Abstract
Anthropogenic impacts are believed to be the primary threats to the eastern Australian population of grey nurse sharks (Carcharias taurus), which is listed as critically endangered, and the most threatened population globally. Analyses of 235 polymorphic amplified fragment length polymorphisms (AFLP) loci and 700 base pairs of mitochondrial DNA control region provide the first account of genetic variation and geographical partitioning (east and west coasts of Australia, South Africa) in C. taurus. Assignment tests, analysis of relatedness and Fst values all indicate that the Australian populations are isolated from South Africa, with negligible migration between the east and west Australian coasts. There are significant differences in levels of genetic variation among regions. Australian C. taurus, particularly the eastern population, has significantly less AFLP variation than the other sampling localities. Further, the eastern Australian sharks possess only a single mitochondrial haplotype, also suggesting a small number of founding individuals. Therefore, historical, rather than anthropogenic processes most likely account for their depauperate genetic variation. These findings have implications for the viability of the eastern Australian population of grey nurse sharks.
Keywords: Carcharias taurus, low genetic variation, migration
1. Introduction
Recent global declines of many shark populations have drawn attention to the devastating effect of over-exploitation on marine ecosystems (Baum et al. 2003). Interpretation, and possible remediation, of these declines requires both demographic and genetic knowledge. Levels of genetic variation may provide good predictors of population persistence, as average heterozygosities are 35% lower in threatened taxa, than in related, non-threatened taxa (Spielman et al. 2004). Isolation and reduction in population size accelerate the erosion of evolutionary potential and escalate the risk of extinction through inbreeding depression (Frankham et al. 2002; Taylor 2003). However, while reduced genetic variation is frequently symptomatic of recent reductions in population size, it may also reflect more ancient events, pre-dating human influences (Menotti-Raymond & O'Brien 1993). Further, low genetic diversity does not necessarily imply loss, as low mutation rate might also be a factor (Martin 1995).
The grey nurse shark (Carcharias taurus) is a large, coastal shark with a widespread but disjunct distribution (Compagno 1984; figure 1). It is listed as globally vulnerable by the World Conservation Union and as critically endangered in eastern Australian waters (Cavanagh et al. 2003). Recreational and commercial fishing and netting have recently severely reduced the eastern Australian population. Quasi-extinction risk is very high unless the population experiences significant immigration (Otway 2004; Otway et al. 2004). Therefore, we have assessed whether eastern Australian grey nurse sharks are genetically isolated, by estimating genetic partitioning and inferred dispersal among populations from the Pacific and Indian Oceans. Intrapopulation estimates of genetic variability from each of these populations provide baseline information for assessing extinction risk.
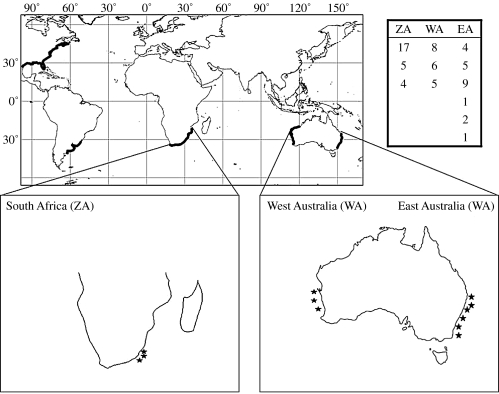
Figure 1.
Distribution of C. taurus (marked black), showing sampling locations (marked with star). Number of individuals sampled at each location from Australian and South African waters (table 1) are arranged in a north–south order.
2. Material and methods
Genomic DNA was extracted from 25 mg of tissue, collected from 67 C. taurus from the eastern and western Australian coasts and South Africa (figure 1), using a QIAmp Tissue Kit (Qiagen Inc., Valencia, CA). AFLP typing of 65 individuals (table 1) involved digestion of 200–400 ng of genomic DNA with the restriction enzymes MseI and EcoRI and generation of profiles using 12 different primer pairs in the final (selective) polymerase chain reaction (PCR). Each primer pair combination had a total of three selective nucleotides on the EcoRI primer (GAC TGC GTA CCA ATT C+ACT, AGT, ATC or AAC) and four selective nucleotides on the MseI primer (GAT GAG TCC TGA GTA A+CAAC, CTGC, CAGC or CTTC). A total of 235 polymorphic AFLP loci were scored. Allele frequencies were estimated with a Bayesian method, assuming a non-uniform prior distribution of allele frequencies (AFLP-SURV v. 1.0; Vekemans 2002). The same software calculated expected heterozygosity (He), pairwise Fst (significance assessed by 1000 permutations) and pairwise relatedness (r). The distance measure, (1−r), was used to generate a neighbourhood joining tree using MEGA 3.0 (Kumar et al. 2004). Genetic subdivision among South African, east and west Australian samples was assessed using STRUCTURE 2.1 (Pritchard et al. 2000; Evanno et al. 2005), assuming no prior information, the admixture model with correlated alleles, and a burn-in phase of 10 000 iterations followed by a run phase of 100 000 iterations. The number of populations (K) ranging from 1 to 3 was tested in three independent runs to establish consistency. The posterior probability was then calculated for each value of K to choose the most likely K. We also used a model with essentially the same parameters as earlier, but providing prior information of population membership, to identify immigrants, or individuals who have recent immigrant ancestry (testing for immigrants over the last two generations; Pritchard et al. 2000).
Table 1.
Genetic diversity at AFLP and mitochondrial markers in C. taurus sampled from east and west Australia and South Africa. (AFLP diversity is estimated by expected heterozygosity (He) and the number of polymorphic loci (PL). Diversity at the mitochondrial control region is described by the number of haplotypes (H), haplotype diversity (Hd) and nucleotide diversity (π). N, number of individuals from which AFLP and mitochondrial data were obtained.)
| AFLP diversity | mitochondrial diversity | ||||||
|---|---|---|---|---|---|---|---|
| sampling region | N | PL | He (±s.e.) | N | H typea,b | Hd (±s.e) | π (±s.e.) |
| eastern Australia | 22 | 59 | 0.103±0.011 | 19 | C19 | 0 | 0 |
| western Australia | 19 | 72 | 0.115±0.011 | 16 | C11 E5 | 0.458±0.024 | 0.0031±0.0001 |
| South Africa | 24 | 167 | 0.145±0.010 | 26 | A3 B8 C11 D4 | 0.717±0.010 | 0.0030±0.0001 |
Haplotypes are coded A–E.
In each region, the number of each of the haplotypes identified (superscript).
For 61 individuals, approximately 700 base pairs of mitochondrial DNA (mtDNA) control region was PCR amplified using primers designed from conserved chondricthyian sequences (forward primer MtGNf (5′-AAY CTG RCA TCT GAT TAA TGC) and reverse primer MtGNr (5′-CATYTTAGCATCTTCAGTGC)). Amplifications (in an MJ Research PTC100 thermocycler) were conducted in 50 μl reactions with initial denaturation for 3 min at 94 °C, followed by six ‘touchdown’ cycles of 94 °C, denaturation for 30 s, annealing temperatures (62, 60, 59, 58, 57 and 55 °C) for 45 s and an extension step of 72 °C for 1 min. On completion of the last touchdown cycle, a further 33 cycles were conducted at 55 °C annealing temperature with a final 72 °C for 3 min. Nucleotide composition was assessed using single-stranded conformational polymorphism analysis (following Sunnucks et al. 2000) and sequencing, which used the forward primer and BigDye termination (Perkin–Elmer Applied Biosystems) and was resolved on an ABI 377 sequencer. The 61 sequences were aligned and edited by eye, leaving 518 sites, of which only five were informative (GeneBank accession numbers: DQ250809–DQ250813). Haplotypic diversity (Hd), nucleotide diversity (π) and theta per DNA sequence (θ) was calculated using the software DnaSP v. 4.10.2 (Rozas et al. 2003).
3. Results
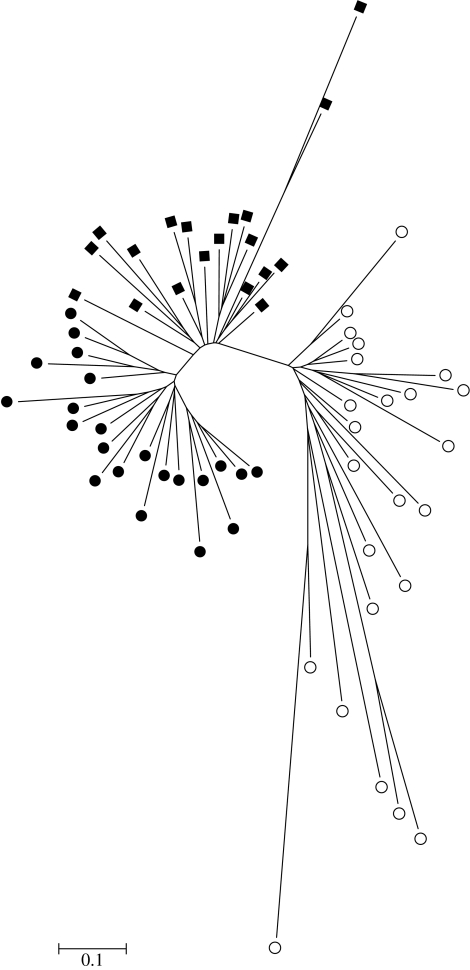
Strong genetic structuring of AFLP's among regions inferred low levels of dispersal. This subdivision was pronounced (Fst: 0.295; p<0.001) with highly significant (p<0.001) Fst values for each pairwise comparison (Fst value between EA and WA=0.243, WA and ZA=0.345 and EA and ZA=0.545). Population subdivision between South Africa and Australia was also strongly supported by analysis with STRUCTURE. However, discrimination between K=2 and 3 was ambiguous (see Pritchard et al. 2000; Evanno et al. 2005 for discussion). Further STRUCTURE analysis, using only Australian samples, clearly resolved two separate populations (eastern and western) thereby supporting K=3. STRUCTURE identified two putative immigrants, or individuals with migrant ancestors (within two generations), within the South African sample (probability of belonging less than 0.050). No immigration was detected within the Australian samples (probability of belonging for each individual greater than 0.999). Clustering of individuals into each sampling region is illustrated by the neighbourhood joining tree of genetic distance (figure 2).
Figure 2.
A neighbourhood joining tree of genetic distance (1−r), clustering individuals sampled from east and west Australia (filled circles and filled squares, respectively) and South Africa (open circles). Note the longer branch lengths (i.e. greater genetic divergence) in the South African samples.
Lower levels of genetic variation in Australian, as compared to South African, populations were supported by both AFLP and mitochondrial data, with the lowest level present in the east (table 1). Comparisons of the number of polymorphic loci present for each of the 12 primer pair combinations further supported this conclusion. The mean number of polymorphic loci (±s.d.) for both the east (4.92±2.82) and west (6.00±2.98) Australian populations were significantly less (Wilcoxon sign; p<0.005) than for the South African sample (13.92±4.87).
4. Discussion
Critically endangered east Australian C. taurus are characterized by less genetic variation than any other population sampled. Genetic variation is highest in the South African population, declining progressively to the east (to western and then eastern Australia; table 1). Limited historical dispersal with ‘sequential founder’ effects, rather than a recent anthropogenic ‘bottleneck’, best explain this pattern. Individuals sampled off Australia are highly divergent from those sampled from South Africa. Further, major differences in genetic structure are evident between eastern and western Australian C. taurus (Fst: 0.243; p<0.001), inferring limited or zero dispersal between these populations. The isolation of Australian C. taurus has important implications for conservation management, since replenishment of the critically endangered eastern and vulnerable western Australian population is unlikely to be achieved via natural migration from more numerous populations elsewhere.
As a species, C. taurus appears to exhibit generally low levels of mitochondrial variation. Although the control region is the most rapidly evolving segment of the mitochondrial genome, little variation was detected (table 1). This level of mtDNA variation in C. taurus may be accounted for by a low rate of molecular evolution in sharks (Martin 1995). Overall, mitochondrial variation detected at the control region in C. taurus (π=0.0025) was lower than reported for some sharks yet similar to others. For example, substantially higher π of 0.0203 was estimated from 95 great white shark (Carcharodon carcharias) samples from Australian and South African waters (calculated from Pardini et al. 2001), while a comparable level of variation was detected in a sample of 323 blacktip sharks (Carcharhinus limbatus), π=0.0021 (Keeney et al. 2005). However, lack of any variation in east Australian C. taurus is notable. Further sampling of the east Australian population is unlikely to yield additional haplotypes (p∼0.1), solved recursively (Hudson 1990) with θ=1.162 calculated from South African and west Australian samples. Genetic variation at nuclear DNA was also lower in east than west Australian C. taurus; however, the magnitude of the difference was greater at the control region. Because effective sizes at mtDNA are approximately one-quarter that of nuclear DNA, differing patterns of variation at nuclear and mtDNA can be expected following population reductions (Fay & Wu 1999).
Low variability in east Australian C. taurus most likely reflects older and slower processes rather than a recent, human-induced population bottleneck. Indeed, human actions leading to the dramatic population size reduction of C. taurus off eastern Australia (particularly over the past 40 years) have not been of sufficient duration to expect substantial erosion of genetic variability, given the species' longevity (25+ years) and the 9–10 years required to reach maturity (Otway 2004). With current population estimates for the Australian east coast of approximately 500 animals (Otway 2004), the effective population size (Ne) is about 50 (Frankham et al. 2002). Even at high rates of decline, for example, if Ne decreased over four generations as follows: 1000, 500, 100 and 50, the expected loss in heterozygosity, as described by is only approximately 3%. In addition, even this negligible loss would be masked to an extent by mainly sampling adults whose genetic constitution was derived from the gene pool available prior to the loss of much diversity.
Here, we provide the first account of genetic structuring in C. taurus, a species in decline throughout much of its distribution, and designated as vulnerable and critically endangered on the west and east coasts of Australia, respectively. Given the ongoing population declines of C. taurus in eastern Australia (Otway 2004; Otway et al. 2004) and negligible migration among Australian populations, extinction is imminent in east Australian waters without urgent conservation efforts.
Acknowledgements
We are grateful to Rory McAuley (WA Fisheries) and the Director and staff of the Natal Sharks Board for collecting samples from West Australia and South Africa, respectively. Phoebe Hill, Sam Hussey and Marita Holley are greatly appreciated for laboratory assistance. We also thank Herman Raadsma at ReproGen—University of Sydney for providing equipment necessary to perform AFLP analysis. Funding was provided by Macquarie University, NSW DPI, Environment Australia, Seaworld Research and Rescue Foundation Inc. (sponsored by Birdseye) and an NRF Mobility Grant to V.P. Finally, we thank Richard Frankham, two anonymous referees and the Editor for valuable suggestions.
References
- Baum J.K, Myers R.A, Kehler D.G, Worm B, Harley S.J, Doherty P.A. Collapse and conservation of shark populations in the northwest Atlantic. Science. 2003;299:389–392. doi: 10.1126/science.1079777. doi:10.1126/science.1079777 [DOI] [PubMed] [Google Scholar]
- Cavanagh, R. D., Kyne, P. M., Fowler, S. L., Musick, J. A., & Bennett, M. B. 2003 The conservation status of Australian chondrichthyans: report of the IUCN Shark Specialist Group Australia and Oceania Regional Red List Workshop, University of Queensland, Brisbane, p. 170.
- Compagno, L. J. V. 1984 Sharks of the world. An annotated and illustrated catalogue of shark species known to date. Part 1. Hexanchiformes to Lamniformes Food and Agriculture Organization (FAO) of the United Nations Species Catalogue, vol. 5. Fisheries Synopsis 125, vol. 4, pp. 1–249. Rome, Italy: FAO.
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 2005;14:74–75. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Fay J, Wu C. A human population bottleneck can account for the discordance between patterns of mitochondrial versus nuclear DNA variation. Mol. Biol. Evol. 1999;16:1003–1005. doi: 10.1093/oxfordjournals.molbev.a026175. [DOI] [PubMed] [Google Scholar]
- Frankham R, Ballou J.D, Briscoe D.A. Introduction to conservation genetics. Cambridge University Press; Cambridge, UK: 2002. [Google Scholar]
- Hudson R.R. Gene genealogies and the coalescent process. Oxford Surv. Evol. Biol. 1990;7:1–45. [Google Scholar]
- Keeney D.B, Heupel M.R, Hueter R.E, Heist E.J. Microsatellite and mitochondrial DNA analyses of the genetic structure of blacktip shark (Carcharhinus limbatus) nurseries in the northwest Atlantic, Gulf of Mexico, and Caribbean Sea. Mol. Ecol. 2005;14:1911–1923. doi: 10.1111/j.1365-294X.2005.02549.x. doi:10.1111/j.1365-294X.2005.02549.x [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. doi:10.1093/bib/5.2.150 [DOI] [PubMed] [Google Scholar]
- Martin A.P. Mitochondrial DNA sequence evolution in sharks: rates, patterns, and phylogenetic inferences. Mol. Biol. Evol. 1995;12:1114–1123. doi: 10.1093/oxfordjournals.molbev.a040285. [DOI] [PubMed] [Google Scholar]
- Menotti-Raymond M, O'Brien S.J. Dating the genetic bottleneck of the African cheetah. Proc. Natl Acad. Sci. USA. 1993;90:3172–3176. doi: 10.1073/pnas.90.8.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otway, N. M. 2004 A grey future for the grey nurse. In Proc. 2004 Conf. of the Australian Association of Veterinary Conservation Biologists (ed. R. Woods), pp. 8–27. Canberra, Australia: Association of Veterinary Conservation Biologists.
- Otway N.M, Bradshaw C.J.A, Harcourt R.G. Estimating the rate of quasi-extinction of the Australian grey nurse shark (Carcharias taurus) population using deterministic age- and stage-classified models. Biol. Conserv. 2004;119:341–350. doi:10.1016/j.biocon.2003.11.017 [Google Scholar]
- Pardini A.T, et al. Sex-biased dispersal of great white sharks. Nature. 2001;412:139–140. doi: 10.1038/35084125. doi:10.1038/35084125 [DOI] [PubMed] [Google Scholar]
- Pritchard J.K, Stephens M, Donelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas J, Sanchez-Delbarrio J.C, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. doi:10.1093/bioinformatics/btg359 [DOI] [PubMed] [Google Scholar]
- Spielman D, Brook B.W, Frankham R. Most species are not driven to extinction before genetic factors impact them. Proc. Natl Acad. Sci. USA. 2004;101:15 261–15 264. doi: 10.1073/pnas.0403809101. doi:10.1073/pnas.0403809101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunnucks P, Wilson A.C, Beheregaray L.B, Zenger K.R, French J, Taylor A.C. SSCP is not so difficult: the application and utility of single stranded conformation polymorphism in evolutionary biology and molecular ecology. Mol. Ecol. 2000;9:1699–1710. doi: 10.1046/j.1365-294x.2000.01084.x. doi:10.1046/j.1365-294x.2000.01084.x [DOI] [PubMed] [Google Scholar]
- Taylor A.C. Assessing the consequences of inbreeding for population fitness: past challenges and future prospects. In: Holt W.V, Pickard A.R, Rodger J, Wildt D.E, editors. Reproductive science and integrated conservation. Cambridge University Press; Cambridge, UK: 2003. pp. 67–81. [Google Scholar]
- Vekemans, X. 2002 AFLP-SURV version 1.0. Distributed by the author. Laboratorie de Génétique et Ecologie Végétale, Université Libre de Bruxelles, Belgium.