Abstract
We tested the hypothesis that malarial parasites (Plasmodium and Haemoproteus) of black-throated blue warblers (Dendroica caerulescens) provide sufficient geographical signal to track population movements between the warbler's breeding and wintering habitats in North America. Our results from 1083 warblers sampled across the species' breeding range indicate that parasite lineages are geographically widespread and do not provide site-specific information. The wide distribution of malarial parasites probably reflects postnatal dispersal of their hosts as well as mixing of breeding populations on the wintering range. When compared to geographically structured parasites of sedentary Caribbean songbirds, patterns of malarial infections in black-throated blue warblers suggest that host–malaria dynamics of migratory and sedentary bird populations may be subject to contrasting selection pressures.
Keywords: Dendroica caerulescens, Haemoproteus, Plasmodium, geographical markers, migratory songbirds, parasites
1. Introduction
Populations of many migratory songbirds are declining precipitously worldwide (Terborgh 1989; Askins et al. 1990), but these changes can vary geographically within a species. To understand the cause of these declines, it may be necessary to identify the wintering locations for each breeding population. Because traditional mark–recapture techniques are relatively inefficient for linking breeding and wintering populations, investigators have concentrated on indirect methods of tracking population movements with genetic (Kimura et al. 2002; Lovette et al. 2003), morphological (AOU 1957) and stable isotope markers (Chamberlain et al. 1997; Rubenstein et al. 2002). Despite some promising results (Hobson 1999; Webster et al. 2002), these efforts have achieved limited success. Recently, it has been suggested that parasites may serve as useful geographical markers for avian populations (Webster et al. 2002).
The black-throated blue warbler, which breeds in eastern North America and winters in the Caribbean basin (Holmes 1994; Graves 1997), has become a model species for studying the connection between breeding and wintering populations (Chamberlain et al. 1997; Rubenstein et al. 2002). One of most common breeding songbirds in the Appalachian Mountains, this species shares similar migration patterns and habitat preferences with some species of conservation concern (Terborgh 1989; Rappole 1995). Based on geographical gradients in stable isotope signatures (hydrogen and carbon) of breeding populations, most black-throated blue warblers wintering in Cuba and Jamaica appear to come from the northern portion of the breeding range, whereas those wintering in Hispaniola and Puerto Rico are from more southerly breeding populations (Rubenstein et al. 2002). However, extensive overlap of isotopic values among populations (Chamberlain et al. 1997; Rubenstein et al. 2002) combined with age-specific and altitudinal effects (Graves et al. 2002) results in poor resolution of connectivity between breeding and wintering areas.
Avian malaria is globally distributed and relatively common in many avian species (Atkinson & Van Riper 1991), though the geographical distribution of genetically distinct parasite lineages is poorly known (Ricklefs & Fallon 2002; Waldenström et al. 2002; Fallon et al. 2005). Although several parasite lineages have extensive geographical distributions in many host species, apparent host specialization of a few malarial parasites (Bensch et al. 2000; Fallon et al. 2005) suggests that at least some Haematozoa may serve as geographical markers for migratory birds. We tested this hypothesis with an analysis of mtDNA lineages (Plasmodium and Haemoproteus spp.) in 1083 male black-throated blue warblers sampled from 32 breeding populations distributed across the species' North American range (figure 1).
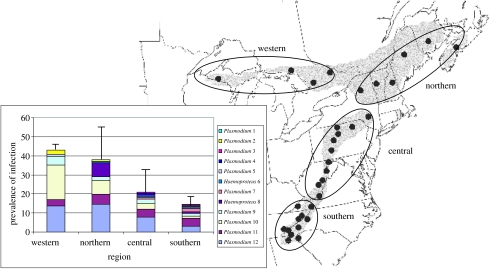
Figure 1.
Breeding distribution and prevalence of infection by geographical region of the black throated blue warbler. Black dots indicate sampling locations. Geographical regions are indicated. Parasite lineages are numbered in order of their abundance from least (1) to most abundant (12). Error bars represent the standard deviation around the mean based on variation in prevalence between populations (n>10) within each region (see electronic supplementary material for sample sizes).
2. Material and methods
(a) Laboratory analysis
Population samples of breeding males were collected across their North American range over a period of 15 years (Graves 1997, 2004; figure 1; electronic supplementary material, table 1). DNA was extracted from blood infused tissue using standard Qiagen extraction kits according to the manufacturer's protocol (Qiagen Inc., Valencia, CA). We screened individuals for parasite infection using a PCR assay based on a conserved RNA region of the parasite's 6 kb mitochondrial genome (Fallon et al. 2003b). Positive and negative control reactions were run with each round of PCR. We genetically typed infections by amplifying and sequencing approximately 320 base pairs of the parasite's cytochrome b gene (Richard et al. 2002; Fallon et al. 2003a). Cyt. b sequences were edited and aligned using Sequencher (Gene Codes Corporation, Ann Arbor, MI) and are available through GenBank (accession numbers DQ321721–DQ321732). Uncorrected sequence divergence of the parasite lineages varied between 1.25 and 12.35%. Values of cyt. b sequence divergence as low as 1.0% have been observed between named species of mammalian malaria parasites (Escalante et al. 1998), and avian parasite lineages differing by as little as 0.5% exhibited complete linkage disequilibrium in one study indicating that lineages described here may constitute full species (Bensch et al. 2004). Sequence divergence below 0.5% was combined to represent single lineages in the cases of P11 and P12. Within lineage variation for these two parasites is 0.3% (1 bp). The remaining lineages have no additional variation.
(b) Statistical and phylogenetic analysis
Heterogeneity of distributions was evaluated using standard goodness of fit tests (G-tests) followed by partitioned analyses (Sokal & Rohlf 1995). We present a 50% majority rule consensus tree based on Bayesian analysis using the general time reversible model (Swofford et al. 1996) with partitioned codon positions and variable sites following a gamma distribution. We ran four Markov chain Monte Carlo chains for 1 000 000 generations, using random trees as starting points, sampling every 100th generation, and discarding the first 1000 trees as burn-in (figure 2). An additional run with 2 000 000 generations, sampling every 1000th generation and discarding the first 500 trees produced an identical topology and posterior clade probabilities. The tree was rooted using a published sequence of Leucocytozoon spp. (Perkins & Schall 2002). We tested for the effect of phylogeny on the abundance and regional geographical distribution of parasite lineages using Phylogenetic Independence (v. 2.0, McGill University, Montreal).
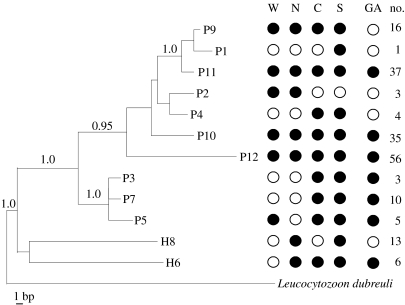
Figure 2.
Phylogenetic relationships of the 12 parasite lineages infecting black throated blue warblers (see §2). Parasite lineage numbers were ranked from least abundant (1) to most abundant (12). Parasite abundance was uncorrelated with phylogeny (Cstat=0.1183, p=0.283). Whether a parasite lineage is geographically widespread or restricted is also not explained by phylogenetic relationships (Cstat=0.0212, p=0.416). The presence of each parasite lineage in four North American locations plus the Greater Antilles is indicated by black solid fill. W, western; N, northern; C, central; S, southern; GA, Greater Antilles; no., number of representative sequences for each lineage from North American sample.
3. Results
We detected 236 infections (21.8% prevalence), 189 of which we genotyped, in the 1083 warblers. Due to the small number of infections in some populations, we grouped our results into four geographical regions for analysis. The prevalence of infection varied regionally with a higher percentage occurring in western (43.1%) and northern (38.3%) populations compared to southern populations (14.5%; G=64.2, d.f.=3, p<0.001, figure 1). We identified 10 genetically distinct Plasmodium lineages and two Haemoproteus lineages. The accumulation of new lineages approached an asymptote after the first 25 infected individuals were genotyped (electronic supplementary material, figure 3) and the estimated species richness of parasites in breeding populations of black-throated blue warblers was less than 13 lineages (Chao 1984).
Three of the 12 parasite lineages accounted for nearly 70% of all infections and were found in each of the four geographical regions (figures 1 and 2). Prevalence of the two most common lineages of Plasmodium varied regionally: lineage ‘P10’ was more common in the west (G=38.3, d.f.=3, p<0.01), and lineage ‘P12’ was more prevalent in the western and central populations (G=22.8, d.f.=3, p<0.01). The third common lineage, Plasmodium ‘P11’, showed no geographical trends in prevalence (G=3.1, d.f.=3, p=0.40).
The remaining lineages were detected at substantially lower frequencies. With the exception of Haemoproteus lineage ‘H8’, which was more common than expected in the north and absent from the central region (G=20.1, d.f.=3, p<0.001), the rarer lineages were also geographically uninformative. The phylogenetic relationships of the 12 parasite lineages provide no additional resolution in terms of distinguishing the parasites' distributions (figure 2).
4. Discussion
Despite predictions that vector-transmitted parasites, such as those that cause avian malaria, might provide site-specific information in migratory birds (Webster et al. 2002), we found no evidence that distinctive lineages of malaria were restricted to a particular breeding population or even a regional subset of the North American distribution of the black-throated blue warbler. All four geographical regions of the breeding range of the black-throated blue warbler had similar parasite ‘signatures’ with the three most common lineages constituting 58–82% of the infections in each region. Populations in the southern Appalachians exhibited the most diverse malarial fauna, probably reflecting the extensive sampling in this region (electronic supplementary material). With the exception of P1 even the rarer lineages were found in more than one region (figure 2), and regardless of minor regional variation in the prevalence of two lineages (P10 and P12), the presence of the three common malarial parasite lineages in each of the regions eliminates their usefulness as geographical markers. Given the spatial distribution of sampling and the large number of specimens assayed thus far, additional sampling is unlikely to discover geographically informative parasite lineages. This indicates that malarial parasites have limited usefulness as geographical markers for individuals of this species.
Although most parasite lineages appeared to be geographically widespread, the prevalence of infection in black-throated blue warblers was correlated with latitude, with populations in the western and northern portions of the breeding range exhibiting the highest percentage of infected individuals (figure 1). Variation in the prevalence of infection within a species has been linked to the abundance of vector species that transmit infections (Sol et al. 2000). However, information on the abundance and species diversity of malarial vectors within the breeding and wintering ranges of the black-throated blue warbler is unavailable. Nevertheless, regional variation in vector ecology, if it exists, has apparently had minimal effect on the geographical distribution of parasite lineages among breeding populations of warblers.
The ubiquitous distribution of common malarial parasite lineages in the migratory black-throated blue warbler stands in distinct contrast to the geographically structured patterns of malarial infections found in the mostly sedentary populations of island birds in the Caribbean region. Over a span of 8° of latitude in the Lesser Antilles, the prevalence of malarial infections as well as the presence or absence of parasite lineages varies from island to island within host species (Fallon et al. 2003a, 2005). Half (n=6) of the parasite lineages detected in black-throated blue warblers also occur in sedentary species in the Greater Antilles, the primary wintering area of the warbler (Fallon et al. 2003a, 2005; Ricklefs et al. 2004; figure 2). Two of the six shared lineages (P11, P12) were recovered from four different host species endemic to the Greater Antilles, indicating transmission of malarial infections on the wintering grounds. One lineage (P11) has been reported from a variety of both sedentary and migratory host species in the Lesser Antilles and on the South American mainland in Venezuela (Fallon et al. 2003a, 2005).
The absence of comparable geographical structuring of malarial parasite lineages within breeding populations of black-throated blue warblers may reflect natal dispersal on the breeding grounds (Graves 1997; Graves et al. 2002), or mixing of breeding populations on the wintering grounds (Chamberlain et al. 1997; Rubenstein et al. 2002), both of which could serve to homogenize parasite communities given transmission in the two regions (Waldenström et al. 2002; Ricklefs et al. 2004). Regardless, malarial parasite lineages clearly do not provide geographical resolution to populations of this model migratory bird. More significantly, our results suggest that host–malaria dynamics in migratory and sedentary species may be subject to contrasting selection pressures leading to variable patterns of infection, including specialization across landscapes (Thompson 1994; Møller & Erritzøe 1998; Fallon et al. 2003a).
Acknowledgments
Funding for this research was provided by the James Bond Fund, the Alexander Wetmore Fund, the Research Opportunities Fund, and the Biodiversity Surveys and Inventory Program of the National Museum of Natural History, Smithsonian Institution. Additional support was provided by the National Institutes of Health (GM63258-02). We would like to thank Robert Ricklefs for helpful comments on an earlier draft of the manuscript. Agencies that granted licences to sample populations are listed in the electronic supplementary material.
Supplementary Material
References
- AOU . 5th edn. American Ornithologists' Union (AOU); Baltimore, MD: 1957. Check-list of North American birds. [DOI] [PubMed] [Google Scholar]
- Askins R.A, Lynch J.F, Greenberg R. Population declines in migratory birds in eastern North America. Curr. Ornithol. 1990;7:1–57. [Google Scholar]
- Atkinson C.T, Van Riper C.I. Pathogenicity and epizootiology of avian haematozoa: Plasmodium, Leucocytozoon and Haemoproteus. In: Loye J.E, Zuk M, editors. Bird–parasite interactions. Ecology, evolution and behavior. Oxford University Press; New York, NY: 1991. pp. 19–48. [Google Scholar]
- Bensch S, Stjernman M, Hasselquist D, Ostman O, Hansson B, Westerdahl H, Pinheiro R.T. Host specificity in avian blood parasites: a study of Plasmodium and Haemoproteus mitochondrial DNA amplified from birds. Proc. R. Soc. B. 2000;267:1583–1589. doi: 10.1098/rspb.2000.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensch S, Perex-Tris J, Waldenstrom J, Hellgren O. Linkage between nuclear and mitochondrial DNA sequences in avian malaria parasites: multiple cases of cryptic speciation? Evolution. 2004;58:1617–1621. doi: 10.1111/j.0014-3820.2004.tb01742.x. [DOI] [PubMed] [Google Scholar]
- Chamberlain C.P, Blum J.D, Holmes R.T, Feng X, Sherry T.W, Graves G.R. The use of isotope tracers for identifying populations of migratory birds. Oecologia. 1997;109:132–141. doi: 10.1007/s004420050067. [DOI] [PubMed] [Google Scholar]
- Chao A. Non-parametric estimation of the number of classes in a population. Scand. J. Stat. 1984;11:265–270. [Google Scholar]
- Escalante A.A, Freeland D.E, Collins W.E, Lal A.A. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc. Natl Acad. Sci. USA. 1998;95:8124–8129. doi: 10.1073/pnas.95.14.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon S.M, Bermingham E, Ricklefs R.E. Island and taxon effects in parasitism revisited: avian malaria in the Lesser Antilles. Evolution. 2003a;57:606–615. doi: 10.1111/j.0014-3820.2003.tb01552.x. [DOI] [PubMed] [Google Scholar]
- Fallon S.M, Ricklefs R.E, Swanson B.L, Bermingham E. Detecting avian malaria: an improved PCR diagnostic. J. Parasitol. 2003b;89:1044–1047. doi: 10.1645/GE-3157. [DOI] [PubMed] [Google Scholar]
- Fallon S.M, Bermingham E, Ricklefs R.E. Host specialization and geographic localization of avian malaria parasites: a regional analysis in the Lesser Antilles. Am. Nat. 2005;165:466–480. doi: 10.1086/428430. [DOI] [PubMed] [Google Scholar]
- Graves G.R. Geographic clines of age ratios of black-throated blue warblers (Dendroica caerulescens) Ecology. 1997;78:2524–2531. [Google Scholar]
- Graves G.R. Testicular volume and asymmetry are age-dependent in black-throated blue warblers (Dendroica caerulescens) Auk. 2004;121:473–485. [Google Scholar]
- Graves G.R, Romanek C.S, Rodriguez Navarro A. Stable isotope signature of philopatry and dispersal in a migratory songbird. Proc. Natl Acad. Sci. USA. 2002;99:8096–8100. doi: 10.1073/pnas.082240899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson K.A. Tracing origins and migration of wildlife using stable isotopes: a review. Oecologia. 1999;120:314–326. doi: 10.1007/s004420050865. [DOI] [PubMed] [Google Scholar]
- Holmes R.T. Black-throated blue warbler (Dendroica caerulescens) In: Poole A, Gill F, editors. The birds of North America, No. 87. The Academy of Natural Sciences; The American Ornithologists' Union; Philadelphia, PA; Washington, DC, USA: 1994. [Google Scholar]
- Kimura M, Clegg S.M, Lovette I.J, Holder K.R, Girman D.J, Milá B, Wade P, Smith T.B. Phylogeographical approaches to assessing demographic connectivity between breeding and overwintering regions in a Nearctic–Neotropical warbler (Wilsonia pusilla) Mol. Ecol. 2002;11:1605–1616. doi: 10.1046/j.1365-294x.2002.01551.x. [DOI] [PubMed] [Google Scholar]
- Lovette I.J, Clegg S.M, Smith T.B. Limited utility of mtDNA markers for determining connectivity among breeding and overwintering locations in three Neotropical migrant birds. Conserv. Biol. 2003;18:156–166. [Google Scholar]
- Møller A.P, Erritzøe J. Host immune defence and migration in birds. Evol. Ecol. 1998;12:945–953. [Google Scholar]
- Perkins S.L, Schall J.J. A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J. Parasitol. 2002;88:972–978. doi: 10.1645/0022-3395(2002)088[0972:AMPOMP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Rappole J.H. Smithsonian Institution Press; Washington, DC: 1995. The ecology of migrant birds. [Google Scholar]
- Richard F.A, Sehgal R.N.M, Jones H.I, Smith T.B. A comparative analysis of PCR-based detection methods for avian malaria. J. Parasitol. 2002;88:819–822. doi: 10.1645/0022-3395(2002)088[0819:ACAOPB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Ricklefs R.E, Fallon S.M. Diversification and host switching in avian malaria parasites. Proc. R. Soc. B. 2002;269:885–892. doi: 10.1098/rspb.2001.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklefs R.E, Fallon S.M, Bermingham E. Evolutionary relationships, cospeciation, and host switching in avian malaria parasites. Syst. Biol. 2004;53:111–119. doi: 10.1080/10635150490264987. [DOI] [PubMed] [Google Scholar]
- Rubenstein D.R, Chamberlain C.P, Holmes R.T, Ayres M.P, Waldbauer J.R, Graves G.R, Tuross N.C. Linking breeding and wintering ranges of a migratory songbird using stable isotopes. Science. 2002;295:1062–1065. doi: 10.1126/science.1067124. [DOI] [PubMed] [Google Scholar]
- Sokal R.R, Rohlf F.J. 3rd edn. W.H. Freeman and Company; New York: 1995. Biometry. [Google Scholar]
- Sol D, Jovani R, Torres J. Geographical variation in blood parasites in feral pigeons: the role of vectors. Ecography. 2000;23:307–314. [Google Scholar]
- Swofford D.L, Olson G.J, Waddell P.J, Hillis D.M. Phylogenetic inference. In: Hillis D.M, Moritz C, Mable B.K, editors. Molecular systematics. 2nd edn. Sinauer; Sunderland, MA: 1996. pp. 430–459. [Google Scholar]
- Terborgh J. Princeton University Press; Princeton, NJ: 1989. Where have all the birds gone? [Google Scholar]
- Thompson J. University of Chicago Press; Chicago, IL: 1994. The coevolutionary process. [Google Scholar]
- Waldenström J, Bensch S, Kiboi S, Hasselquist D, Ottosson U. Cross-species infection of blood parasites between resident and migratory songbirds in Africa. Mol. Ecol. 2002;11:1545–1554. doi: 10.1046/j.1365-294x.2002.01523.x. [DOI] [PubMed] [Google Scholar]
- Webster M.S, Marra P.P, Haig S.M, Bensch S, Holmes R.T. Links between worlds: unraveling migratory connectivity. Trends Ecol. Evol. 2002;17:76–83. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.