase inhibitor 4-guanidino-Neu5Ac2en leads to instability of the enzyme. Virology 225:240-242. [DOI] [PubMed] [Google Scholar]
- 22.McKimm-Breschkin, J. L., A. Sahasrabudhe, T. J. Blick, M. McDonald, P. M. Colman, G. J. Hart, R. C. Bethell, and J. N. Varghese. 1998. Mutations in a conserved residue in the influenza virus neuraminidase active site decreases sensitivity to Neu5Ac2en-derived inhibitors. J. Virol. 72:2456-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Potier, M., L. Mameli, M. Belisle, L. Dallaire, and S. B. Melancon. 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-d-N-acetylneuraminate) substrate. Anal. Biochem. 94:287-296. [DOI] [PubMed] [Google Scholar]
- 24.Roberts, N. A. 2001. Treatment of influenza with neuraminidase inhibitors: virological implications. Philos. Trans. Biol. Sci. 356:1897.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Staschke, K. A., J. M. Colacino, A. J. Baxter, G. M. Air, A. Bansal, W. J. Hornback, J. E. Munroe, and W. G. Laver. 1995. Molecular basis for the resistance of influenza viruses to 4-guanidino-Neu5Ac2en. Virology 214:642-646. [DOI] [PubMed] [Google Scholar]
- 26.Varghese, J. N., and P. M. Colman. 1991. Three-dimensional structure of the neuraminidase of influenza virus A/Tokyo/3/67 at 2.2 Å resolution. J. Mol. Biol. 221:473-486. [DOI] [PubMed] [Google Scholar]
- 27.von Itzstein, M., W. Y. Wu, G. B. Kok, M. S. Pegg, J. C. Dyason, B. Jin, T. Van Phan, M. L. Smythe, H. F. White, S. W. Oliver, et al. 1993. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature 363:418-423. [DOI] [PubMed] [Google Scholar]
- 28.Wetherall, N., T. Trivedi, J. Zeller, C. Hodges-Savola, J. L. McKimm-Breschkin, M. Zambon, and F. G. Hayden. 2003. Evaluation of neuraminidase enzyme assays using different substrates to measure susceptibility of influenza clinical isolates to neuraminidase inhibitors: report of the Neuraminidase Inhibitor Susceptibility Network. J. Clin. Microbiol. 41:742-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zambon, M., and F. G. Hayden. 2001. Position statement: global neuraminidase inhibitor susceptibility network. Antivir. Res. 49:147-156. [DOI] [PubMed] [Google Scholar]
- 30.Ziegler, T., M. L. Hemphill, M. L. Ziegler, G. Perez-Oronoz, A. I. Klimov, A. W. Hampson, H. L. Regnery, and N. J. Cox. 1999. Low incidence of rimantadine resistance in field isolates of influenza A viruses. J. Infect. Dis. 180:935-939. [DOI] [PubMed] [Google Scholar]