Abstract
Integrins are cell adhesion molecules that play critical roles in development, wound healing, hemostasis, immunity and cancer. Advances in the past two years have shed light on the structural basis for integrin regulation and signaling, especially on how global conformational changes between bent and extended conformations relate to the inter-domain and intra-domain shape shifting that regulates affinity for ligand. The downward movements of the C-terminal helices of the α I and β I domains and the swing-out of the hybrid domain play pivotal roles in integrin conformational signaling. Experiments have also shown that integrins transmit bidirectional signals across the plasma membrane by coupling extracellular conformational change with an unclasping and separation of the α and β transmembrane and cytoplasmic domains.
Introduction
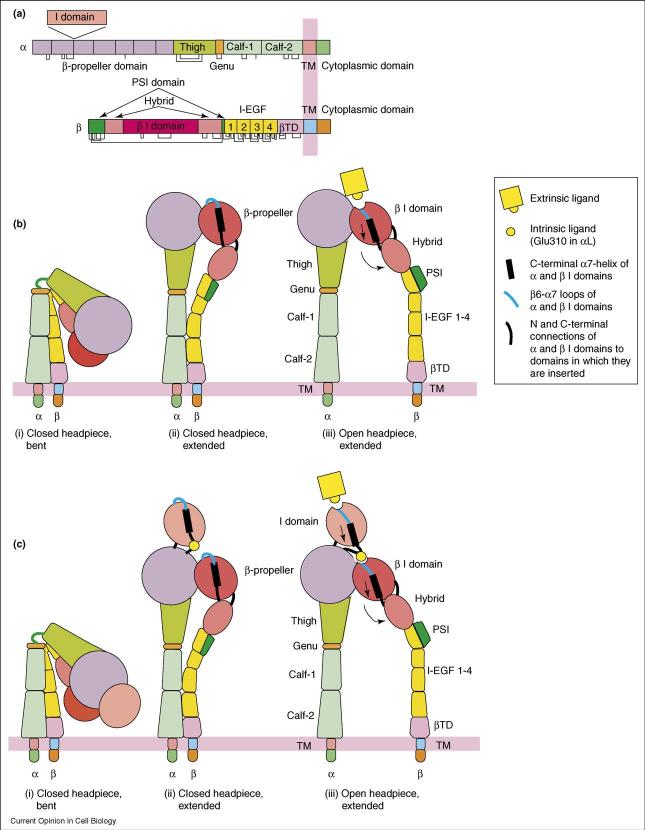
Integrins are cell adhesion molecules that mediate cell–cell, cell–extracellular matrix and cell–pathogen interactions. They transmit signals bidirectionally across the plasma membrane and regulate many biological functions, including wound healing, cell differentiation and cell migration. Integrins contain two non-covalently associated, type I transmembrane (TM) glycoprotein α and β subunits with large extracellular domains, single-spanning TM domains and short cytoplasmic domains (Figure 1a). The structures of the extracellular fragment of integrin αVβ3 revealed an unexpected, compact, V-shaped conformation, with each leg bent (Figure 1b) [1,2]. Recently, an increasing number of studies have together established that the bent conformation represents the physiological low-affinity state, whereas priming and ligand binding induce a large-scale conformational rearrangement in which the integrin extends with a ‘switchblade’-like motion (Figure 1b and c) [3-5,6••]. In this review, we focus on recent progress on how signals are communicated between the ligand binding domains and the plasma membrane at the molecular and atomic level.
Figure 1.
Integrin architecture and conformational changes associated with affinity regulation. (a) Organization of domains within the primary structures. (b,c) Conformational change of integrins lacking an I domain (b) or containing an α I domain (c). The domains are shown with the same color scheme as in (a).
Integrin ectodomain crystal structures
The integrin β-subunits contain very sophisticated domain insertions: the β I domain is inserted in the hybrid domain, which is in turn inserted in the PSI (for plexins, semaphorins, and integrins) domain (Figure 1a) [6••]. These domain insertions play a critical role in integrin signal transmission. The β I domain directly binds ligands in integrins that lack α I domains, and indirectly regulates ligand binding by integrins that contain α I domains. The structure of the β I domain was first solved in the context of αVβ3 extracellular domains in the absence of ligand [1]. The β I domain is structurally homologous to integrin α subunit I domains, which have been solved only as isolated domains, and are described in more detail below. In α I domains, rearrangements in loops surrounding the metal-ion-dependent adhesion site (MIDAS) increase affinity for ligand, and are linked to downward displacement of the α7-helix. Soaking of a ligand-mimetic Arg-Gly-Asp-containing cyclic peptide into the integrin αVβ3 crystals revealed that the Arg binds the αV β-propeller domain and the Asp binds a metal ion held in the MIDAS of the β3 I domain [2]. Movements of residues near the MIDAS in the β1-α1 loop, α1-helix, and β6-α7 loop were seen that enabled ligand binding in the closed state (Figure 2a). However, downward displacement of the α7-helix was not seen (Figure 2a), and it was therefore suggested that the α I and the β I domains are activated by distinct mechanisms [2]. However, subsequent mutagenesis studies [7-9,10•,11] and the structure of the αIIbβ3 headpiece co-crystallized with different ligands [6••] revealed downward α7-helix displacement in the open, high-affinity state of the β I domain (Figure 2a), and marked structural similarity between α I and β I domain allostery.
Figure 2.
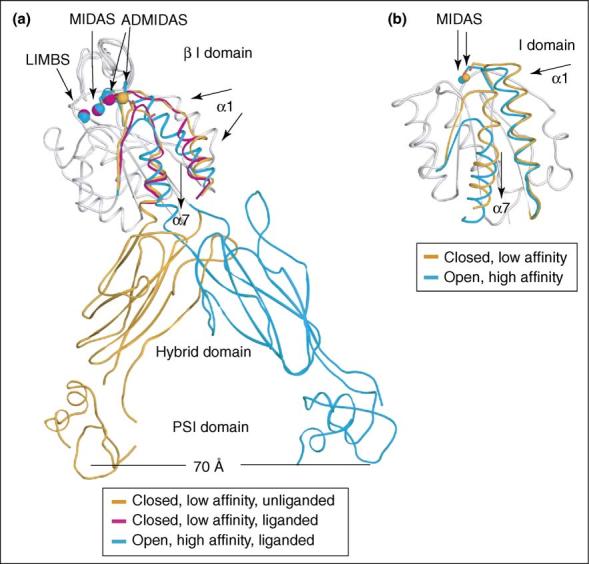
Conformational regulation in integrin headpiece domains. (a) Overview of the movements of the β I, hybrid, and PSI domains. Non-moving segments of the β I backbone are shown as a grey worm. Moving segments are color-coded. The downward movement of the α7 helix is coupled to the swing-out of the hybrid domain, which in turn plays a critical role in transmitting signals between the ligand-binding headpiece and the integrin legs. (b) Conformational change of the α I domain. Non-moving segments of the backbone are shown as a grey worm. The moving segments, shown as Cα-traces, of the closed (gold) and open (cyan) αM I domains and their MIDAS metal ions are shown, and direction of movement is shown with arrows. The downward movement of the α7 helix plays a critical role in transmitting signals between the α I domain and the β I domain.
The β3 subunit I, hybrid, and PSI domains from the closed, low-affinity unliganded αVβ3 structure, the closed, low-affinity liganded (ligand soaked) αVβ3 structure, and the open, high-affinity liganded (ligand co-crystallized) αIIbβ3 structure are compared in Figure 2a. The liganded, high-affinity αIIbβ3 headpiece structure enables atomic-level understanding of the mechanism of integrin activation [6••]. In the high-affinity, liganded β I domain compared with the low-affinity, unliganded β I domain, there are concerted movements of the β1-α1 and β6-α7 loops surrounding the ligand-binding pocket and of the α1 and α7 helices (Figure 2). Coordination of the Met335 backbone carbonyl in the β6-α7 loop to the ADMIDAS (adjacent to MIDAS) Ca2+ in the low-affinity, unliganded conformation is broken in the high-affinity, liganded conformation. This enables the ADMIDAS metal, and residues in the β1-α1 loop that coordinate to both the ADMIDAS and the MIDAS metals, to shift markedly, remodeling the ligand binding site and increasing affinity for ligand. Movements of the α1-helix, β6-α7 loop and α7-helix are tightly coupled, so that reshaping to the high-affinity ligand-binding site is allosterically linked to downward movement of the α7-helix. This linkage is critical for propagation of conformational signals from the ligand-binding pocket to the other integrin domains and vice versa (Figure 2a).
The orientation between the β I and hybrid domains appears to be the critical ‘translator’ converting global conformational change into local intradomain conformational changes that regulate affinity (Figure 2a). The piston-like displacement of the β I domain α7-helix in the high-affinity, liganded crystal structure results in complete remodeling of the interface between these domains, leading to the swing-out of the hybrid domain (Figure 1b, panel iii and Figure 2a) [6••]. Relative to the closed conformation, the hybrid domain swings out about 60°, causing the knees of the α and β subunits to separate by 70 Å
Structures of the β3 and β2 integrin PSI domains and β2 I-EGF1 domain [6••,12,13•] revised the connectivity of the previously identified long-range disulfide bond, which is now shown to link β3 Cys-13 to Cys-435 (or β2 Cys-11 to Cys-425). The structures show that this long-range disulfide bond occurs within the PSI domain, and therefore the hybrid domain is inserted in the PSI domain (Figure 1a). During the rearrangement of the headpiece between the closed and open conformations, there is no change in the hybrid/PSI domain interface. Therefore, this rigid interface, which is reinforced by the two polypeptide chain connections, nearby disulfide bonds and an Arg deeply buried in the interface, enables the PSI domain to amplify the leg separation triggered by the swing-out of the hybrid domain [6••,13•] (Figure 2a). The PSI and I-EGF1 domains are also shown to be intimately associated so that the hybrid and PSI/I-EGF1 domains move as a rigid unit [13•]. Some activating antibodies bind to the PSI domain and induce the high affinity state [14,15].
The knee of the β subunit is located between the PSI/I-EGF1 and I-EGF2 domains; the knee or genu of the α subunit is a small Ca2+-binding loop between the thigh and calf-1 domains. Work using an αL antibody that reports extension and maps to the inner face of the thigh domain, and which requires the genu and a Ca2+-coordinating residue donated by the calf-1 domain, suggests that integrin α-subunit extension occurs by movement of the thigh–genu linker (αV residues 594-595), a conclusion that is supported by structural inspection [16].
A large range of studies support the importance of integrin extension and hybrid-domain swing-out in integrin activation. Perhaps most definitive are the crystal structures of four independent examples of the αIIbβ3 head-piece, in two different crystal forms, with three different ligands or a pseudo ligand bound, all of which reveal similar hybrid domain swing-out [6••]. The swing-out of the hybrid domain necessitates the existence of the extended conformation, because the hybrid domain is central in the interfaces that are buried in the bent conformation; these interfaces are completely broken by hybrid domain swing-out [6••].
Integrin electron microscopy studies
Early electron microscopy (EM) studies revealed extended conformations. A later EM study revealed the bent conformation and showed it had low affinity for ligand and was stabilized by Ca2+ and close association of the α and β subunits near their junction with the membrane, and that activation with Mn2+ or breaking the juxtamembrane clasp favored extension [3]. In integrins on the cell surface, stabilizing the bent conformation with mutationally introduced disulfide bonds inhibits ligand binding [3]. Both closed and open headpiece conformations, i.e. with the hybrid domains swung in or out, respectively (Figure 1b), were seen in unclasped and Mn2+-treated extended integrins; however, cyclic Arg-Gly-Asp not only induced extension but also specifically stabilized the open headpiece conformation [3]. Recent studies with the I-domain-containing integrins αLβ2 and αXβ2 reveal the same three overall conformational states. Furthermore, Fab fragments of antibodies known to induce or report the active conformation bind exclusively to the extended conformation, with both open and closed headpiece conformations present, definitively establishing that extension is sufficient to activate integrins on the cell surface, and that physiologic agonists such as chemoattractants and agents such as phorbol myristate acetate and the talin head domain induce cell surface integrin extension [17••]. Electron tomography of negatively stained, active detergent-soluble αIIbβ3 purified on an Arg-Gly-Asp peptide affinity column reveals an extended conformation with >90% of particles showing an open headpiece structure that matches perfectly [18•] the open, liganded αIIbβ3 headpiece crystal structure [6••].
Two notable studies differ in their conclusions from those described above. Cryo-EM reconstructions of detergent-soluble αIIbβ3 molecules revealed a conformation that is intermediate between bent and extended conformations [19]. However, for particles the size of integrins, cryo-EM cannot distinguish between a particle in two different orientations or two different conformations [20]. Since preparations of integrins, including αIIbβ3, often contain a mixture of particles with different conformations [3], the intermediate αIIbβ3 conformation may have resulted from averaging together particles in extended and bent conformations. Other observations support this speculation, since to fit an atomic model into the intermediate αIIbβ3 EM density, marked changes in orientation at the β-propeller/thigh interface and calf-1/calf-2 interface were required [19] that are inconsistent with recent EM studies [3,4,17••,18•].
A negative stain study of integrin αVβ3 in 0.2 mM Mn2+ or 0.2 mM Mn2+ with a fibronectin fragment revealed a bent conformation [21], whereas a study of αVβ3 with 1mM Mn2+, or cyclic Arg-Gly-Asp in the presence of either 1 mM Mn2+ or 5 mM Ca2+, revealed predominantly extended conformations [3]. These differences might reflect the different ligands or Mn2+ concentrations used. Another important difference is the extensive aggregation present in the one field view shown in Adair et al. [21] but not in the eight field views shown in Takagi et al. [3], which led Adair et al. [21] to state that, “We cannot exclude the possibility that unsampled regions on the grid might have preferentially arisen from aggregated extended forms.”
It is interesting to note the differing methodologies employed in 3D reconstructions of negatively stained integrins [20,22]. In the random conical tilt [4] and tomography studies [18•], each particle was imaged at two or 23 different tilt angles, respectively, and 3D electron density maps were then computed independently of any crystal structure information. The EM density showed excellent agreement with crystal structures that were currently [4,18•] or only subsequently [4] available. In the angular reconstitution study [21], each particle was imaged at a single angle. Reconstruction used resolution-filtered crystal structures as starting models, and particles were automatically selected for use in reconstruction if they were similar to 2D projections of these models. The final models are similar to the starting atomic models, except that in the liganded model several of the fibronectin domains are no longer present and density for I-EGF domains 1 and 2, which was absent from the starting model, was acquired; however, in the unliganded model, density for I-EGF domains 1 and 2 remains missing in the final model.
Other studies on ectodomain conformation
Aside from structural work [3,4,6••,17••,18•,23] integrin extension and hybrid domain swing-out are supported by a wide range of other studies. Stabilizing the open head-piece by mutationally introducing an N-glycosylation site into the hybrid-β I domain interface increases ligand-binding affinity [24,25]. An allosteric β1 antibody that inhibits ligand binding has been shown by epitope mapping and EM to restrict the swing-out of the hybrid domain [25]. The functional properties of a β2 mAb suggest it also inhibits by blocking signal transmission at the β I-hybrid domain interface [26]. Activation-dependent mAbs that map to the inner face of the hybrid domain support conformational change between the β I and hybrid domains [9,27]. Epitope exposure suggests that ligand binding and a mutation of the β I domain α7 helix that stabilizes the high affinity state induce hybrid domain swing-out, confirming the relationship between movement of the α7 helix and hybrid domain swing-out [9]. Integrin extension on the cell surface was confirmed by studies using fluorescence resonance energy transfer (FRET) between fluorescent ligand-bound integrins and lipophilic probes [28].
As an alternative or supplement to integrin extension and hybrid domain swing-out, a ‘deadbolt’ model has been proposed in which interaction at a very small 60 Å2 interface between the β-tail domain CD loop (the dead-bolt) and the β I domain α7 helix is critical for stabilizing integrins in the low affinity state [29]. Since hybrid domain swing-out requires β6-α7 loop and α7-helix displacement, mutation of the β-tail domain CD loop is required to test this model. We found that deleting β3 integrin CD loop residues 672–674 or mutating these residues to Ala has no effect on ligand binding or activation epitope exposure by integrins αIIbβ3 and αVβ3 (our unpublished data). Therefore, the β-tail CD loop does not function as a deadbolt.
Integrins containing an α I domain
Compared to the integrins lacking an α I domain, conformational regulation of integrins containing an α I domain requires the additional step of transmission of allostery from the β I domain to the α I domain (Figure 1c). Crystal structures of α I domains reveal three distinct conformations, namely closed, intermediate and open [30,31]. They differ not only in the coordination of residues with the MIDAS, but in the structure of surrounding loops and in the positions of the β6-α7 loop and α1 and α7 helices (Figure 2b). Introducing pairs of cysteines to stabilize the β6-α7 loop in the intermediate and open conformations led to 500- and 10,000-fold higher affinity to ICAM-1, respectively [31]. Molecular dynamic studies showed that the intermediate conformation was on the pathway from the closed to the open conformation of the αL and αM I domains, but not the α1 and α2 I domains [32]. The study provides strong support for the idea that the intermediate conformation could be of physiologic importance for fine regulation of integrin affinity.
EM studies of αXβ2 and αLβ2 integrins reveal no activation-dependent change in α I domain orientation relative to the β propeller domain analogous to that observed between the β I domain and hybrid domain [17••]. This is consistent with a proposal that the α I domain α7-helix transmits allostery between the α I MIDAS and the β I MIDAS. That is, it is proposed that in the active state, downward movement of the α I domain α7-helix enables an invariant Glu residue that is present a few residues after the α7-helix to act as an ‘intrinsic ligand’ and engage the β I MIDAS [33,34•]. Yang et al. showed that individual mutation of the αL linker residue Glu-310 or β2 MIDAS residues Ala-210 or Tyr-115 to cysteine abolishes I domain activation, whereas the double mutations of αL-E310C with either β2-A210C or β2-Y115C form disulfide bonds that constitutively activate ligand binding [34•]. The activation effect of the disulfide mutant is susceptible to small molecule antagonists that bind underneath the I domain α7-helix and certain allosteric antagonistic antibodies. This study provides direct evidence for an activating interaction between αL residue Glu-310 and the β2 MIDAS (Figure 1c, panel iii), and suggests that the α7-helix and its linker are better modeled as a pull spring than a bell rope [34•].
Conformational change in integrin cytoplasmic and transmembrane domains
Recently, the basis for integrin activation across the plasma membrane has also been studied. Separation of integrin legs results in integrin activation [5,35,36,37•], suggesting that association of the integrin TM and cytoplasmic domains stabilizes integrins in the low affinity state. FRET shows that in the resting state the integrin α and β subunit cytoplasmic domains are close to one another, but undergo significant spatial separation upon inside-out activation induced by G-protein-coupled receptors, phorbol ester or talin head domain, or upon outside-in signaling induced by ligand binding [5]. NMR studies of the integrin cytoplasmic tails suggest that their association is weak, with significant differences between published structures of associated cytoplasmic domains [38•,39,40], or with association between α and β subunit cytoplasmic domains being undetectable [41]. How the talin head domain and filamin bind to the integrin β cytoplasmic domain and activate integrins has been revealed by NMR structures [42-45].
Disulfide scanning of the exofacial portions of the TM domains showed a specific α-helical interface between the α and β? TM domains in the resting state [37•]. The two TM domains separate rather than rearrange after activation of integrins from inside the cell. Introduction of disulfide bridges to prevent or reverse separation abolished the activating effect of cytoplasmic mutations [37•]. Several other mutagenesis studies also suggest that a specific interface stabilizes integrins in the resting state [46,47,48•]. Modeling of the integrin TM domain interface, with or without experimental data, has resulted in models with different interfaces [37•,48•,49,50]. Further experimental data and more comprehensive analysis are required. Homomeric TM domain association following heterodimeric TM dissociation has been proposed [51], but in subsequent studies it has been shown that this does not occur as a consequence of integrin activation by α and β subunit TM separation, although it might occur after binding to multivalent ligands [47,52]. Thus, numerous studies from different labs suggest that integrin bidirectional signaling across the plasma membrane is accomplished by coupling extracellular conformational change to an unclasping and separation of the α and β TM and cytoplasmic domains.
Conclusions
Recent structural, biochemical and biophysical studies have greatly advanced our understanding of the mechanisms underlying integrin bidirectional signaling across the plasma membrane. We should always consider that integrins are in dynamic equilibrium among many different conformational states, rather than locked in one specific state. As reviewed above, intracellular signals and ligand binding act by shifting the equilibrium and altering the population of the different conformational states. It will be of great interest to use biophysical methods to probe the dynamics of integrin signaling under physiological conditions.
Acknowledgements
The work was supported by NIH grants (HL48675, CA31798 and AI72765 to T.A.S.) and American Heart Association grant (0535403T to B.H.L.).
References and recommended reading
Papers of particular interest, published within the 2-year period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Xiong J-P, Stehle T, Diefenbach B, Zhang R, Dunker R, Scott DL, Joachimiak A, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin αVβ3. Science. 2001;294:339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiong JP, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin αVβ3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- 3.Takagi J, Petre BM, Walz T, Springer TA. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 2002;110:599–611. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 4.Takagi J, Strokovich K, Springer TA, Walz T. Structure of integrin α5β1 in complex with fibronectin. EMBO J. 2003;22:4607–4615. doi: 10.1093/emboj/cdg445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim M, Carman CV, Springer TA. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 2003;301:1720–1725. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- 6••.Xiao T, Takagi J, Wang J-h, Coller BS, Springer TA. Structural basis for allostery in integrins and binding of ligand-mimetic therapeutics to the platelet receptor for fibrinogen. Nature. 2004;432:59–67. doi: 10.1038/nature02976. The crystal structure of the αIIbβ3 open headpiece sheds light on allosteric regulation of the conformation and affinity for ligand of the integrin ectodomain, and how fibrinogen-mimetic therapeutics bind to αIIbβ3. The paper reveals the atomic basis for the conformational regulation of the β I domain and inter-domain communication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo B-H, Takagi J, Springer TA. Locking the β3 integrin I-like domain into high and low affinity conformations with disulfides. J Biol Chem. 2004;279:10215–10221. doi: 10.1074/jbc.M312732200. [DOI] [PubMed] [Google Scholar]
- 8.Yang W, Shimaoka M, Chen JF, Springer TA. Activation of integrin β subunit I-like domains by one-turn C-terminal α-helix deletions. Proc Natl Acad Sci USA. 2004;101:2333–2338. doi: 10.1073/pnas.0307291101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mould AP, Barton SJ, Askari JA, McEwan PA, Buckley PA, Craig SE, Humphries MJ. Conformational changes in the integrin βA domain provide a mechanism for signal transduction via hybrid domain movement. J Biol Chem. 2003;278:17028–17035. doi: 10.1074/jbc.M213139200. [DOI] [PubMed] [Google Scholar]
- 10•.Barton SJ, Travis MA, Askari JA, Buckley PA, Craig SE, Humphries MJ, Mould AP. Novel activating and inactivating mutations in the integrin β1 subunit A domain. Biochem J. 2004;380:401–407. doi: 10.1042/BJ20031973. Mutagenesis study of the β1 β I domains suggests that the regulation of the β I domain involves concerted movement of the α1 and α7 helices. Mutating residues of these two helices either increases or decreases integrin ligand binding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jannuzi AL, Bunch TA, West RF, Brower DL. Identification of integrin β subunit mutations that alter heterodimer function in situ. Mol Biol Cell. 2004;15:3829–3840. doi: 10.1091/mbc.E04-02-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiong JP, Stehle T, Goodman SL, Arnaout MA. A novel adaptation of the integrin PSI domain revealed from its crystal structure. J Biol Chem. 2004;279:40252–40254. doi: 10.1074/jbc.C400362200. [DOI] [PubMed] [Google Scholar]
- 13•.Shi M, Sundramurthy K, Liu B, Tan SM, Law SK, Lescar J. The crystal structure of the plexin-semaphorin-integrin domain/hybrid domain/I-EGF1 segment from the human integrin β2 subunit at 1,8-A resolution. J Biol Chem. 2005;280:30586–30593. doi: 10.1074/jbc.M502525200. A high-resolution structure of a β2 integrin fragment containing the PSI, hybrid and I-EGF1 domains is solved. The structure suggests that these three domains may move as a rigid body during integrin activation. [DOI] [PubMed] [Google Scholar]
- 14.Honda S, Tomiyama Y, Pelletier AJ, Annis D, Honda Y, Orchekowski R, Ruggeri Z, Kunicki TJ. Topography of ligand-induced binding sites, including a novel cation-sensitive epitope (AP5) at the amino terminus, of the human integrin β3 subunit. J Biol Chem. 1995;270:11947–11954. doi: 10.1074/jbc.270.20.11947. [DOI] [PubMed] [Google Scholar]
- 15.Mould AP, Travis MA, Barton SJ, Hamilton JA, Askari JA, Craig SE, Macdonald PR, Kammerer RA, Buckley PA, Humphries MJ. Evidence that monoclonal antibodies directed against the integrin β subunit plexin/semaphorin/integrin domain stimulate function by inducing receptor extension. J Biol Chem. 2005;280:4238–4246. doi: 10.1074/jbc.M412240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie C, Shimaoka M, Xiao T, Schwab P, Klickstein LB, Springer TA. The integrin α subunit leg extends at a Ca2+-dependent epitope in the thigh/genu interface upon activation. Proc Natl Acad Sci U S A. 2004;101:15422–15427. doi: 10.1073/pnas.0406680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17••.Nishida N, Xie C, Shimaoka M, Cheng Y, Walz T, Springer TA. Three distinctive ectodomain conformations and their interconversion in integrins that contain I domains. Immunity. 2006 doi: 10.1016/j.immuni.2006.07.016. Detailed EM images of αLβ2 and αXβ2 with bound Fab and allosteric antagonists definitively establish the relation between integrin activation on cell surfaces and integrin extension. [DOI] [PubMed] [Google Scholar]
- 18•.Iwasaki K, Mitsuoka K, Fujiyoshi Y, Fujisawa Y, Kikuchi M, Sekiguchi K, Yamada T. Electron tomography reveals diverse conformations of integrin αIIbβ3 in the active state. J Struct Biol. 2005;150:259–267. doi: 10.1016/j.jsb.2005.03.005. Electron tomography of αIIbβ3 in the active state shows that 90% of the particles are in the extended conformation with an open headpiece. [DOI] [PubMed] [Google Scholar]
- 19.Adair BD, Yeager M. Three-dimensional model of the human platelet integrin αIIbβ3 based on electron cryomicroscopy and x-ray crystallography. Proc Natl Acad Sci U S A. 2002;99:14059–14064. doi: 10.1073/pnas.212498199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification - powerful tools in modern electron microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adair BD, Xiong JP, Maddock C, Goodman SL, Arnaout MA, Yeager M. Three-dimensional EM structure of the ectodomain of integrin αVβ3 in a complex with fibronectin. J Cell Biol. 2005;168:1109–1118. doi: 10.1083/jcb.200410068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng Y, Wolf E, Larvie M, Zak O, Aisen P, Grigorieff N, Harrison SC, Walz T. Single particle reconstructions of the transferrin-transferrin receptor complex obtained with different specimen preparation techniques. J Mol Biol. 2006;355:1048–1065. doi: 10.1016/j.jmb.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 23.Mould AP, Symonds EJ, Buckley PA, Grossmann JG, McEwan PA, Barton SJ, Askari JA, Craig SE, Bella J, Humphries MJ. Structure of an integrin-ligand complex deduced from solution x-ray scattering and site-directed mutagenesis. J Biol Chem. 2003;278:39993–39999. doi: 10.1074/jbc.M304627200. [DOI] [PubMed] [Google Scholar]
- 24.Luo B-H, Springer TA, Takagi J. Stabilizing the open conformation of the integrin headpiece with a glycan wedge increases affinity for ligand. Proc Natl Acad Sci U S A. 2003;100:2403–2408. doi: 10.1073/pnas.0438060100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo B-H, Strokovich K, Walz T, Springer TA, Takagi J. Allosteric β1 integrin antibodies that stabilize the low affinity state by preventing the swing-out of the hybrid domain. J Biol Chem. 2004;279:27466–27471. doi: 10.1074/jbc.M404354200. [DOI] [PubMed] [Google Scholar]
- 26.Tng E, Tan SM, Ranganathan S, Cheng M, Law SK. The integrin αLβ2 hybrid domain serves as a link for the propagation of activation signal from its stalk regions to the I-like domain. J Biol Chem. 2004;279:54334–54339. doi: 10.1074/jbc.M407818200. [DOI] [PubMed] [Google Scholar]
- 27.Tang RH, Tng E, Law SK, Tan SM. Epitope mapping of monoclonal antibody to integrin αLβ2 hybrid domain suggests different requirements of affinity states for intercellular adhesion molecules (ICAM)-1 and ICAM-3 binding. J Biol Chem. 2005;280:29208–29216. doi: 10.1074/jbc.M503239200. [DOI] [PubMed] [Google Scholar]
- 28.Chigaev A, Buranda T, Dwyer DC, Prossnitz ER, Sklar LA. FRET detection of cellular α4-integrin conformational activation. Biophys. J. 2003;85:3951–3962. doi: 10.1016/S0006-3495(03)74809-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnaout MA, Mahalingam B, Xiong JP. Integrin structure, allostery, and bidirectional signaling. Annu Rev Cell Dev Biol. 2005;21:381–410. doi: 10.1146/annurev.cellbio.21.090704.151217. [DOI] [PubMed] [Google Scholar]
- 30.Lee J-O, Bankston LA, Arnaout MA, Liddington RC. Two conformations of the integrin A-domain (I-domain): a pathway for activation? Structure. 1995;3:1333–1340. doi: 10.1016/s0969-2126(01)00271-4. [DOI] [PubMed] [Google Scholar]
- 31.Shimaoka M, Xiao T, Liu J-H, Yang Y, Dong Y, Jun C-D, McCormack A, Zhang R, Joachimiak A, Takagi J, et al. Structures of the αL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation. Cell. 2003;112:99–111. doi: 10.1016/s0092-8674(02)01257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin M, Andricioaei I, Springer TA. Conversion between three conformational states of integrin I domains with a C-terminal pull spring studied with molecular dynamics. Structure. 2004;12:2137–2147. doi: 10.1016/j.str.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Alonso JL, Essafi M, Xiong JP, Stehle T, Arnaout MA. Does the integrin αA domain act as a ligand for its βA domain? Curr Biol. 2002;12:R340–R342. doi: 10.1016/s0960-9822(02)00852-7. [DOI] [PubMed] [Google Scholar]
- 34•.Yang W, Shimaoka M, Salas A, Takagi J, Springer TA. Intersubunit signal transmission in integrins by a receptor-like interaction with a pull spring. Proc Natl Acad Sci U S A. 2004;101:2906–2911. doi: 10.1073/pnas.0307340101. Individual mutation of αL linker residue Glu-310 or β2 MIDAS residues Ala-210 or Tyr-115 to cysteine abolishes I domain activation, whereas the double mutation of αL-E310C with either β2-A210C or β2-Y115C forms a disulfide bond that constitutively activates ligand binding. Thus the study provides strong evidence that the αL Glu310 acts as a intrinsic ligand for the β I domain. Inter-communication between the I domain and the β I domain plays a key role in I domain-containing integrin activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takagi J, Erickson HP, Springer TA. C-terminal opening mimics ‘inside-out’ activation of integrin α5β1. Nature Struct. Biol. 2001;8:412–416. doi: 10.1038/87569. [DOI] [PubMed] [Google Scholar]
- 36.Xiong YM, Chen J, Zhang L. Modulation of CD11b/CD18 adhesive activity by its extracellular, membrane-proximal regions. J Immunol. 2003;171:1042–1050. doi: 10.4049/jimmunol.171.2.1042. [DOI] [PubMed] [Google Scholar]
- 37•.Luo B-H, Springer TA, Takagi J. A specific interface between integrin transmembrane helices and affinity for ligand. PLoS Biol. 2004;2:776–786. doi: 10.1371/journal.pbio.0020153. Disulfide bond scanning of the exofacial regions of αIIβ and β3 TM domains shows a specific α-helical interface in the resting state. The interface is lost rather than rearranged upon integrin activation by extra-cellular mutations. Introduction of disulfide bridges to prevent transmembrane separation abolishes inside-out activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.Vinogradova O, Vaynberg J, Kong X, Haas TA, Plow EF, Qin J. Membrane-mediated structural transitions at the cytoplasmic face during integrin activation. Proc Natl Acad Sci U S A. 2004;101:4094–4099. doi: 10.1073/pnas.0400742101. This NMR study of the α and β cytoplasmic domains was carried out in the presence of diphosphatidylcholine micelles. The results show that there are significant interactions between the lipid and α and β cytoplasmic domains. The structures might represent more physiologic conformations of the integrin cytoplasmic domains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vinogradova O, Velyvis A, Velyviene A, Hu B, Haas TA, Plow EF, Qin J. A structural mechanism of integrin αIIbβ3 ‘inside-out’ activation as regulated by its cytoplasmic face. Cell. 2002;110:587–597. doi: 10.1016/s0092-8674(02)00906-6. [DOI] [PubMed] [Google Scholar]
- 40.Weljie AM, Hwang PM, Vogel HJ. Solution structures of the cytoplasmic tail complex from platelet α IIb- and β 3-subunits. Proc Natl Acad Sci USA. 2002;99:5878–5883. doi: 10.1073/pnas.092515799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ulmer TS, Yaspan B, Ginsberg MH, Campbell ID. NMR analysis of structure and dynamics of the cytosolic tails of integrin α IIb beta 3 in aqueous solution. Biochemistry. 2001;40:7498–7508. doi: 10.1021/bi010338l. [DOI] [PubMed] [Google Scholar]
- 42.Ulmer TS, Calderwood DA, Ginsberg MH, Campbell ID. Domain-specific interactions of talin with the membrane-proximal region of the integrin β3 subunit. Biochemistry. 2003;42:8307–8312. doi: 10.1021/bi034384s. [DOI] [PubMed] [Google Scholar]
- 43.Kiema T, Lad Y, Jiang P, Oxley CL, Baldassarre M, Wegener KL, Campbell ID, Ylanne J, Calderwood DA. The molecular basis of filamin binding to integrins and competition with talin. Mol. Cell. 2006;21:337–347. doi: 10.1016/j.molcel.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 44.Garcia-Alvarez B, de Pereda JM, Calderwood DA, Ulmer TS, Critchley D, Campbell ID, Ginsberg MH, Liddington RC. Structural determinants of integrin recognition by talin. Mol Cell. 2003;11:49–58. doi: 10.1016/s1097-2765(02)00823-7. [DOI] [PubMed] [Google Scholar]
- 45.Tadokoro S, Shattil SJ, Eto K, Tai V, Liddington RC, de Pereda JM, Ginsberg MH, Calderwood DA. Talin binding to integrin β tails: a final common step in integrin activation. Science. 2003;302:103–106. doi: 10.1126/science.1086652. [DOI] [PubMed] [Google Scholar]
- 46.Li W, Metcalf DG, Gorelik R, Li R, Mitra N, Nanda V, Law PB, Lear JD, Degrado WF, Bennett JS. A push-pull mechanism for regulating integrin function. Proc. Natl. Acad. Sci U S A. 2005;102:1424–1429. doi: 10.1073/pnas.0409334102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo B-H, Carman CV, Takagi J, Springer TA. Disrupting integrin transmembrane domain heterodimerization increases ligand binding affinity, not valency or clustering. Proc Natl Acad Sci U S A. 2005;102:3679–3684. doi: 10.1073/pnas.0409440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48•.Partridge AW, Liu S, Kim S, Bowie JU, Ginsberg MH. Transmembrane domain packing stabilizes integrin aIIbb3 in the low affinity state. J Biol Chem. 2005;280:7294–7300. doi: 10.1074/jbc.M412701200. By random mutagenesis of the TM and cytoplasmic domains of β3, new mutations that activate integrin ligand binding are identified. The study suggests that disrupting the interface between two TM domains activates integrins. [DOI] [PubMed] [Google Scholar]
- 49.Gottschalk KE, Kessler H. Evidence for hetero-association of transmembrane helices of integrins. FEBS Lett. 2004;557:253–258. doi: 10.1016/s0014-5793(03)01443-1. [DOI] [PubMed] [Google Scholar]
- 50.Gottschalk KE. A coiled-coil structure of the αIIbβ3 integrin transmembrane and cytoplasmic domains in its resting state. Structure. 2005;13:703–712. doi: 10.1016/j.str.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 51.Li R, Mitra N, Gratkowski H, Vilaire G, Litvinov SV, Nagasami C, Weisel JW, Lear JD, DeGrado WF, Bennett JS. Activation of integrin αIIbβ3 by modulation of transmembrane helix associations. Science. 2003;300:795–798. doi: 10.1126/science.1079441. [DOI] [PubMed] [Google Scholar]
- 52.Kim M, Carman CV, Yang W, Salas A, Springer TA. The primacy of affinity over clustering in regulation of adhesiveness of the integrin αLβ2. J Cell Biol. 2004;167:1241–1253. doi: 10.1083/jcb.200404160. [DOI] [PMC free article] [PubMed] [Google Scholar]