Abstract
Poly C binding protein 1 (PCBP) is involved in the transcriptional regulation of neuronal mu-opioid receptor gene. In this study, we examined the molecular basis of PCBP cellular/nuclear localization in neuronal cells using EGFP fusion protein. PCBP, containing three KH domains and a variable domain, distributed in cytoplasm and nucleus with a preferential nuclear expression. Domain-deletional analyses suggested the requirement of variable and KH3 domains for strong PCBP nuclear expression. Within the nucleus, a low nucleolar PCBP expression was observed, and PCBP variable domain contributed to this restricted nucleolar expression. Furthermore, the punctate nuclear pattern of PCBP was correlated to its single-stranded (ss) DNA binding ability, with both requiring cooperativity of at least three sequential domains. Collectively, certain PCBP domains thus govern its nuclear distribution and transcriptional regulatory activity in the nucleus of neurons, whereas the low nucleolar expression implicates the disengagement of PCBP in the ribosomal RNA synthesis.
Keywords: poly C binding protein 1, KH domains and variable domain, cellular expression, nucleolar and nucleoplasmic regions, a punctate nuclear pattern, neuronal cells
Introduction
Opioids, such as morphine, elicit several pharmacological effects, such as analgesia, euphoria and respiratory depression. Prolonged opioid administration results in the tolerance and dependence [1-3]. Based on pharmacological studies and the cloning of opioid receptors, three types of opioid receptors, mu (MOR), kappa (KOR), and delta (DOR) have been identified, all of which are G-protein coupled receptors. Of these, MOR is proven to be the main receptor mediating the morphine-induced analgesia, tolerance and addiction [4]. The first cloned MOR-1 gene has proven to be the prominent MOR protein among several different splice variants [5-9].
The mouse MOR genome contains three different promoters designated as proximal, distal, and far upstream promoter. The far upstream promoter is located approximately 10 Kb upstream of the MOR-1 translation start site [9]. The distal promoter is located 794 bp upstream of the translation start site [10]. The proximal promoter initiates MOR transcription from four transcription initiation sites that are located in a region spanning from 291 to 268 bp upstream of the translation start site [5]. Among these three promoters, the MOR transcription is mainly initiated by the proximal promoter in the adult mouse brain as well as during development [11-12].
Several transcription factors, such as Sp factors (double-stranded DNA binding protein) and single-stranded DNA binding proteins, have been shown to interact with the MOR proximal promoter region and regulate MOR gene expression [2,13-16]. Recently, poly C binding protein 1 (PCBP), a single-stranded (ss) DNA binding protein, was cloned from a mouse brain cDNA library using the yeast one-hybrid screening system [16]. PCBP can regulate the MOR gene expression via binding to the ssDNA element in the proximal promoter region [16-19]. PCBP belongs to the K Homology (KH) domain family, which contains over 20 known proteins involved in many aspects of gene expression. Members of this family can contribute to the formation of a complex surrounding nascent pre-mRNA, thereby affecting RNA processing [20], such as SRC pyrimidine-binding protein (SPy) [21]. The prototypic example of this KH domain family, heterogeneous nuclear ribonucleoprotein (hnRNP) K, is known to participate in transcriptional regulation [22] and translational regulation [23-24]. Similarly, PCBP is known to be involved in RNA stabilization [25], translational activation and translational silencing [26-27]. For example, PCBP can bind to human α-globulin mRNA 3'UTR and form an α complex [25,28]. This type of mRNA stabilization does not appear to be limited to α-globulin; data has suggested that binding of PCBP to 3'UTR pyrimidine rich determinants may represent a general mechanism for stabilization of long lived cellular mRNA [29]. In addition, PCBP can function as a transcriptional regulator in MOR gene expression [16-19].
To further understand the multiple functional roles of PCBP in neuronal cells, we investigated the relationship of these roles to its cellular and sub-nuclear distribution in neurons. A compartmentalization has been observed for several protein factors, such as Ikaros that contributes to a repressor complex [30], and the AIRE protein, a transcription factor that serves as a component of a protein complex on the target site of chromatin [31]. In addition, the ATRX protein was found to be associated with the nuclear matrix during mitosis [32], and proliferating cell nuclear antigen helps to make up a DNA replisome complex [33]. Cellular compartmentalization may represent an efficient way of organizing a factor topologically for its physiological function roles. In this study, we therefore examined the cellular and nuclear distribution of PCBP and in particular the molecular basis underlying PCBP nuclear distribution in neuronal cells.
Materials and Methods
Plasmid construction
The pEGFP-C1 mammalian expression vector (Clontech Laboratories), driven by the CMV promoter, was used to construct the full length and various truncated poly C binding proteins 1 (PCBP) fused with EGFP. The DNA fragments of full length or various deletion fragments of PCBP (K12v, K2v3, K12, K2v and vk3) were obtained via the restriction enzyme digestion of the plasmid of pcDNA3-PCBP or the plasmid containing various truncated PCBPs [19]. These DNA fragments were purified and then inserted into the multiple cloning sites of pEGFP-C1 vector. The resulting plasmids were subjected to DNA sequencing to ensure that the coding sequences of the fusion proteins were in frame. For construction of the truncated PCBP constructs (K123 and K23), PCR amplification was carried out using the full length PCBP as the template with appropriate primer set to amplify DNA fragments of K12, K2 and K3. PCR amplification was carried out over 25 cycles with each round consisting of 1 minute at 95 °C, 30 seconds at 68 °C and 60 seconds (K12) or 30 seconds at 72 °C. The resulting PCR products were cloned into pCR II vector (Invitrogen), and then were subjected to the DNA sequencing. The DNA fragments containing the correct sequences were digested with restriction enzymes, purified and cloned into the multiple cloning sites of pEGFP-C1 vector. The resulting plasmids were subjected to the DNA sequencing to ensure the coding sequences of these fusion proteins were in frame.
Cell culture
Mouse neuroblastoma neuro2a cells (American Type Culture Collection) were grown in Dulbecco's Modified Eagle Medium (DMEM) with 10 % heat-inactivated fetal calf serum in an atmosphere of 10 % CO2 and 95 % air at 37 °C.
Transient transfection
Cells were grown overnight on the coverslips pre-coated with poly-L lysine (Sigma). Cells at approximately 40% confluence were transfected with various plasmids individually using the Superfect (Qiagene) lipofection method. Forty-eight hours after transfection, cells were washed twice with phosphate buffered saline (PBS) and ready for fixation.
Cell fixation and propidium iodide staining
Cells were fixed with 4% paraformaldehyde solution in PBS, and washed twice with PBS. The cells were subjected to perforation using a 0.1% Triton X100 solution containing 4% paraformaldehyde. Cells were washed with PBS, treated with RNase at 37 °C, and washed with PBS again before staining using 0.03 μg/ml propidium iodide. The cells were washed again with PBS solution, before these coverslips were placed onto a clean glass slide with a drop of anti-fade reagent (Vector Labs).
The staining of nucleolus
Cells were fixed with 4% paraformaldehyde solution in PBS, and washed twice with PBS. The cells were perforated using a 0.2% Triton X100 solution containing 4% paraformaldehyde. Cells were washed three times with PBS, then incubated with the blocking solution, containing 2% goat serum and 1% BSA in PBS, for 30 min at R. T. Cells were then incubated with anti-nucelolin (C23) antibody conjugated rhodamine (Santa Cruz Biotech.) overnight at 4 °C, and then were washed again with PBS solution. The coverslips were placed onto a glass slide with a drop of anti-fade reagent.
Confocal Microscopy
The cell image was viewed using a laser scanning confocal microscope (Fluoview 1000, Olympus) with multi-line argon laser lines at emissions of 457nm, 488nm, and 514nm, and green NeHe laser lines at an emission of 543nm. The cells were viewed under phase contrast at 10X and fluorescence light using oil immersion at 40X for image capture.
Results
Cellular distribution of the full length PCBP protein in neuronal cells
The full-length PCBP expression construct was generated by fusing the enhanced green fluorescence protein (EGFP) with PCBP using the mammalian expression vector driven by the CMV promoter. The resulting pEGFP-PCBP plasmid was transfected into neuronal neuro2a cells via the transient transfection. The transfected cells were fixed, and laser scanning confocal microscopy was used to examine the cellular distribution of PCBP.
As shown in Fig. 1A, the expression of PCBP protein (indicated by the green color) is observed throughout the nucleus and cytoplasm of neuronal cells. However, the nuclear distribution of PCBP appeared relatively stronger than its cytosolic expression. The nuclear hotspot (punctate) pattern was also observed, as indicated by arrows (Fig. 1A). To verify the existence of a higher amount of PCBP in nucleus region than in the cytosol, we subsequently performed the nuclear staining using propidium iodide (indicated as PI, by the red color), which interacted with DNA. The co-localization of PCBP and the nucleus is indicated by the yellow color of the merged image (indicated as Merge) in Fig. 1A, which confirmed a higher PCBP expression in the nucleus region of the neuronal cells. In summary, these results demonstrated that PCBP is distributed in both cytoplasm and nucleus regions with a higher expression in the nucleus of neuronal cells.
Fig. 1.
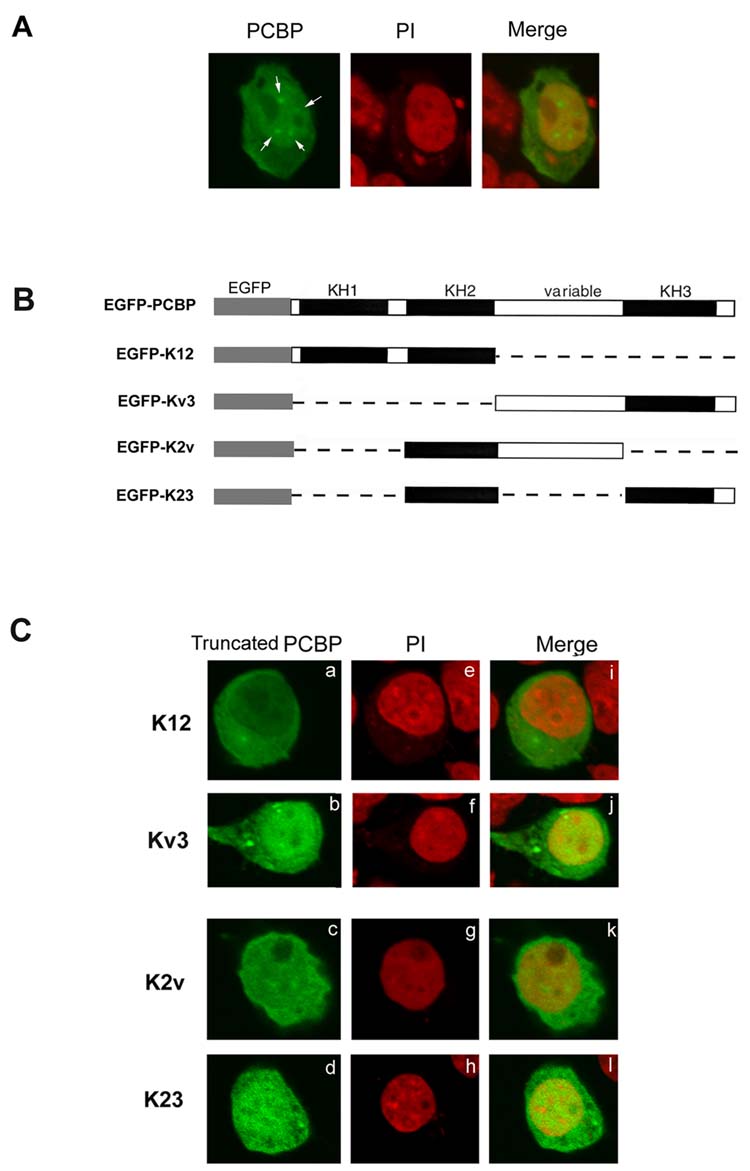
Sub-cellular localization of the full length or truncated poly C binding protein 1 (PCBP)
A, Cellular distribution of poly C binding protein 1 (PCBP) in the neuronal cells. Neuro2A cells were transiently transfected with the pEGFP-PCBP plasmid, in which the full length of PCBP was fused with EGFP. Transfected cells were fixed and viewed using a laser scanning confocal microscope. PCBP: The cellular distribution of the full length PCBP (indicated by the green color). Arrows point to the punctate nuclear pattern. PI: Nuclear region is shown in red color, which was stained using propidium iodide. Merge: The merged image of PCBP and PI, showing co-localization (indicated as the yellow color) of the green fluorescence labeled PCBP and PI nuclear staining. B, Schematic diagram represents the full length and various two-domain deletions of PCBPs, which were fused with the EGFP of the pEGFP-C1 vector. C, Effects of two-domain deletion of PCBPs on the cellular distribution. Neuronal cells were transfected with various two-domain constructs of PCBP plasmids. Cells were fixed and viewed using a confocal microscope. Truncated PCBP: The subcellular distributions (indicated by the green color) of two-domain PCBPs, including K12 (a), Kv3 (b), K2v (c) and K23 (d). PI (e-h), Nuclear region shown in red color was stained using propidium iodide. Merge (i-l), The merged image (indicated as the yellow color) of truncated PCBP and PI, showing co-localization of the green fluorescence labeled truncated PCBP and PI nuclear staining.
Both variable and KH3 domain are required for directing the preferential PCBP nuclear expression in neuronal cells
We next investigated the molecular basis underlying high expression of PCBP in nucleus of neurons. PCBP belongs to the KH domain family and contains three KH domains (Fig. 1B). The KH1 domain of PCBP located at the N-terminus is followed by the KH2 domain and then the variable domain with the KH3 domain located at the C-terminus. We therefore examined the physiological impact of PCBP domains on its nucleus expression. Two-domain deletions of PCBPs were constructed, but not three-domain deletions, to avoid the possibility of generating a small protein diffusing in and out of nucleus. The full length PCBP was divided into two parts, one containing the KH12 domains and the other containing the variable and KH3 domains (Fig. 1B). These two-domain of PCBPs, K12 and Kv3, were fused with EGFP, and the resulting plasmids were transfected into neuronal cells. As shown in Fig. 1C, the KH12 protein (a) was expressed more in the cytosolic portion than in the nuclear region, whereas, Kv3 protein (b) was expressed more in the nuclear than in cytosolic region. These results indicated that variable and KH3 domain may be involved in directing PCBP nuclear localization in neuronal cells. The observations were further verified via the merged images (i for KH12 and j for Kv3) of green fluorescence images with the propidium iodide stainings (e for KH12 and f for Kv3). Thus either one or both of the variable and KH3 domains were important for nucleus localization in neuronal cells.
To address this question further, other two-domain PCBP constructs, including K2v or K23 (containing only the variable domain or KH3 domain), were prepared. As shown in Fig. 1C, K2v (c) and K23 (d) displayed no preferential nuclear expression of PCBP as compared to that of Kv3 protein (b), as was further verified via the merged images (k for K2v and l for K23) of green fluorescence images with the propidium iodide stainings (g for K2v and h for K23). These results suggested that both variable domain and KH3 domain were necessary for a preferential PCBP nucleus expression in neuron.
This conclusion was further confirmed by the results of one-domain deletions (Fig. 2A). As shown in Fig. 2B, the K2v3 protein (a), containing both variable and KH3 domains, was expressed more strongly in the nucleus. The K12v protein (b), containing only the variable domain, did not display a stronger nuclear expression as compared to its cytosolic expression. Furthermore, the K123 protein (c), lacking the variable domain, displayed a preferential cytosolic expression with hardly a nuclear distribution. These observations were further confirmed via the merged images (g for K2v3, h for K12v and i for K123) of green fluorescence images with the propidium iodide stainings (d for K2v3, e for K12v and f for K123).
Fig. 2.
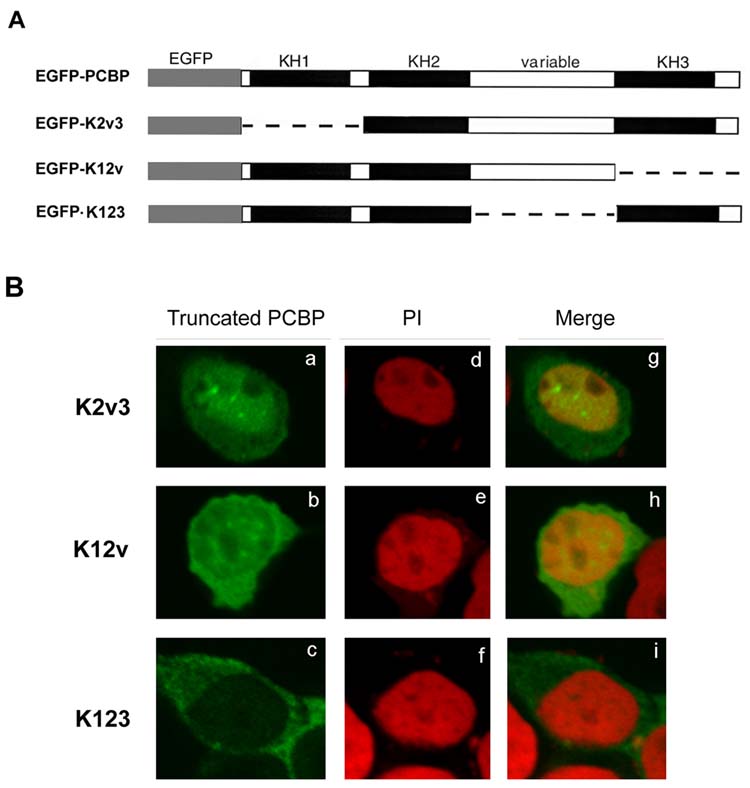
Sub-cellular localization of the one-domain deletion PCBP
A, Schematic representation of one-domain deletions of PCBPs, including K2v3, K12v and K123, which were fused with the EGFP of the pEGFP-C1 mammalian expression vector. B, Effect of one-domain deletion of PCBP on the cellular distribution in neuronal cells. Neuronal cells were transiently transfected with one-domain deletion of PCBP plasmids. The transfected cells were fixed and viewed using a confocal microscope. Truncated PCBP: the subcellular distributions (indicated by the green color) of one-domain deletion of PCBPs, including K2v3 (a), K12v (b) and K123 (c). PI (d-f): Nuclear region shown in red color was stained using propidium iodide. Merge (g-i): The merged image of truncated PCBP and PI, showing co-localization (indicated as the yellow color) of the truncated PCBP and PI nuclear staining.
Taken together, the above results suggested that both variable and KH3 domains containing the nuclear localization signals are required for directing a strong PCBP nuclear localization in neuronal cells.
Low nucleolus expression of PCBP in the nucleus of neuronal cells
In addition to its generally stronger nuclear expression, an uneven nuclear distribution of PCBP in neuronal cells was also noticed. The sub-nuclear distribution of PCBP was then examined. The nucleus can be divided into the nucleoplasmic and nucleolar regions. The nucleolus usually appears as an oval or round shape in the nucleus. A nucleolar marker, nucleolin, was selected, and the nucleolus was visualized with the anti-nucleolin antibody conjugated with rhodamine. Neuronal cells transfected with the pEGFP-PCBP plasmid were fixed and permeabilized using Triton X-100. As shown in Fig. 3A, the regions of low nuclear PCBP expression (a-b, indicated by arrow heads) coincided with the area staining with antibody against the C23-nucleolin (c-d, indicated by arrows). The merged images (e-f) further confirmed this conclusion. Results therefore suggested that PCBP was expressed less in the nucleolus than in the nucleoplasmic area within the nucleus of neuronal cells.
Fig. 3.
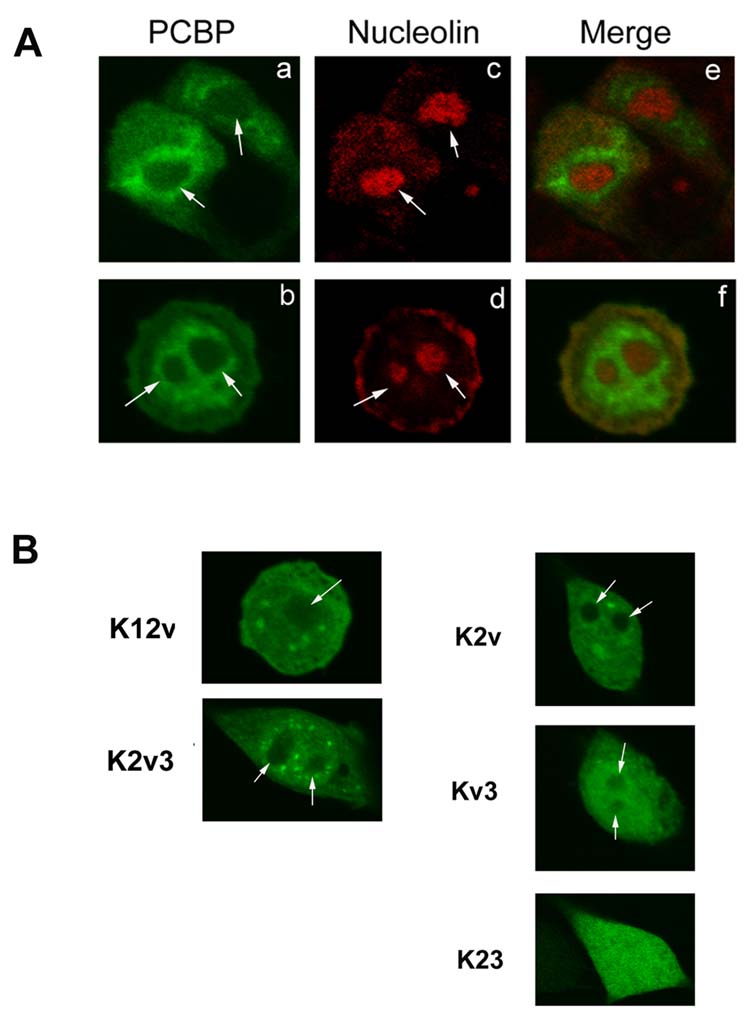
Effect of domain deletions of PCBP on its nucleolar expression in neuronal cells
A, Low nucleolar expression of PCBP in the nucleus of neuronal cells. Neuronal cells were transiently transfected with the pEGFP-PCBP plasmid, fixed, perforated using Triton X-100 and viewed using a confocal microscope. PCBP (a-b): The cellular distribution of the full length PCBP. Arrows showed the low PCBP expression inside the nucleus which is a round or oval shape. Nucleolin (c-d): The oval or round shape of nucleolar region (indicated by arrows) is shown in red color, which was stained using anti-nucleolin antibody conjugated with rhodamine. Merge (e-f): The merged image of PCBP and nucleolus, showing that there was barely a co-localization of the green fluorescence labeled PCBP and nucleolin staining in the nucleolus region. B, Effect of domain deletion of PCBP on its nucleolar distribution in neuronal cells. Neuronal cells were transfected with one-domain deletion of PCBP plasmids, pEGFP-K12v or pEGFP–K2v3, or two-domain deletion of PCBP plasmids, pEGFP-K2v, pEGFP-Kv3 or pEGFP-K23. The sub-nuclear distributions of various truncated PCBPs shown in green color. Arrows indicate the round or oval shape of nucleolar region inside the nucleus of neuron cells.
The functional role of the variable domain of PCBP and its low nucleolar expression in neuronal cells
To understand the structural basis underlying the low nucleolar expression of PCBP in neuronal cells, the physiological impact of specific PCBP domains was determined. One-domain truncated PCBPs, K12v and K2v3, fused with EGFP were transfected into neuronal cells. As shown in Fig. 3B, both K12v and K2v3 still displayed the low nucleolar expression of PCBP (as indicated by arrows). This result implicated that the KH2 and/or variable domain might be related to the low nucleolar expression of PCBP.
Two-domain deletions of PCBPs containing either KH2 or variable domain or both domains were then examined. As shown in Fig. 3B, K2v and Kv3 still displayed the low PCBP expression in nucleolus (indicated by arrows). Both truncated proteins contained the variable domain as a common feature, suggesting the possible involvement of variable domain in directing low nucleolus PCBP expression. This conclusion was then further confirmed using K23 protein (Fig. 3B), which displayed no clear distinction between nucleolar and nucleoplasmic regions.
Collectively, these results suggested that the variable domain of PCBP by itself contained a signal to restrict its expression in nucleolus.
The punctate nuclear pattern of PCBP in neuronal cells is correlated to its single-stranded DNA binding capability
Hotspots of PCBP (Fig. 4A, a-b, indicated by arrows) inside the nucleus of neuronal cells were also observed. This punctate nuclear pattern may result from the protein as a component of a complex binding to its target sites on chromatin [30-33]. Since PCBP is known to participate in MOR gene regulation and binds to the single-stranded (ss) DNA element in the MOR proximal promoter region in vivo [16-19], we questioned if the formation of nuclear hotspots of PCBP resulted from its DNA binding capability. Our previous electrophoretic mobility shift assay study [19] using the MOR ssDNA element as the DNA binding target had demonstrated that the single-domain deletions of PCBPs, K2v3 and K12v, possess a stronger ssDNA binding ability (approximately 65% of that of the full length of PCBP) than those of two-domain deletions of PCBP (approximately 20% or less than that of the full length of PCBP).
Fig. 4.
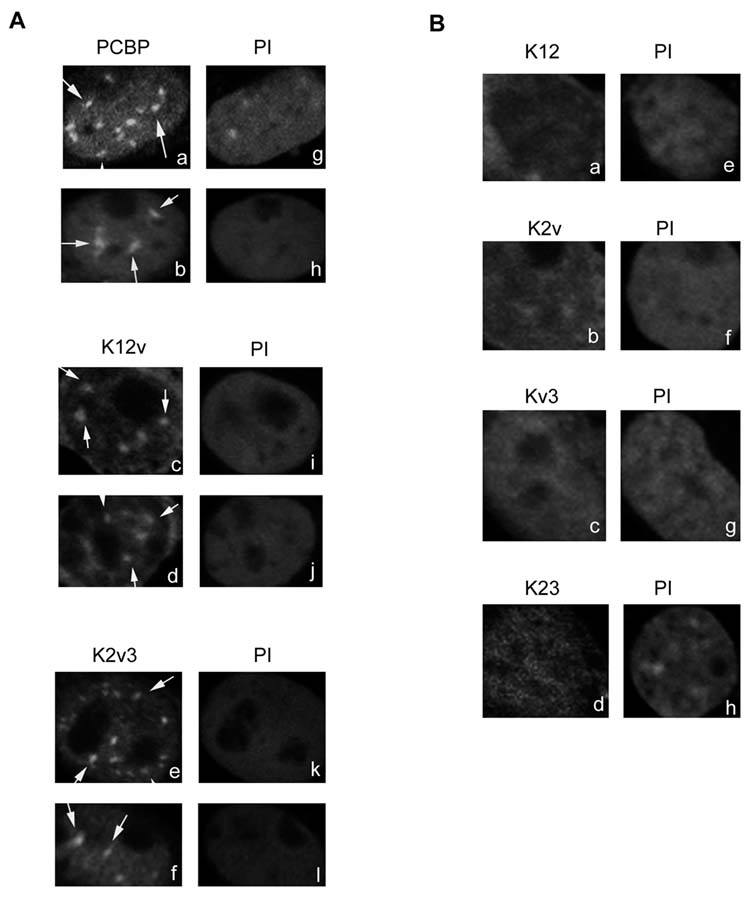
Effect of domain deletion of PCBP on its punctate nuclear pattern in neuronal cells
A, The punctate nuclear pattern of full-length and one-domain deletion of PCBP in neuronal cells. Neuronal cells were transfected with the full length or one-domain deletion of PCBP plasmids, pEGFP-K12v or pEGFP-K2v3. Transfected cells were fixed and viewed using a confocal microscope. The punctate nuclear pattern of the full length (a-b) or various truncated PCBPs (c-f) are shown. Arrows point to hotspots in the nucleus. PI (g-l), Nuclear region is stained using propidium iodide. B, No obvious punctate nuclear pattern of two-domain deletion of PCBP was observed in neuronal cells. Neuronal cells were transfected with two-domain deletion of PCBP plasmids, pEGFP-K12, pEGFP-K2v, pEGFP-Kv3 or pEGFP-K23. The nuclear distribution patterns of various truncated PCBPs are shown in a-d. PI (e-h), Nuclear region is stained using propidium iodide.
We therefore examined if expression of truncated K2v3 and K12v protein could display the punctate nuclear pattern. As shown in Fig. 4A, both truncated PCBPs, K12v (c-d) and K2v3 (e-f), displayed the punctate nuclear patterns (indicated by arrows), which were similar to that (a-b) of the full length of PCBP. The nuclear localization of these hotspots was further confirmed by the propidium iodide stainings (g-l). These results showed that nuclear hotspot patterns of K12v and K2v3 were similar to that of PCBP.
To further investigate if lowering the ssDNA binding ability of PCBP would result in the reduction of nuclear hotspots, various two-domain deletion of PCBP constructs were used and their nuclear expression patterns were examined. As shown in Fig. 4B (a-d), a drastic decrease of nuclear hotspots was visualized, and the nuclear location of the cell was confirmed via the propidium iodide staining (e-h). These results suggested that lowering the ssDNA MOR binding ability of PCBP resulted in a significant loss of its nuclear punctate pattern.
In summary, data suggested that the punctate nuclear pattern of PCBP was correlated with its ssDNA binding ability, with a significant loss of the punctate nuclear pattern associated with constructs exhibiting decreased ssDNA binding ability.
Discussion
Poly C binding protein 1 (PCBP) is a well-known RNA binding protein involved in various forms of gene regulation. Previous studies have also demonstrated that PCBP can serve as a transcription regulator and is involved in the regulation of mu-opioid receptor (MOR) gene expression [16-19]. In order to regulate the MOR gene expression in the nervous system, PCBP must enter the nucleus and bind to the target gene. In here, we therefore examined the compartmentalization of PCBP and determined the molecular basis underlying its nuclear distribution in neuronal cells.
Our results demonstrated that PCBP was distributed throughout the nucleus and cytoplasm in neuronal cells (Fig. 1A). It is reasonable to detect the cytosolic distribution of PCBP, because protein synthesis of PCBP takes place in the cytosol. However, PCBP displayed a higher nuclear localization with some hot spots (punctate pattern) in the nucleus of neuronal cells (Fig. 1A), supporting its important functional roles in the nucleus of neuronal cells, such as serving as a transcriptional regulator of neuronal MOR gene expression.
PCBP belongs to the KH domain family and contains four domains; a variable region and three KH domains (KH1, KH2 and KH3). We therefore performed an in-depth study to determine functional roles of PCBP domains on its nuclear localization in the neuron. PCBP domain-deletional analyses (Fig. 1C and Fig. 2B) suggested a primary role of the variable domain and KH3 domain in the preferential nuclear localization of PCBP in neuronal cells. When both variable domain and KH3 domain were present, the nuclear distribution was dominant (Fig. 1C, Kv3 and Fig. 2B, K2v3); whereas when both domains were deleted, the nuclear distribution was drastically reduced (Fig. 1C, K12). In addition, when only KH3 or variable domain was present, some degree of nuclear distribution was restored (Fig. 1C, K2v and K23 as well as Fig. 2B, K12v), but not as strongly as when both domains were present (Fig. 1C, Kv3 and Fig. 2B, K2v3). Moreover, results from deletion of variable domain (Fig. 2B, K123) further explicate the critical role of variable domain in directing PCBP nuclear expression in neuron. In conclusion, results demonstrated the critical functional role of variable domain in directing PCBP nuclear localization of neuronal cells, with the presence of KH3 domain to enhance PCBP preferential nuclear localization.
The necessity of the variable and KH3 domain for the preferential PCBP nuclear expression in neuronal cells suggested that both domains may contain the nuclear localization signals (NLS) for neuronal cells. Generally, it is accepted that large proteins require a NLS to be actively transported into nucleus through the nuclear pore complex (NPC) [34]. The NLS of the KH domain family has been elucidated, with the NLS of the heterogeneous nuclear ribonucleoprotein K (hnRNP K) serving as a model for the entire family [35]. NLS is categorized as monopartite, bipartite and a M9 sequence. Monopartite NLSs contained a single cluster of basic residues and were first identified in the SV40 large T antigen [36]. Bipartite NLSs contain two clusters of residues separated by a linker of 5-20 unconserved amino acids [37]. A third class of NLS, named the M9 sequence, has been identified for the nuclear RNA binding proteins [38]. The M9 sequence alone allows for nuclear localization, suggesting that it may direct contact with the nuclear pore complex [35]. No such consensus NLS is found in the sequence of PCBP; however, this lack of sequence similarities may imply a different nuclear import pathway. In addition, no obliteration of cytosolic expression was observed among all truncated PCBPs (Fig.1C and Fig. 2B), implicating that the variable and KH3 domains of PCBP may function to shift the equilibrium between its cytosolic and nuclear localization, instead of directing the complete nuclear translocation of PCBP. Therefore future investigation of the nucleus import pathway of PCBP in neuron will be useful in advancing understanding of the role of multi-functional PCBP in MOR gene regulation.
There are several possibilities why both variable domain and KH3 domain are required for the preferential PCBP nuclear localization. One possibility is that NLS of the KH3 domain, which alone is not able to confer the preferential nuclear distribution of PCBP, may interact directly with the NLS of the variable domain, enhancing the effects of the latter on directing the nuclear trafficking of PCBP in neuronal cells, because the KH3 domain alone is not able to confer the preferential nuclear distribution of PCBP without the presence of the variable domain. A similar notion is observed in a PDZ-domain protein, L-periaxin, which contains three essential domains for its nuclear distribution [39]. The first domain is essential for nuclear localization, and additional domains, containing weaker nuclear localization signals, can enhance its nuclear expression. A second possibility is that the NLS residing in the KH3 domain contains a weak NLS signal, which may not be able to interact directly with the NLS of the variable domain, but it may interact with other nuclear transport related proteins and therefore enhance the PCBP nuclear localization. This notion is supported by the example of hnRNP type I protein, which belongs to the same KH domain family, and interact of which with importin-α is correlated to its nuclear expression [40]. The third possibility is that the KH3 domain may interact with the other proteins which can be translocated into the nucleus, and therefore enhance the PCBP nuclear localization. This is supported by the example of the IκBα protein. One domain of IκBα protein serves as the dominant NLS of the protein, but two other domains contain weak NLS signals that are able to direct some proteins into the nucleus [41], and it has been hypothesized that the second and third domains control nuclear import by interactions with different cytoplasmic proteins, including hnRNP A1 [42]. It is possible that the KH3 domain of PCBP contains a weaker signal, similar to those domains observed in IκBα, that interacts with proteins such as hnRNP A1 to enhance the effects of the variable domain on the nuclear localization of PCBP.
In addition to PCBP displaying a higher nucleus expression in neuronal cells, an uneven nuclear expression of PCBP was also observed. A low nucleolar PCBP expression was confirmed by staining the nucleolus using anti-nucleolin antibody conjugated with rhodamine (Fig. 3A), and our results further suggested that the variable domain of PCBP may contribute to this restricted nucleolar expression (Fig. 3B). Though the detailed mechanism governing this low level of nucleolar PCBP expression is unclear, it implicates the disengagement of PCBP with the ribosomal RNA synthesis, processing and pre-ribosome storage [43].
The punctate nuclear pattern of PCBP showed another interesting feature of the sub-nuclear compartmentalization of PCBP in neuronal cells (Fig. 4A). A punctate nuclear pattern often indicates that the protein is a component of a nuclear complex [30-33]. PCBP has previously been demonstrated to interact directly with the MOR proximal promoter in vivo by chromatin immunoprecipitation (ChIP) assay [18]. Furthermore, our domain-deletion study provided a further insight into the molecular basis of punctate nuclear PCBP expression, showing that the pattern is correlated to its ssDNA binding ability of PCBP. The two-domain PCBPs, possessing low ssDNA binding capability, did not display significant hotspots in the nuclear region of neuronal cells (Fig. 4B), whereas three-domain PCBPs, which possessed significant ssDNA binding [19], were required to exhibit the punctate nuclear pattern (Fig. 4A), similar to that of the full length PCBP (Fig. 4A). This correlation of binding with the punctate nuclear pattern of PCBP therefore further supports that PCBP can be a component of nuclear complexes, which may interconnect to the transcription machinery and may be involved in the transcriptional regulation of other genes, in addition to the mu-opioid receptor gene.
It is also interesting that PCBP displayed two different types of nuclear topologies in neuronal cells (Fig. 1A and 3A): a punctate nuclear pattern and a dispersed pattern (PCBP was expressed in the entire nucleoplasmic region). The topologic organization of a factor can provide an efficient way for a factor to exert its physiological functions in close proximity to a specific effector. Since multiple steps along the pathway of transcription and gene regulation are regulated, different topological distribution of PCBP may represent different functional roles in the neuron. In addition to its expression patterns, there are subtle differences found between neuronal and non-neuronal cells in regard to PCBP sub-cellular distribution and the functional contribution of PCBP domain. First, in non-neuronal cells (HeLa cells), PCBP is nearly exclusively expressed in the nucleus, with cytosolic expression was barely visualized [49]; whereas in neuronal cells, the cytosolic expression of PCBP is clearly seen (though the PCBP nuclear expression is relatively higher). The expression of PCBP in neuronal processes has also been clearly observed (data not shown). Second, the PCBP variable domain is a solo determinant for its nucleus localization in HeLa cells [44], where as both variable domain and KH3 domain are necessary for directing PCBP nuclear preferential expression in neurons.
These results clearly suggested that PCBP is a fine tuned structure, which requires the complex coordination of multiple domains to confer its specific cellular and nuclear compartmentalization in neuron. Because MOR gene is mainly expressed in the central nervous system, MOR promoter activity must be modulated by tissue/cell-specific factors [45-46]. With PCBP as a factors involved in the MOR gene expression in brain, the interaction of various factors with different PCBP domains as well as the post-translation modification of PCBP via different signal transduction pathways may further provide a means of fine tuning the MOR gene expression in neuron.
Acknowledgements
We thank Dr. Joshua Berlin for the use of the confocal microscope in the beginning of early experiments. The acquisition of Fluoview 1000 confocal microscope is supported by the MRI grant of NSF. We thank Drs. Linda Hsu, Hsien-Ching Liu and Andrew P. Smith for their intellectual contributions and helping preparation of the manuscript. This research was supported by NIH research grant DA-016673.
Abbreviations used
- MOR
mu-opioid receptor
- PCBP
poly C binding protein 1
- ss
single-stranded
- KH domain
K homology domain
- hnRNP K
heterogeneous nuclear ribonucleoprotein K
- NLS
nuclear localization signal
References
- 1.Kieffer BL, Evan CL. Opioid tolerance–in search of the holy grail. Cell. 2002;108:587–590. doi: 10.1016/s0092-8674(02)00666-9. [DOI] [PubMed] [Google Scholar]
- 2.Law PY, Loh HH, Wei LN. Insight into the receptor transcription and signaling: implications in opioid tolerance and dependence. Neuropharm. 2004;47:300–311. doi: 10.1016/j.neuropharm.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 3.Van Ree JM, Gerrits M, Vanderschuren L. Opioids, reward and addiction: An encounter of biology, psychology, and medicine. Pharmacol. Rev. 1999;51:341–396. [PubMed] [Google Scholar]
- 4.Kieffer BL, Gaveriaux-Ruff C. Exploring the opioid system by gene knockout. Prog. Neurobio. 2002;66:285–306. doi: 10.1016/s0301-0082(02)00008-4. [DOI] [PubMed] [Google Scholar]
- 5.Min BH, Augustin LB, Felsheim RF, Fuchs JA, Loh HH. Genomic structure analysis of promoter sequence of a mouse mu opioid receptor gene. Proc. Natl. Acad. Sci. USA. 1994;91:9081–9085. doi: 10.1073/pnas.91.19.9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bare LA, Mansson E, Yang D. Expression of two variants of the human mu-opioid receptor mRNA in SK-N-SH cells and human brain. FEBS. 1994;354:213–216. doi: 10.1016/0014-5793(94)01129-x. [DOI] [PubMed] [Google Scholar]
- 7.Koch T, Schulz S, Pfeiffer M, Klutzny M, Schroder H, Kahl E, Hollt V. Carboxyl-terminal splicing of the rat mu opioid receptor modulates agonist-mediated internalization and receptor resensitization. J. Biol. Chem. 1998;273:13652–13657. doi: 10.1074/jbc.273.22.13652. [DOI] [PubMed] [Google Scholar]
- 8.Pan YX, Xu J, Mahurter L, Bolan E, Xu M, Pasternak GW. Generation of the mu opioid receptor (MOR-1) protein by three new splice variants of the Oprm gene. Proc. Natl. Acad. Sci. USA. 2001;98:14084–14089. doi: 10.1073/pnas.241296098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan YX. Identification and characterization of a novel promoter of the mouse mu opioid receptor gene (Oprm) that generates eight splice variants. Gene. 2002;295:97–108. doi: 10.1016/s0378-1119(02)00825-9. [DOI] [PubMed] [Google Scholar]
- 10.Liang Y, Mestek A, Yu L, Carr LG. Cloning and characterization of the promoter region of the mouse mu opioid receptor gene. Brain Res. 1995;679:82–88. doi: 10.1016/0006-8993(95)00222-c. [DOI] [PubMed] [Google Scholar]
- 11.Ko JL, Minnerath SR, Loh HH. Dual promoters of mouse mu-opioid receptor gene. Biochem. Biophy. Res. Commun. 1997;234:351–357. doi: 10.1006/bbrc.1997.6640. [DOI] [PubMed] [Google Scholar]
- 12.Ko JL, Chen HC, Loh HH. Differential promoter usage of mouse mu-opioid receptor gene during development. Mol. Brain Res. 2002;104:184–193. doi: 10.1016/s0169-328x(02)00357-1. [DOI] [PubMed] [Google Scholar]
- 13.Ko JL, Liu HC, Minnerath SR, Loh HH. Transcriptional regulation of mouse mu-opioid receptor gene. J. Biol. Chem. 1998;273:27678–27685. doi: 10.1074/jbc.273.42.27678. [DOI] [PubMed] [Google Scholar]
- 14.Ko JL, Liu HC, Loh HH. Role of an AP-2-like element in transcriptional regulation of mouse mu-opioid receptor gene. Mol. Brain Res. 2003;112:153–162. doi: 10.1016/s0169-328x(03)00086-x. [DOI] [PubMed] [Google Scholar]
- 15.Ko JL, Loh HH. Single-stranded DNA-binding complex involved in transcriptional regulation of mouse mu-opioid receptor gene. J. Biol. Chem. 2001;276:788–795. doi: 10.1074/jbc.M004279200. [DOI] [PubMed] [Google Scholar]
- 16.Ko JL, Loh HH. Poly C Binding Protein, a Single-Stranded DNA Binding Protein, Regulates Mouse mu-Opioid Receptor Gene Expression. J. Neurochem. 2005;93:749–761. doi: 10.1111/j.1471-4159.2005.03089.x. [DOI] [PubMed] [Google Scholar]
- 17.Kim SS, Pandey KK, Choi HS, Law PY, Wei LN, Loh HH. Poly C binding protein family is a transcription factor in mu-opioid receptor gene expression. Mol. Pharm. 2005;68:729–36. doi: 10.1124/mol.105.012245. [DOI] [PubMed] [Google Scholar]
- 18.Rivera-Gines A, Cook RJ, Loh HH, Ko JL. Interplay of Sps and Poly C binding protein 1 on the mu-opioid receptor gene expression. Biochem. Biophys. Res. Comm. 2006;345:530–537. doi: 10.1016/j.bbrc.2006.04.117. [DOI] [PubMed] [Google Scholar]
- 19.Malik AK, Flock KE, Godavarithi CL, Loh HH, Ko JL. Molecular basis underlying the poly C binding protein 1 as a regulator of the proximal promoter of mouse mu-opioid receptor gene. Brain Res. 2006 doi: 10.1016/j.brainres.2006.07.019. In press. [DOI] [PubMed] [Google Scholar]
- 20.Chen H, Hewison M, Hu B, Adams JS. Heterogeneous nuclear ribonucleoprotein (hnRNP) binding to hormone response elements: A cause of vitamin D resistance. Proc. Natl. Acad. Sci. USA. 2003;100:6109–6114. doi: 10.1073/pnas.1031395100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ritchie S, Pasha M, Batten D, Sharma R, Olson D, Ross A, Bonham K. Identification of the SRC pyrimidine-binding protein (Spy) as hnRNP K: implications in the regulation of SRC1A transcription. Nucleic Acids Res. 2003;31:1502–1513. doi: 10.1093/nar/gkg246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michelotti EF, Michelotti GA, Aronsohn AI, Levens D. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol. Cell. Biol. 1996;16:2350–2360. doi: 10.1128/mcb.16.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ostarek DH, Ostarek-Lederer A, Wilm M, Thiele BJ, Mann M, Hentze MW. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3' end. Cell. 1997;89:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- 24.Ostarek-Lederer A, Ostarek DH, Cans C, Neubauer G, Bomsztyk K, Superti-Furga G, Hentze M. C-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Mol. Cell. Biol. 2002;22:4535–4543. doi: 10.1128/MCB.22.13.4535-4543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss IM, Liebhaber SA. Erythroid cell-specific mRNA stability elements in the alpha 2-globin 3' nontranslated region. Mol. Cell. Biol. 1995;15:2457–2465. doi: 10.1128/mcb.15.5.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blyn L, Towner J, Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virology. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makeyev A, Liebhaber S. The polyI-binding proteins: a multiplicity of functions and a search for mechanisms. RNA. 2002;8:265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waggoner SA, Liebhaber SA. Identification of mRNAs associated with alphaCP2-containing RNP complexes. Mol. Cell. Biol. 2003;23:7055–7067. doi: 10.1128/MCB.23.19.7055-7067.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holcick M, Liebhaber SA. Four highly stable eukaryotic mRNAs assemble 3' untranslated region RNA–protein complexes sharing cis and trans components. Proc. Natl. Acad. Sci. USA. 1997;94:2410–2414. doi: 10.1073/pnas.94.6.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cobb B, Morales-Alcelay S, Kleiger G, Brown K, Fisher A, Smale S. Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. Genes and Dev. 2000;14:2146–2160. doi: 10.1101/gad.816400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riderle C, Christensen H, Schweiger S, Lehrach H, Yaspo M. AIRE encodes a nuclear protein co-localizing with cytoskeletal filaments: altered sub-cellular distribution of mutants lacking the PHD zinc fingers. Hum. Mol. Gen. 1998;8:277–290. doi: 10.1093/hmg/8.2.277. [DOI] [PubMed] [Google Scholar]
- 32.Berube N, Smeenk C, Picketts D. Cell cycle-dependent phosphorylation of the ATRX protein correlates with changes in nuclear matrix and chromatin association. Hum, Mol. Gen. 2000;9:539–547. doi: 10.1093/hmg/9.4.539. [DOI] [PubMed] [Google Scholar]
- 33.Leonhardt H, Rahn H, Weinzierl P, Sporbert A, Cremer T, Zink D, Cardoso C. Dynamics of DNA replication factories in living cells. J. Cell. Biol. 2000;149:271–279. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gasiorowski J, Dean D. Mechanisms of nuclear transport and interventions. Adv. Drug Deliv. Rev. 2003;55:703–716. doi: 10.1016/s0169-409x(03)00048-6. [DOI] [PubMed] [Google Scholar]
- 35.Michael W, Eder P, Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalderon D, Richardson W, Markham A, Smith A. Sequence requirements for nuclear localization of simian virus large-T antigen. Nature. 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- 37.Robbins J, Dilworth S, Laskey R, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 38.Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J. Cell. Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sherman D, Brophy P. A tripartite nuclear localization signal in the PDZ-domain protein Lperiaxin. J. Biol. Chem. 2000;275:4537–4540. doi: 10.1074/jbc.275.7.4537. [DOI] [PubMed] [Google Scholar]
- 40.Romanelli M, Morandi C. Importin-α binds to an unusual bipartite nuclear localization signal in the heterogeneous ribonucleoprotein type I. Eur. J. Biochem. 2002;269:2727–2734. doi: 10.1046/j.1432-1033.2002.02942.x. [DOI] [PubMed] [Google Scholar]
- 41.Sachdev S, Hoffman A, Hannink M. Nuclear localization of IκBα is mediated by the second ankyrin repeat: the IκBα ankyrin repeats define a novel class of cis-acting nuclear import sequences. Mol. Cell. Biol. 1998;18:2524–2534. doi: 10.1128/mcb.18.5.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hay D, Kemp G, Dargemont C, Hay R. Interaction between hnRNPA1 and IκBα is required for maximal activation of NF-κB-dependent transcription. Mol. Cell. Biol. 2001;2:3482–3490. doi: 10.1128/MCB.21.10.3482-3490.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kinderman NB, Harrington CA, Drengler SM, Jones KJ. Ribosomal RNA transcriptional activation and processing in hamster facial motoneurons: effects of axotomy with or without exposure to testosterone. J. Comp. Neurol. 1998;401:205–216. [PubMed] [Google Scholar]
- 44.Chkheidze AN, Liebhaber SA. A novel set of nuclear localization signals determine distributions of the αCP RNA-binding proteins. Mol. Cell. Biol. 2003;23:8405–8415. doi: 10.1128/MCB.23.23.8405-8415.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schoenherr CJ, Anderson DJ. Silencing is golden: negative regulation in the control of neuronal gene transcription. Current Opinion Neurobio. 1995;5:566–571. doi: 10.1016/0959-4388(95)80060-3. [DOI] [PubMed] [Google Scholar]
- 46.Kim CS, Hwang CK, Choi HS, Song KY, Law PY, Wei LN, Loh HH. Neuron-restrictive silencer factor (NRSF) functions as a repressor in neuronal cells to regulate the mu opioid receptor gene. J. Biol. Chem. 2004;279:46464–46473. doi: 10.1074/jbc.M403633200. [DOI] [PubMed] [Google Scholar]


