Abstract
Sixty-five clinical isolates of Streptococcus pneumoniae, all collected in Italy between 1999 and 2002 and resistant to both tetracycline (MIC, ≥8 μg/ml) and erythromycin (MIC, ≥1 μg/ml), were investigated. Of these strains, 11% were penicillin resistant and 23% were penicillin intermediate. With the use of the erythromycin-clindamycin-rokitamycin triple-disk test, 14 strains were assigned to the constitutive (cMLS) phenotype of macrolide resistance, 44 were assigned to the partially inducible (iMcLS) phenotype, 1 was assigned to the inducible (iMLS) phenotype, and 6 were assigned to the efflux-mediated (M) phenotype. In PCR assays, 64 of the 65 strains were positive for the tetracycline resistance gene tet(M), the exception being the one M isolate susceptible to kanamycin, whereas tet(K), tet(L), and tet(O) were never found. All cMLS, iMcLS, and iMLS isolates had the erythromycin resistance gene erm(B), and all M phenotype isolates had the mef(A) or mef(E) gene. No isolate had the erm(A) gene. The int-Tn gene, encoding the integrase of the Tn916-Tn1545 family of conjugative transposons, was detected in 62 of the 65 test strains. Typing assays showed the strains to be to a great extent unrelated. Of 16 different serotypes detected, the most numerous were 23F (n = 13), 19A (n = 10), 19F (n = 9), 6B (n = 8), and 14 (n = 6). Of 49 different pulsed-field gel electrophoresis types identified, the majority (n = 39) were represented by a single isolate, while the most numerous type included five isolates. By high-resolution restriction analysis of PCR amplicons with four endonucleases, the tet(M) loci from the 64 tet(M)-positive pneumococci were classified into seven distinct restriction types. Overall, a Tn1545-like transposon could reasonably account for tetracycline and erythromycin resistance in the vast majority of the pneumococci of cMLS, iMcLS, and iMLS phenotypes, whereas a Tn916-like transposon could account for tetracycline resistance in most M phenotype strains.
In Streptococcus pneumoniae, tetracycline resistance is predominantly due to ribosomal protection, i.e., the production of cytoplasmic proteins—-encoded by tet(M) or, less often, other tet genes—capable of interacting with the ribosome and making it insensitive to tetracycline inhibition (3, 38). An efflux-mediated mechanism reducing the intracellular tetracycline concentration to subtoxic levels is less common in streptococci, where this mechanism is due to membrane proteins encoded by the gene tet(K) or tet(L) (3).
Pneumococcal resistance to macrolides is due to either target site modification or active efflux (40). The former, prevalent mechanism usually depends on a posttranscriptional, methylase-mediated modification of 23S rRNA encoded by the erm(B) gene (19, 40). erm(B)-associated coresistance to macrolide, lincosamide, and streptogramin B (MLS) antibiotics can be expressed either constitutively, with high-level resistance to all MLS antibiotics (cMLS phenotype), or inducibly (iMLS phenotype). More often, pneumococcal strains appear inducibly resistant to only 16-membered macrolides and constitutively resistant to lincosamides (iMcLS phenotype) (22). Another methylase first described (31) and then found to be extensively present (15, 17) in Streptococcus pyogenes, mediated by a gene originally called erm(TR) and now designated erm(A) according to current nomenclature (30), has only occasionally been reported in S. pneumoniae (2, 35). Recently, mutations in 23S rRNA or ribosomal proteins leading to macrolide resistance in clinical isolates of S. pneumoniae have also been described (10, 37). A macrolide efflux mechanism, first demonstrated with S. pneumoniae and S. pyogenes and then with other streptococci, is associated with a resistance pattern (M phenotype) characterized by low-level resistance to only 14- and 15-membered macrolides among MLS antibiotics (34). M phenotype resistance is mediated by mef genes, two variants of which, mef(A) and mef(E)—originally discovered in S. pyogenes (4) and S. pneumoniae (36), respectively—have 90% identity (36) and have been regarded as a single gene class designated mef(A) (30). However, the two variants have recently been shown to be carried by different genetic elements in S. pneumoniae (9), and due to a number of important differences in the properties of mef(A)- and mef(E)-carrying pneumococci (9, 22), it has been recommended that the distinction between the two genes be maintained. A novel erythromycin efflux system, not associated with mef(A) or with other known macrolide efflux genes, has lately been described in erm(A)-positive strains of S. pyogenes with inducible, high-level resistance to erythromycin (14). However, its presence in S. pneumoniae and other streptococci has not yet been addressed.
In streptococci, drug resistance determinants occur more frequently on conjugative transposons than on plasmids. In particular, in S. pneumoniae the association of erythromycin resistance and tetracycline resistance may be due to Tn1545 and related conjugative transposons, which encode erythromycin resistance via erm(B) and tetracycline resistance via tet(M) and also kanamycin resistance via aphA3 (8). These transposons belong to a larger class of conjugative transposons, typically represented by Tn916, which encode tet(M)-mediated resistance to tetracycline but not resistance to erythromycin or kanamycin (6). An integrase gene usually called int-Tn, related to the second of the 24 open reading frames of Tn916, is characteristic of the Tn916-Tn1545 family of conjugative transposons (6).
In this study, a collection of clinical S. pneumoniae isolates resistant to both tetracycline and erythromycin were typed and investigated for a number of phenotypic and genotypic characteristics inherent to either resistance.
MATERIALS AND METHODS
Bacterial strains.
Sixty-five clinical isolates of S. pneumoniae, all resistant to both tetracycline (MIC, ≥8 μg/ml) and erythromycin (MIC, ≥1 μg/ml), were tested. All strains, collected from several Italian laboratories between 1999 and 2002, were isolated from upper respiratory tract specimens (the vast majority), sputum, blood, or cerebrospinal fluid. Strain identification was confirmed in our laboratory by conventional tests, such as susceptibility to optochin and solubility in bile, and by the API system (Biomérieux, Marcy-l'Etoile, France).
Antibiotics and susceptibility tests.
Tetracycline, erythromycin, minocycline, and penicillin were purchased from Sigma Chemical Co., St. Louis, Mo. Broth microdilution MICs were determined and MIC breakpoints of tetracycline and erythromycin were interpreted as recommended by the National Committee for Clinical Laboratory Standards (24). Mueller-Hinton II broth (BBL Microbiology Systems, Cockeysville, Md.) supplemented with 3% lysed horse blood was used as the test medium. The inoculum was 5 × 105 CFU/ml, and S. pneumoniae ATCC 49619 was used for quality control. Kanamycin susceptibility was determined by a standard agar diffusion test (25) using 30-μg commercial disks (Oxoid Ltd., Basingstoke, United Kingdom) with the following zone diameter breakpoints: susceptible, ≥18 mm; intermediate, 14 to 17 mm; resistant, ≤13 mm.
Determination of the macrolide resistance phenotype.
Test strains were assigned to the constitutive (cMLS), the partially inducible (iMcLS), the inducible (iMLS), or the efflux-mediated (M) macrolide resistance phenotype on the basis of the triple-disk (erythromycin plus clindamycin and rokitamycin) test, as described previously (23).
Gene detection by PCR.
Tetracycline resistance genes tet(K) and tet(L) were detected by using the primer pairs described by Trzcinski et al. (39), and tet(M) and tet(O) were detected by using those described by Corso et al. (7) and Olsvik et al. (26), respectively. Erythromycin resistance genes erm(A) and erm(B) were detected by using the oligonucleotide primers designated III8 and III10 by Seppälä et al. (31) and the primer pair developed by Sutcliffe et al. (33), respectively. The mef gene was detected by using the primer pair described by Sutcliffe et al. (33); mef(A) and mef(E) were then distinguished by PCR restriction fragment length polymorphism analysis of the 348-bp amplicon with BamHI (New England Biolabs, Beverly, Mass.), which has no restriction site in mef(E) and one in mef(A), generating two fragments of 284 and 64 bp (22). The integrase gene int-Tn, associated with the Tn916-Tn1545 family of conjugative transposons, was detected by using the primer pair described by Poyart-Salmeron et al. (27). DNA preparation and amplification and electrophoresis of PCR products were carried out by adapting established methods (16) to the procedures described for the individual primer pairs.
Serotyping.
All isolates were serotyped by the capsular swelling test using specific antisera (Statens Seruminstitut, Copenhagen, Denmark). Serotypes were indicated with conventional capsular type designations.
PFGE.
SmaI macrorestriction fragment patterns were analyzed by pulsed-field gel electrophoresis (PFGE); macrorestriction and PFGE were performed, and the relevant patterns were analyzed and compared as recently described elsewhere (29). For clusters with at least two isolates, types were designated with lowercase letters in order of size.
HRRA.
High-resolution restriction analysis (HRRA) of the tet(M) gene was carried out essentially as described by Doherty et al. (12). Briefly, a 10-μl aliquot of the PCR product obtained by using the primer pair described by Corso et al. (7) from each tet(M)-positive isolate was digested with the following restriction endonucleases: AciI, MseI, RsaI, and TaqI (New England Biolabs). Restriction fragments were separated by agarose (4%) gel electrophoresis and visualized by staining with ethidium bromide. The molecular size marker (100-bp DNA ladder) was from M-Medical Genenco, Florence, Italy. Each restriction pattern yielded by each endonuclease was assigned a number; restriction genotypes were determined on the basis of the combined restriction patterns of all four enzymes and lettered with capitals in order of size.
RESULTS
Antibiotic susceptibility.
The distribution of MICs of tetracycline, erythromycin, and penicillin for the 65 tetracycline- and erythromycin-resistant isolates of S. pneumoniae is summarized in Table 1. Forty-three strains were susceptible to penicillin (MIC, <0.12 μg/ml), 15 were intermediate, and seven were resistant (the MIC for five of these strains was 2 μg/ml and that for the other two was 4 μg/ml). All the strains but one were resistant to kanamycin according to the results of the disk tests.
TABLE 1.
Distribution of MICs of tetracycline, erythromycin, and penicillin for 65 tetracycline- and erythromycin-resistant isolates of S. pneumoniae
| Antibiotic | No. of strains inhibited by MIC (μg/ml) ofa:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <0.12 | 0.12 | 0.25 | 0.5 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | >128 | |
| Tetracycline | 5 | 13 | 15 | 13 | 2 | 17 | ||||||
| Erythromycin | 1 | 3 | 1 | 5 | 2 | 4 | 7 | 42 | ||||
| Penicillin | 43 | 5 | 9 | 1 | 5 | 2 | ||||||
No strain was inhibited by a MIC of 1 μg/ml.
Macrolide resistance phenotypes.
On the basis of the erythromycin-clindamycin-rokitamycin triple-disk test, 14 of the 65 test strains were assigned to the cMLS phenotype, 44 were assigned to the iMcLS phenotype, 1 was assigned to the iMLS phenotype, and 6 were assigned to the M phenotype. These findings, together with the ranges of MICs of erythromycin, are summarized in Table 2.
TABLE 2.
Macrolide resistance phenotypes of 65 tetracycline- and erythromycin-resistant isolates of S. pneumoniae
| Macrolide resistance phenotype | No. (%) of strains | Range of MICs (μg/ml) of erythromycin |
|---|---|---|
| cMLS | 14 (21.6) | ≥128 |
| iMcLS | 44 (67.7) | 4->128 |
| iMLS | 1 (1.5) | >128 |
| M | 6 (9.2) | 2-16 |
Genotypic strain characterization.
As shown in Table 3, the tetracycline resistance genes tet(K), tet(L), and tet(O) were not found. In contrast, 64 of the 65 test strains were positive for tet(M), the exception being one M phenotype isolate (MIC of penicillin, 0.25 μg/ml; MIC of tetracycline, 16 μg/ml; MIC of minocycline, 0.25 μg/ml; MIC of erythromycin, 16 μg/ml), the only isolate susceptible to kanamycin, which was negative for all four tetracycline resistance genes tested. All cMLS and iMcLS isolates as well as the iMLS isolate had the erm(B) gene, and all six M phenotype isolates had the mef gene [four with mef(E) and two with mef(A)]. The mef gene was also found in six isolates with the erm(B) gene: five [four with mef(E) and one with mef(A)] of the iMcLS phenotype and one [with mef(A)] of the cMLS phenotype. No isolate had the erm(A) gene. Of the 59 strains carrying both tet(M) and erm(B), i.e., all the strains of the cMLS, iMcLS, and iMLS phenotypes, 57 were also positive for the int-Tn gene (all except a cMLS isolate and an iMcLS isolate). Of the six M phenotype strains, which carried mef(A) or mef(E) as the only erythromycin resistance determinant, the five with the tet(M) gene were also int-Tn positive, whereas the isolate lacking tet(M) as well as the other tetracycline resistance genes tested was also int-Tn negative.
TABLE 3.
Macrolide resistance phenotypes and their association with tetracycline and erythromycin resistance genes, the int-Tn gene, serotypes, PFGE types, and restriction types of the tet(M) gene in 65 tetracycline- and erythromycin-resistant isolates of S. pneumoniae
| Macrolide resistance phenotype (n) | Resistance genea
|
int-Tn gene | No. of strains | Serotype(s) (no. of strains)b | PFGE type(s) (no. of strains)b,c | Restriction type of tet(M) loci (no. of strains)b,d | |||
|---|---|---|---|---|---|---|---|---|---|
| tet(M) | erm(B) | mef(A) | mef(E) | ||||||
| cMLS (14) | + | + | − | − | + | 12 | 23F (3), 19F (3), 14 (2), 3, 6B, 9A, 15A | b (2), d (2), c, Ost (7) | A (4), E (3), G (3), B (2) |
| + | + | + | − | + | 1 | 19A | Ost | C | |
| + | + | − | − | − | 1 | 6B | Ost | A | |
| iMcLS (44) | + | + | − | − | + | 38 | 19A (8), 23F (7), 19F (5), 6B (3), 3 (2), 6A (2), 10A (2), 10F (2), 14 (2), 15A, 33F, 36 | a (4), c (2), e (2), f (2), i (2), b, h, j, Ost (23) | A (22), C (5), D (4), B (3), F (3), E |
| + | + | − | + | + | 4 | 6B, 7F, 19F, 23F | a, b, g, h | A, B, C, D | |
| + | + | + | − | + | 1 | 3 | g | A | |
| + | + | − | − | − | 1 | 6B | Ost | A | |
| iMLS (1) | + | + | − | − | + | 1 | 23F | Ost | A |
| M (6) | + | − | − | + | + | 4 | 6B, 11A, 19A, 23F | j, Ost (3) | B (3), D |
| + | − | + | − | + | 1 | 14 | Ost | B | |
| − | − | + | − | − | 1e | 14 | Ost | ||
tet(K), tet(L), tet(O), and erm(A) were not detected in any test strain. +, present; −, absent.
The number of strains is indicated for types represented by more than one strain.
Ost, one-strain type.
Restriction profiles were as follows (different profiles yielded by the same endonucleases are distinguished by subscript numerals): A, RsaI1, MseI, TaqI, AciI1; B, RsaI2, MseI, TaqI, AciI2; C, RsaI2, MseI, TaqI, AciI3; D, RsaI1, MseI, TaqI, AciI3; E, RsaI1, MseI, TaqI, AciI4; F, RsaI2, MseI, TaqI, AciI1; G, RsaI1, MseI, TaqI, AciI5.
This strain was the only kanamycin-susceptible isolate and was also found to be susceptible to minocycline.
Typing.
Sixteen different serotypes, of which 11 were represented by at least two isolates, were detected. The most numerous was 23F (n = 13), followed by 19A (n = 10), 19F (n = 9), 6B (n = 8), and 14 (n = 6).
Forty-nine PFGE types, of which 10 were represented by at least two isolates, were detected. Type a, the most numerous, included five isolates.
The distribution of serotypes and PFGE types among the 65 erythromycin- and tetracycline-resistant S. pneumoniae isolates and their associations with the characteristics described above (macrolide resistance phenotypes, tetracycline and erythromycin resistance genes, and the int-Tn gene) are summarized in Table 3.
Restriction types of the tet(M) gene.
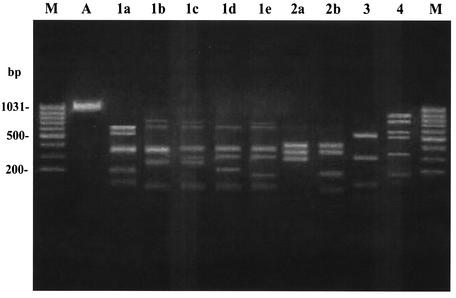
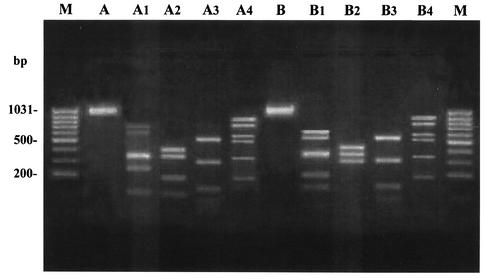
With HRRA of allelic variation within tet(M), endonucleases MseI and TaqI each yielded consistently the same fingerprinting profiles from the PCR amplicons of all strains, whereas AciI and RsaI produced five and two distinct profiles, respectively (Fig. 1). Altogether, the tet(M) loci from the 64 tet(M)-positive pneumococci analyzed fell into seven restriction types (A to G). Their distribution is reported in Table 3. Over half (31 of 59) of the strains with the erm(B) gene (cMLS, iMcLS, and iMLS phenotypes) had restriction type A, whereas four of the five strains with the mef gene (M phenotype) exhibited restriction type B. Restriction types A and B are shown in Fig. 2.
FIG. 1.
Different fingerprinting profiles obtained by digesting the tet(M) amplicons from 64 tet(M)-positive pneumococci with four endonucleases. Lane M, molecular size marker (100-bp ladder). Lane A, undigested tet(M) amplicon. Lanes 1a to 1e, different AciI profiles (AciI1 to AciI5). Lanes 2a and 2b, different RsaI profiles (RsaI1 and RsaI2). Lane 3, MseI profile. Lane 4, TaqI profile.
FIG. 2.
HRRA patterns of two tet(M)-positive pneumococci with restriction types A and B. Lane M, molecular size marker (100-bp ladder). Lane A, undigested tet(M) amplicon of the strain exhibiting restriction type A; lanes A1 to A4, restriction profiles yielded by endonucleases AciI, RsaI, MseI, and TaqI, respectively. Lane B, undigested tet(M) amplicon of the strain exhibiting restriction type B; lanes B1 to B4, restriction profiles yielded by endonucleases AciI, RsaI, MseI, and TaqI, respectively.
DISCUSSION
In clinical isolates of S. pneumoniae, tetracycline resistance is frequently associated with erythromycin resistance. In one large U.S. survey carried out from 1999 to 2000, >60% of multiresistant pneumococci exhibiting erythromycin resistance were also resistant to tetracycline (11); in Europe, associations of >80% in erythromycin-resistant pneumococci isolated in Spain (32) and Italy (21) have recently been reported. This association may reflect the widespread presence in pneumococcal populations of transposons, typified by Tn1545, thought to result from the insertion over time of resistance determinants, such as erm(B) for erythromycin and aphA3 for kanamycin, into primitive gram-positive conjugative transposons carrying tet(M) and the integrase gene int-Tn, typified by Tn916 (3, 6).
Among the 65 tetracycline- and erythromycin-resistant clinical strains of S. pneumoniae investigated in this study, the presence of Tn1545-like transposons could reasonably account for tetracycline and erythromycin resistance in the vast majority (at least 57 of 59) of the strains of the cMLS, iMcLS, and iMLS phenotypes, i.e., those sharing tet(M), erm(B), kanamycin resistance, and the int-Tn gene. The remaining two isolates shared tet(M), erm(B), and kanamycin resistance but were int-Tn negative. However, tet(M) was also associated with int-Tn in five of the six strains of the M phenotype, i.e., strains lacking erm genes and expressing erythromycin resistance due to mef-mediated active efflux. Unlike erm(B) in pneumococci with constitutive or inducible MLS resistance, the mef gene in M phenotype pneumococci is not known to be linked to tetracycline resistance, which in these strains could be due to a Tn916-like transposon. Similar findings—i.e., M phenotype pneumococci carrying mef(A) or mef(E), tet(M), and int-Tn—have recently been reported in Spain (32) and Scotland (1). Interestingly, the same tet(M) allele (type II) was identified by HRRA in four of our five M phenotype strains carrying mef(A) or mef(E), tet(M), and int-Tn. In contrast, the prevalent tet(M) allele (type I) of the seven detected in the 64 tet(M)-positive pneumococci was identified in isolates of all phenotypes (cMLS, iMcLS, and iMLS) with erm(B)-mediated erythromycin resistance but in no strain with mef(A) or mef(E)-mediated resistance (M phenotype). The sixth M phenotype pneumococcus was the only test strain susceptible to kanamycin and the only one lacking tet(M). Since this strain was also negative for tet(O), tet(K), and tet(L), it is possible that some less common tet gene capable of conferring tetracycline resistance on streptococci (3, 5) was involved; however, its susceptibility to minocycline suggests an efflux mechanism (3).
It is worth noting that in none of our tetracycline- and erythromycin-resistant S. pneumoniae isolates did we detect the tet(O) gene, whose presence in this species has occasionally been reported in limited numbers of South African (41), North American (20), and German (28) isolates. Conversely, it has very recently been shown that tet(O) largely accounts for tetracycline resistance in tetracycline- and erythromycin-resistant isolates of S. pyogenes, in which it is typically associated with erm(A) or mef(A) (E. Giovanetti, A. Brenciani, R. Lupidi, M. C. Roberts, and P. E. Varaldo, Program Abstr. 12th Eur. Congr. Clin. Microbiol. Infect. Dis., abstr. P472, p. 80, 2002). There are, however, few reports of erm(A) in S. pneumoniae isolates (2, 35); in particular, we have never found this gene in Italian pneumococci. As regards mef(A), while only true mef(A) is found in S. pyogenes isolates (E. Giovanetti and P. E. Varaldo, unpublished results), both mef(A) and the mef(E) variant can be found in M phenotype erythromycin-resistant pneumococci (9, 22). However, in the case of associated tetracycline resistance, whereas in S. pyogenes isolates the mef gene is associated with tet(O) (Giovanetti et al., 12th, ECCMID), this study shows that in pneumococci it is associated with tet(M).
The 65 tetracycline- and erythromycin-resistant S. pneumoniae strains tested were distributed over 16 serotypes and 49 PFGE types. These findings, in addition to the other phenotypic and genotypic differences, indicate that the test strains were substantially unrelated. This suggests a spread of resistance due to horizontal transfer of transposons or resistance determinants rather than to clonal dissemination. The most numerous serotypes (23F, 19A, 19F, 6B, 14, etc.) are essentially those that have more frequently been seen to combine macrolide and erythromycin resistance in other studies in Italy (18, 21) and in other European countries (1, 28). The rates of both penicillin-resistant (11%) and penicillin-intermediate (23%) isolates in our collection of tetracycline- and erythromycin-resistant pneumococci were higher than those recently reported among clinical S. pneumoniae isolates in Italy (13, 21).
Acknowledgments
This work was supported in part by a grant from the Italian Ministry of Education, University and Research.
REFERENCES
- 1.Amezaga, M. R., P. E. Carter, P. Cash, and H. McKenzie. 2002. Molecular epidemiology of erythromycin resistance in Streptococcus pneumoniae isolates from blood and noninvasive sites. J. Clin. Microbiol. 40:3313-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betriu, C., M. Redondo, M. L. Palau, A. Sánchez, M. Gómez, E. Culebras, A. Boloix, and J. J. Picazo. 2000. Comparative in vitro activities of linezolid, quinupristin-dalfopristin, moxifloxacin, and trovafloxacin against erythromycin-susceptible and -resistant streptococci. Antimicrob. Agents Chemother. 44:1838-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clancy, J., J. Petitpas, F. Dib-Hajj, W. Yuan, M. Cronan, A. V. Kamath, J. Bergeron, and J. A. Retsema. 1996. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol. Microbiol. 22:867-879. [DOI] [PubMed] [Google Scholar]
- 5.Clermont, D., O. Chesneau, G. de Cespédès, and T. Horaud. 1997. New tetracycline resistance determinants coding for ribosomal protection in streptococci and nucleotide sequence of tet(T) isolated from Streptococcus pyogenes A498. Antimicrob. Agents Chemother. 41:112-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clewell, D. B., S. E. Flannagan, and D. D. Jaworski. 1995. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 7.Corso, A., E. P. Severina, V. F. Petruk, Y. R. Mauriz, and A. Tomasz. 1998. Molecular characterization of penicillin-resistant Streptococcus pneumoniae isolates causing respiratory disease in the United States. Microb. Drug Resist. 4:325-337. [DOI] [PubMed] [Google Scholar]
- 8.Courvalin, P., and C. Carlier. 1987. Tn1545: a conjugative shuttle transposon. Mol. Gen. Genet. 206:259-264. [DOI] [PubMed] [Google Scholar]
- 9.Del Grosso, M., F. Iannelli, C. Messina, M. Santagati, N. Petrosillo, S. Stefani, G. Pozzi, and A. Pantosti. 2002. Macrolide efflux genes mef(A) and mef(E) are carried by different genetic elements in Streptococcus pneumoniae. J. Clin. Microbiol. 40:774-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Depardieu, F., and P. Courvalin. 2001. Mutation in 23S rRNA responsible for resistance to 16-membered macrolides and streptogramins in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:319-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doern, G. V., K. P. Heilmann, H. K. Huynh, P. R. Rhomberg, S. L. Coffman, and A. B. Brueggemann. 2001. Antimicrobial resistance among clinical isolates of Streptococcus pneumoniae in the United States during 1999-2000, including a comparison of resistance rates since 1994-1995. Antimicrob. Agents Chemother. 45:1721-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doherty, N., K. Trzcinski, P. Pickerill, P. Zawadzki, and C. G. Dowson. 2000. Genetic diversity of the tet(M) gene in tetracycline-resistant clonal lineages of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2979-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felmingham, D., R. R. Reinert, Y. Hirakata, and A. Rodloff. 2002. Increasing prevalence of antimicrobial resistance among isolates of Streptococcus pneumoniae from the PROTEKT surveillance study, and comparative in vitro activity of the ketolide, telithromycin. J. Antimicrob. Chemother. 50:25-37. [DOI] [PubMed] [Google Scholar]
- 14.Giovanetti, E., A. Brenciani, R. Burioni, and P. E. Varaldo. 2002. A novel efflux system in inducibly erythromycin-resistant strains of Streptococcus pyogenes. Antimicrob. Agents Chemother. 46:3750-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giovanetti, E., M. P. Montanari, M. Mingoia, and P. E. Varaldo. 1999. Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains in Italy and heterogeneity of inducibly resistant strains. Antimicrob. Agents Chemother. 43:1935-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hynes, W. L., J. J. Ferretti, M. S. Gilmore, and R. A. Segarra. 1992. PCR amplification of streptococcal DNA using crude cell lysates. FEMS Microbiol. Lett. 94:139-142. [DOI] [PubMed] [Google Scholar]
- 17.Kataja, J., P. Huovinen, the Macrolide Resistance Study Group, and H. Seppälä. 2000. Erythromycin resistance genes in group A streptococci of different geographical origins. J. Antimicrob. Chemother. 46:789-792. [DOI] [PubMed] [Google Scholar]
- 18.Latini, L., M. P. Ronchetti, R. Merolla, F. Guglielmi, S. Bajaksouzian, M. P. Villa, M. R. Jacobs, and R. Ronchetti. 2002. Prevalence of mefE, erm and tet(M) genes in Streptococcus pneumoniae strains from Italy. Int. J. Antimicrob. Agents 13:29-33. [DOI] [PubMed] [Google Scholar]
- 19.Leclercq, R. 2002. Mechanisms of resistance to macrolides and lincosamides: nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34:482-492. [DOI] [PubMed] [Google Scholar]
- 20.Luna, V. A., and M. C. Roberts. 1998. The presence of the tetO gene in a variety of tetracycline-resistant Streptococcus pneumoniae serotypes from Washington State. J. Antimicrob. Chemother. 42:613-619. [DOI] [PubMed] [Google Scholar]
- 21.Marchese, A., E. Tonoli, G. Balistreri, E. Debbia, and G. C. Schito. 2000. Antibiotic susceptibility patterns and serotypes of antibiotic resistant and/or invasive Streptococcus pneumoniae strains circulating in Italy. Microb. Drug Resist. 6:163-170. [DOI] [PubMed] [Google Scholar]
- 22.Montanari, M. P., M. Mingoia, I. Cochetti, and P. E. Varaldo. 2003. Phenotypes and genotypes of erythromycin-resistant pneumococci in Italy. J. Clin. Microbiol. 41:428-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montanari, M. P., M. Mingoia, E. Giovanetti, and P. E. Varaldo. 2001. Differentiation of resistance phenotypes among erythromycin-resistant pneumococci. J. Clin. Microbiol. 39:1311-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 25.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial disk susceptibility tests, 7th ed. Approved standard M2-A7. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 26.Olsvik, B., I. Olsen, and F. C. Tenover. 1995. Detection of tet(M) and tet(O) using the polymerase chain reaction in bacteria isolated from patients with periodontal disease. Oral Microbiol. Immunol. 10:87-92. [DOI] [PubMed] [Google Scholar]
- 27.Poyart-Salmeron, C., P. Trieu-Cuot, C. Carlier, and P. Courvalin. 1989. Molecular characterization of two proteins involved in the excision of the conjugative transposon Tn1545: homologies with other site-specific recombinases. EMBO J. 8:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reinert, R. R., S. Simic, A. Al-Lahham, S. Reinert, M. Lemperle, and R. Lütticken. 2001. Antimicrobial resistance of Streptococcus pneumoniae recovered from outpatients with respiratory tract infections in Germany from 1998 to 1999: results of a national surveillance study. J. Clin. Microbiol. 39:1187-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ripa, S., C. Zampaloni, L. A. Vitali, E. Giovanetti, M. P. Montanari, M. Prenna, and P. E. Varaldo. 2001. SmaI macrorestriction analysis of Italian isolates of erythromycin-resistant Streptococcus pyogenes and correlations with macrolide-resistance phenotypes. Microb. Drug Resist. 7:65-71. [DOI] [PubMed] [Google Scholar]
- 30.Roberts, M. C., J. Sutcliffe, P. Courvalin, L. B. Jensen, J. Rood, and H. Seppälä. 1999. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob. Agents Chemother. 43:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seppälä, H., M. Skurnik, H. Soini, M. C. Roberts, and P. Huovinen. 1998. A novel erythromycin resistance methylase gene (ermTR) in Streptococcus pyogenes. Antimicrob. Agents Chemother. 42:257-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seral, C., F. J. Castillo, M. C. Rubio-Calvo, A. Fenoll, C. Garca, and R. Gomez-Lus. 2001. Distribution of resistance genes tet(M), aph3′-III, catpC194 and the integrase gene of Tn1545 in clinical Streptococcus pneumoniae harbouring erm(B) and mef(A) genes in Spain. J. Antimicrob. Chemother. 47:863-866. [DOI] [PubMed] [Google Scholar]
- 33.Sutcliffe, J., T. Grebe, A. Tait-Kamradt, and L. Wondrack. 1996. Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sutcliffe, J., A. Tait-Kamradt, and L. Wondrack. 1996. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob. Agents Chemother. 40:1817-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Syrogiannopoulos, G. A., I. N. Grivea, A. Tait-Kamradt, G. D. Katopodis, N. G. Beratis, J. Sutcliffe, P. C. Appelbaum, and T. A. Davies. 2001. Identification of an erm(A) erythromycin resistance methylase gene in Streptococcus pneumoniae isolated in Greece. Antimicrob. Agents Chemother. 45:342-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tait-Kamradt, A., J. Clancy, M. Cronan, F. Dib-Hajj, L. Wondrack, W. Yuan, and J. Sutcliffe. 1997. mefE is necessary for erythromycin-resistant M phenotype in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 41:2251-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tait-Kamradt, A., T. Davies, P. C. Appelbaum, F. Depardieu, P. Courvalin, J. Petipas, L. Wondrack, A. Walker, M. R. Jacobs, and J. Sutcliffe. 2000. Two new mechanisms of macrolide resistance in clinical strains of Streptococcus pneumoniae from Eastern Europe and North America. Antimicrob. Agents Chemother. 44:3395-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor, D. E., and A. Chau. 1996. Tetracycline resistance mediated by ribosomal protection. Antimicrob. Agents Chemother. 40:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trzcinski, K., B. S. Cooper, W. Hryniewicz, and C. G. Dowson. 2000. Expression of resistance to tetracyclines in strains of methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 45:763-770. [DOI] [PubMed] [Google Scholar]
- 40.Weisblum, B. 2000. Resistance to the macrolide-lincosamide-streptogramin antibiotics, p. 694-710. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 41.Widdowson, C. A., K. P. Klugman, and D. Hanslo. 1996. Identification of the tetracycline resistance gene, tet(O), in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 40:2891-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]