Abstract
Hepatitis delta virus (HDV) produces two essential forms of the sole viral protein from the same open reading frame by using host RNA editing activity at the amber/W site in the antigenomic RNA. The roles of these two forms, HDAg-S and HDAg-L, are opposed. HDAg-S is required for viral RNA replication, whereas HDAg-L, which is produced as a result of editing, inhibits viral RNA replication and is required for virion packaging. Both the rate and amount of editing are important because excessive editing will inhibit viral RNA replication, whereas insufficient editing will reduce virus secretion. Here we show that for HDV genotype III, which is associated with severe HDV disease, HDAg-L strongly inhibits editing of a nonreplicating genotype III reporter RNA, while HDAg-S inhibits only when expressed at much higher levels. The different inhibitory efficiencies are due to RNA structural elements located ca. 25 bp 3′ of the editing site in the double-hairpin RNA structure required for editing at the amber/W site in HDV genotype III RNA. These results are consistent with regulation of amber/W editing in HDV genotype III by a negative-feedback mechanism due to differential interactions between structural elements in the HDV genotype III RNA and the two forms of HDAg.
RNA editing at the viral amber/W site plays a central role in the replication cycle of hepatitis delta virus (HDV), a subviral human pathogen that increases the risk of severe liver disease in those infected with its helper, hepatitis B virus (HBV) (37). The virus uses adenosine-to-inosine RNA editing activity of the host cell, catalyzed by the cellular enzyme ADAR1 (17, 35, 36, 40), to produce two forms of the sole viral protein, hepatitis delta antigen (HDAg), from the same open reading frame (4, 26, 35). These forms, HDAg-S and HDAg-L, differ by the presence of an additional 19 to 20 amino acids at the C terminus of HDAg-L. During the course of virus replication, the UAG (amber) stop codon of HDAg-S is changed to a UGG (trp) codon as a result of editing at adenosine 1012 (the amber/W site) in the antigenome (7, 35). This change directs the translation of the C-terminal 19 to 20 amino acids unique to HDAg-L and alters the form and function of the viral protein (42, 44). HDAg-S is required for RNA replication, whereas HDAg-L is required for packaging the viral RNA with the envelope of hepatitis B surface antigen but inhibits RNA replication (2, 9, 18, 39, 44).
Varying the extent of editing at the amber/W site, either by altering ADAR expression or by site-directed mutagenesis near the site, can have important consequences for HDV replication and virus production (3, 16). Excessive editing at the amber/W site results in reduced levels of RNA replication and reduced production of viable virions because edited antigenomes encode HDAg-L, which is a trans-dominant inhibitor of HDV RNA replication (13, 14). Insufficient editing can lead to increased intracellular HDV RNA replication but inhibits virion production (3). Thus, the kinetics and extent of editing are likely regulated during HDV replication to maximize the rate of virus production. We previously showed that expression of HDAg-S, which binds HDV RNA (10-12, 22, 23, 33), can strongly inhibit editing of a nonreplicating HDV genotype I reporter RNA in transfected cells and may play a role in regulating editing during the HDV replication cycle (36).
Editing is highly specific for the HDV amber/W site and requires specific structural elements in the RNA (3, 4, 36). For HDV genotype I, the structure required for editing is part of the unbranched rod structure characteristic of HDV RNA (4, 7). However, editing at the HDV genotype III amber/W site requires a double-hairpin structure that deviates substantially from the unbranched rod, with nearly 80 bp rearranged (3). Of the three known HDV genotypes, type III is associated with the most severe disease and is the most distantly related genetically (∼40% divergent) (5, 8). It is found exclusively in northern South America and occurs in association with HBV genotype F (8), the most distantly related of the HBV genotypes (29). The use of very different structures for editing in genotypes I and III further emphasizes the differences between these genotypes that are apparent in phylogenetic analysis of RNA sequences (5, 8, 28) and in the genotype-specific support of replication by genotypes I and III HDAg (6).
Here we examine the inhibitory effects of genotype III HDAg on editing of a nonreplicating HDV genotype III reporter RNA. We find that, for genotype III RNA, HDAg-L inhibits editing more strongly than does HDAg-S and that this differential inhibitory activity requires particular elements of the branched double-hairpin RNA structure required for genotype III amber/W site editing (3).
MATERIALS AND METHODS
Plasmid constructs.
The nonreplicating HDV RNA expression construct pHDV-III-NR has been described previously (6, 7). This construct produces antigenomic-sense HDV genotype III RNA, from which internal segments have been deleted. The deletions do not affect the ability of the RNA to form structures required for amber/W RNA editing but do eliminate HDAg synthesis and the ability of the RNA to replicate, even if HDAg is provided in trans from another expression construct. The replication-competent RNA expression construct pHDV-III(−) produces genomic replicating HDV genotype III RNA after transfection of Huh-7 cells. pHDV-III(−)Ag(−) is a previously described (6) site-directed mutant of pHDV-III(−) that produces HDV genotype III RNA that is defective for HDAg synthesis and can replicate only if HDAg is supplied in trans (e.g., by a cotransfected HDAg expression construct). The expression constructs for genotype III HDAg, pHDAg-S-III and pHDAg-L-III, have also been described (6). The plasmid pcDNAneoAgS-III was created by inserting the 779-bp BamHI-SmaI (positions 90 to 988; nucleotide numbering according to reference 5) HDAg-S-coding fragment from pHDV-III(−) between the BamHI and EcoRV sites of pcDNAIneo (Invitrogen, Carlsbad, Calif.).
Construction of mutations.
Mutations M24 and M25 were created by using the PCR primers 5′-GTAAACCCATACTATGGGAAGCTGGGCACGAAGCCC-3′, and 5′-AATCCCGGGCCCCCTCCCAGGACTGGTCCCGATAGGGG-3′, respectively. Sequence positions 1022 to 1090 and 539 to 612 (numbering is on the genomic strand according to references 5 and 43) were deleted from M24 and M25, respectively. Fragments were cloned into the nonreplicating type III antigenomic RNA expression construct pHDV-III-NR by using restriction sites PflMI and XhoI (M24) or ApaI and HindIII (M25). The combined mutant M24/25 contained both mutations. Sequences of cloned fragments were verified for the presence of the desired mutation and the absence of unintended mutations by sequence analysis (MWG Biotech, High Point, N.C.).
Transfections.
Huh-7 cells were plated in 12-well dishes and transfected with a total of 1 μg of plasmid DNA per well by using Lipofectamine Plus (Invitrogen) according to the manufacturer's recommended protocol. In cases in which the total amount of transfected expression plasmids was <1 μg, the parental expression construct pCMV3 with no HDV insert (7) was added to bring the total amount of transfected plasmid DNA to 1 μg. All experiments were repeated at least once and included duplicate transfections. The cell line Huh-AgSIII was created by transfecting Huh-7 cells with pcDNAneoAgS-III; cells were grown in medium containing 500 μg of G418 (Invitrogen)/ml to select for HDAg-S-expressing cells. Expression was verified by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and immunoblotting with a monoclonal antibody specific for HDAg (32).
RNA analysis.
Cellular RNA was harvested at the indicated times posttransfection by using the RNEasy kit and Qiashredder (Qiagen) according to the manufacturer's recommended procedures. RNA editing was analyzed by StyI digestion of reverse transcription-PCR (RT-PCR) products as described previously (3, 7, 35). Editing at the amber/W site creates a StyI restriction site in PCR amplification products derived from edited RNAs that does not exist in products derived from unedited RNAs; the extent of editing can therefore be measured by StyI digestion of PCR products. Editing was quantified by electrophoresis of 32P-labeled, StyI-digested PCR products, followed by radioanalytic imaging (InstantImager; Packard Instruments, Meriden, Conn.). The PCR primers used for replicating HDV RNA were 5414 and 5415, as described previously (7). For analysis of editing in cells cotransfected with pHDV-III-NR and HDAg expression constructs, the primers used were specific for the nonreplicating RNA, and the annealing temperature was raised to 60°C to avoid amplifying HDAg mRNA. The primers used for nonreplicating genotype III RNA were 5′-GGGGATCTCGAGAAGGAT-3′ (IIINR-1) and 5′-GTGAGTTCTATTGCCCTATAGTT-3′ (IIINR-2). Using these primers and annealing conditions, no PCR products were obtained from cells transfected with HDAg expression constructs in the absence of nonreplicating RNA expression constructs.
Northern blot analysis of HDV RNA.
RNA was electrophoresed through 1.2% agarose gels containing 2.2 M formaldehyde, transferred onto Immobilon-Ny+ nylon membrane (Millipore), and hybridized with a genomic-sense [32P]CTP-labeled probe as described previously (36). Relative RNA levels were determined by using a PhosphorImager (Molecular Dynamics).
Immunoblot analysis.
Transfected cells were rinsed twice with cold 1× phosphate-buffered saline (PBS) and harvested at the indicated times. After collection by a brief spin, cells were suspended in lysis buffer (150 mM NaCl, 50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 0.2% NP-40, 1% Triton X-100, 0.1% SDS, 5 μl of protease inhibitor cocktail/ml). Equivalent fractions of cell lysates were denatured in 2% SDS sample buffer and analyzed by electrophoresis and immunoblotting, with human anti-HDAg monoclonal antibody T1/39 (32), as described previously (6, 36). Immunoblot detection was done by using horseradish peroxidase-conjugated goat anti-human immunoglobulin G (H+L; KPL, Gaithersburg, Md.) and a chemiluminescence kit (LumiGLO; KPL).
RESULTS
Differential inhibition of HDV genotype III amber/W editing by HDAg-S and HDAg-L.
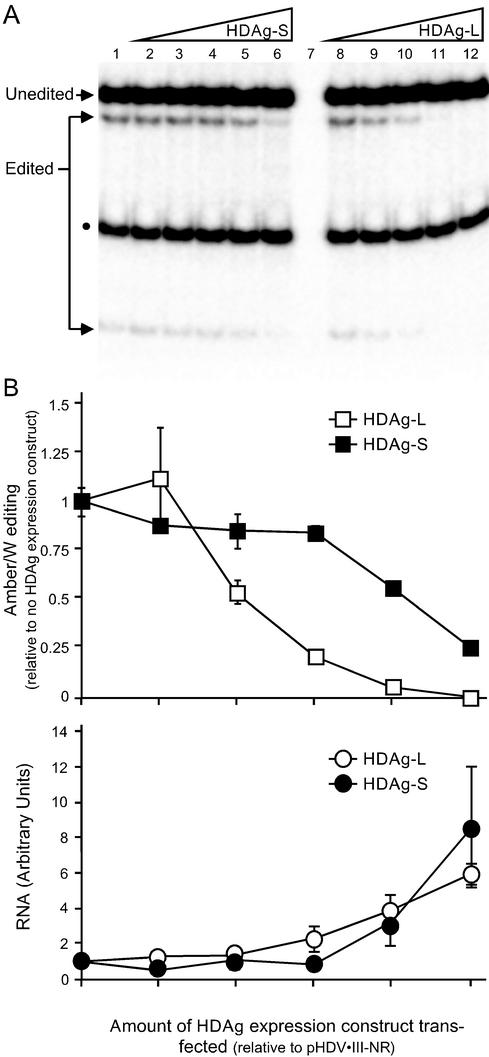
Previous analysis of editing in HDV genotype I showed that amber/W site editing in a nonreplicating reporter RNA could be inhibited by HDAg expression (36). Whereas the unbranched rod characteristic of HDV is required for editing in HDV genotype I (4), a branched double-hairpin structure is required for editing in genotype III (3). We wanted to know whether the different RNA structure required for genotype III amber/W editing would alter the inhibitory effect of HDAg on editing. We therefore performed a similar analysis of the inhibition of genotype III amber/W RNA editing by genotype III HDAg-S and HDAg-L. Huh-7 cells were cotransfected with pHDV-III-NR, an expression construct for nonreplicating HDV genotype III RNA, and different amounts of genotype III HDAg-S and HDAg-L expression constructs. RNAs were harvested 2 days posttransfection, and editing was analyzed by RT-PCR followed by StyI digestion (Fig. 1). This assay detects amber/W editing by the appearance of a StyI restriction site that occurs only in PCR products derived from edited RNAs (7, 16, 35, 36). The percent editing is given by dividing the sum of the intensities of the StyI digestion bands (“Edited” in Fig. 1) by the sum of the edited and unedited bands (16, 35, 36). The primers used in this assay were specific for the nonreplicating editing reporter RNA and did not detect HDAg mRNA derived from the HDAg-S and HDAg-L expression constructs (Fig. 1A, lane 7).
FIG. 1.
Inhibition of RNA editing at the HDV genotype III amber/W site by HDAg-S and HDAg-L. (A) Huh-7 cells in 12-well plates were cotransfected, as described in Materials and Methods, with 500 ng of the nonreplicating genotype III antigenomic RNA expression construct pHDV-III-NR and various amounts of the genotype III HDAg-S or HDAg-L expression constructs pHDAg-S-III (lanes 2 to 6) and pHDAg-L-III (lanes 8 to 12). The amounts of HDAg expression construct cotransfected were as follows: lane 1, none; lanes 2 and 8, 0.05 ng; lanes 3 and 9, 0.5 ng; lanes 4 and 10, 5 ng; lanes 5 and 11, 50 ng; lanes 6 and 12, 500 ng. As a negative control for the RT-PCR, cells were transfected with 500 ng each of pHDAg-S-III and pHDAg-L-III (lane 7). The RNA was harvested 2 days posttransfection and analyzed for editing at the amber/W site by RT-PCR-StyI digestion as described previously (36), except that the primers used were IIINR-1 and IIINR-2, as described in Materials and Methods. Editing creates a StyI restriction digestion site that is not present in unedited RNA. Thebands due to edited and unedited RNA are labeled; the smaller band indicated by a black dot is due to a preexisting StyI site in the cDNA that is not affected by editing. Results shown are representative of three independent experiments. (B) Effect of amount of HDAg expression construct cotransfected on editing and nonreplicating RNA levels.In the upper part of panel B is a graphical representation of results from panel A combined with the results from two independent experiments. Because of variations in the amount of editing observed in the different experiments (between 5% and 10%), the results for each experiment were first normalized to the level of editing observed in the absence of a cotransfected HDAg expression construct. Error bars indicate standard deviations; for some points the error bars are not visible because they are smaller than the symbols used for the data. Symbols: ▪, cotransfection with pHDAg-S-III; □, cotransfection with pHDAg-L-III. In the lower part of panel B is a graph of Northern blot analysis of the antigenomic pHDV-III-NR RNA. RNA levels are presented as the fold increase relative to the amount of nonreplicating RNA in cells with no HDAg cotransfected.
We observed that both HDAg-S and HDAg-L inhibit editing in a nonreplicating genotype III editing reporter RNA but that HDAg-L was a much more effective inhibitor of amber/W site editing than HDAg-S (Fig. 1). Analysis of HDAg expression levels by immunoblot indicated that for each amount of HDAg expression plasmid transfected, HDAg-S and HDAg-L were equivalently expressed, and the expression levels correlated well with the amount of transfected DNA (not shown). Nearly 50% inhibition was achieved at HDAg-L expression levels 100-fold lower than that of HDAg-S; moreover, at expression levels for which HDAg-S inhibited editing by not quite 50%, inhibition by HDAg-L was >90% (Fig. 1).
HDAg has been shown to bind the unbranched rod structure of HDV RNA and can stabilize nonreplicating RNAs (such as that analyzed here) that can form the unbranched rod structure (6, 21). To determine whether the observed inhibition of editing could be an indirect effect due to stabilization of the RNA and saturation of the editing enzyme by increased levels of nonreplicating RNA, we analyzed RNA levels by Northern blot hybridization. Consistent with our previous analysis of the inhibition of genotype I amber/W editing by HDAg (36), we found that cotransfection with either HDAg-S or HDAg-L increased the level of nonreplicating RNA, most likely by stabilizing the RNA against degradation (21). Although the effects of HDAg-S and HDAg-L on amber/W editing were remarkably different, their effects on the level of nonreplicating RNA were essentially identical (Fig. 1B). Importantly, the effect of HDAg-L on editing did not correlate with its effect on the level of nonreplicating RNA (Fig. 1B); indeed, the level of nonreplicating HDV RNA was nearly unchanged over the concentration range of HDAg-L that yielded the bulk of the inhibitory effect on editing. Thus, the effect of HDAg-L on genotype III amber/W site editing is not due to an indirect effect of increased levels of HDV RNA and is not related to the ability of HDAg-L to stabilize HDV RNA.
Destabilizing the genotype III RNA unbranched rod structure does not affect the differential inhibition of genotype III amber/W editing by HDAg-S and HDAg-L.
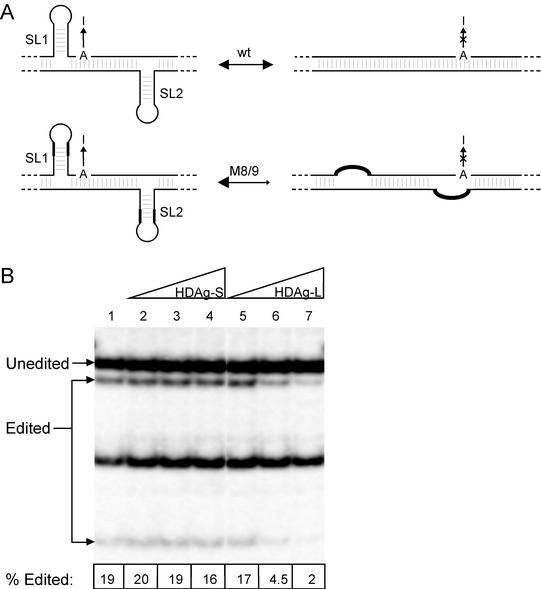
HDV genotype III RNA assumes at least two mutually exclusive conformations in the vicinity of the amber/W editing site: the unbranched rod structure, which is required for RNA replication, and the branched double-hairpin structure, which is required for amber/W editing (3) (Fig. 2A, top). Altering the distribution of the RNA between these structures could affect editing by either increasing or decreasing the amount of RNA in the conformation necessary for editing. Indeed, mutations that favor the double-hairpin structure in a nonreplicating RNA increase the extent of editing, most likely by shifting the distribution toward the branched double-hairpin structure (3). Thus, in the “stem-flip” mutant construct M8/9 (3), 18 and 16 transversion mutations were introduced into stem-loops SL1 and SL2, respectively, such that the stability of the double-hairpin structure was unaffected but the stability of the unbranched rod was substantially reduced (Fig. 2A); these mutations increased editing in a nonreplicating reporter RNA by about fourfold (3).
FIG. 2.
Inhibition of genotype III RNA editing by HDAg is not affected by mutations that stabilize the branched double-hairpin RNA structure relative to the unbranched rod structure. (A) Schematic of the branched double-hairpin structure required for genotype III RNA editing (left) and the unbranched rod structure. In the “stem-flip” mutant M8/9 (3) the 8 or 9 bp nearest the loops of stem-loops SL1 and SL2 are reversed, destabilizing the unbranched rod structure but leaving the predicted stability of the double-hairpin structure unaffected. In this mutant the distribution of the RNA between the unbranched rod and branched double-hairpin structures is probably altered, as indicated by the arrows. The amber/W site adenosine is indicated by “A,” and the editing event is indicated by a vertical arrow extending from “A” to “I”; an “X” is placed on this arrow above the unbranched rod structure because editing does not occur in this conformation (3). (B) Inhibition of amber/W editing in M8/9 mutant RNA by genotype III HDAg-S and HDAg-L. Huh-7 cells were cotransfected with the nonreplicating genotype III RNA expression construct pHDV-III-NR-M8/9 (3), and various amounts of the genotype III HDAg-S or HDAg-L expression constructs pHDAg-S-III (lanes 2 to 4) and pHDAg-L-III (lanes 5 to 7). Amounts of HDAg expression construct cotransfected were: lane 1, none; lanes 2 and 5, 0.5 ng; lanes 3 and 6, 5 ng; lanes 4 and 7, 50 ng. RNA was harvested and editing analyzed as in Fig. 1. The numbers at the bottom of the panel are the average editing values obtained from duplicate transfections.
To determine whether the different inhibitory effects of HDAg-S and HDAg-L on genotype III amber/W editing could be due to different effects on the distribution of genotype III RNA between the unbranched rod and double-hairpin structures, Huh-7 cells were transfected with an expression construct for a nonreplicating HDV genotype III RNA containing the M8/9 mutation and different amounts of HDAg-S and HDAg-L expression constructs. We observed that the inhibitory effects of HDAg-S and HDAg-L on amber/W editing in the M8/9 mutation were indistinguishable from their effects on wild-type nonreplicating HDV genotype III RNA (Fig. 2). Although the M8/9 mutation increased editing approximately fourfold, editing was still strongly inhibited by HDAg-L. Moreover, because the inhibition of editing occurred at similar HDAg levels for nonreplicating RNAs containing either wild-type sequence or the M8/9 mutations, the inhibition is not simply due to interference with editing in a poor editing substrate. Thus, inhibition of genotype III RNA editing by HDAg does not appear to involve shifting the distribution of the RNA between different conformations that have different editing capacities.
Roles of stem-loops SL1 and SL2 in genotype III RNA editing and its regulation.
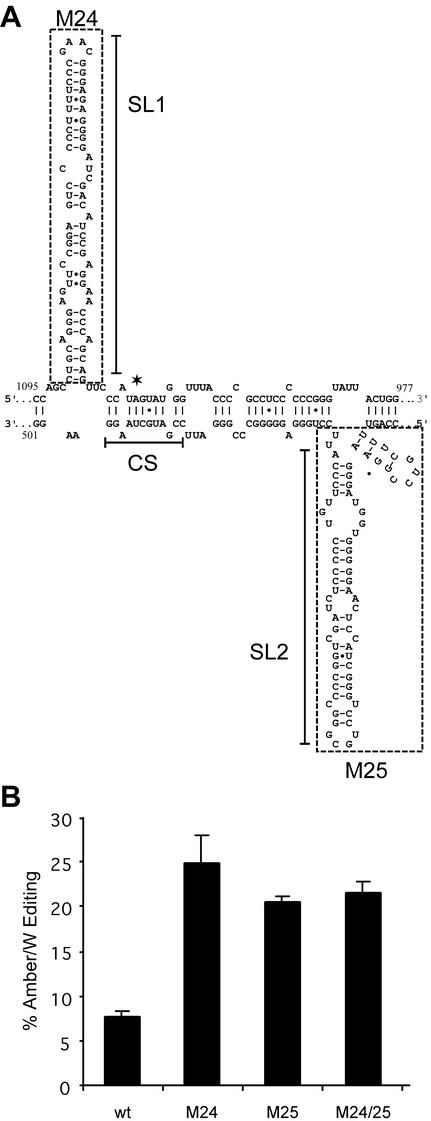
To determine the potential roles of the stem-loops SL1 and SL2 of the branched double-hairpin structure in the differential effects of HDAg-S and HDAg-L on genotype III amber/W editing, we deleted sequences corresponding to these structures in the nonreplicating RNA expression construct pHDV-III-NR (Fig. 3). Mutation M24 lacks SL1 and mutation M25 lacks SL2; these two mutations are combined in the double mutant M24/25, which lacks both SL1 and SL2. Analysis of predicted RNA structure stabilities of RNAs containing these mutations with the program mfold (48) indicated that the M24, M25, and M24/25 deletions are similar to the M8/9 mutations in that they are predicted to favor an RNA conformation in which the amber/W site is base paired with U509, as in the double-hairpin structure, rather than mismatched with C578, as in the unbranched rod. Note that RNA containing the M24/25 mutations, with both SL1 and SL2 deleted, is predicted to form an unbranched rod structure that differs from that formed by wild-type RNA; in particular, the base pairs in the vicinity of the amber/W site in this mutant structure are the same as in the wild-type double-hairpin structure and different from the wild-type unbranched rod.
FIG. 3.
Deletion of stem-loops SL1 and SL2 from the branched double-hairpin structure increases genotype III amber/W RNA editing in a nonreplicating RNA. (A) Schematic of the branched double-hairpin RNA structure. The amber/W site is indicated by an asterisk; SL1 and SL2 denote stem-loops 1 and 2. CS denotes sequences com-plementary to sequences in the immediate vicinity of the amber/W site; base pairing between these sets of sequences is essential for amber/W editing (3). Sequences deleted in mutants M24 (nucleotides 1022 to 1090) and M25 (nucleotides 539 to 612) are shown boxed by dashed lines. (B) Huh-7 cells were transfected with the nonreplicating genotype III antigenomic RNA expression construct pHDV-III-NR containing either wild-type sequences or the indicated mutations. Editing was analyzed as in Fig. 1. The values shown are the average of editing in three independent samples from one experiment and are representative of other experiments that gave similar results. Standard deviations are indicated by vertical lines.
Similar to the “stem-flip” mutant M8/9 (3), in the absence of HDAg, all three of the stem-loop deletion mutations—M24, M25, and M24/25—exhibited substantially (four- to fivefold) higher amber/W editing than did wild-type genotype III RNA (Fig. 3B). These increases are consistent with the predicted effect of these mutations (i.e., shifting the distribution of the RNA toward the conformation compatible with editing). Furthermore, these results strongly confirm our previous identification of the branched double-hairpin structure as the substrate for genotype III amber/W editing and, in particular, the base pairs formed around the amber/W site in this structure. The high level of editing observed in RNAs containing the M25 mutation unequivocally rules out the involvement of the genotype III unbranched rod structure in amber/W site editing because the sequences from the noncoding side of the HDV RNA that base pair with the amber/W site region in the unbranched rod structure are deleted in this mutation. It is also important that the high level of editing observed for the M24/25 mutant, which forms a variant of the unbranched rod, demonstrates that SL1 and SL2 themselves are not required for HDV genotype III amber/W editing.
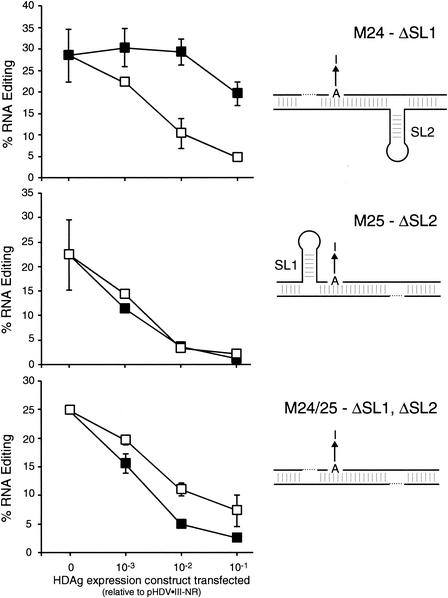
To specifically investigate the roles of the stem-loops SL1 and SL2 in the differential inhibitory effects of HDAg-S and HDAg-L on editing, Huh-7 cells were transfected with pHDV-III-NR mutant M24, M25, or M24/25 and with different amounts of genotype III HDAg-S and HDAg-L expression constructs. RNAs were harvested 2 days posttransfection, and editing was analyzed as in Fig. 1. Deletion of SL1 (mutant M24) produced an effect similar to that observed with the M8/9 mutation: editing was increased relative to nonreplicating wild-type RNAs, but there was no effect on the inhibitory effects of HDAg-S and HDAg-L (Fig. 4, top). However, deletion of SL2 (mutant M25), which affected editing in the absence of HDAg similarly to deleting SL1 (Fig. 3), improved the inhibitory efficiency of HDAg-S such that the difference between HDAg-S and HDAg-L was eliminated (Fig. 4, middle). This result indicates that the sequences deleted in the M25 mutant, principally SL2, reduce the effectiveness of genotype III HDAg-S as an inhibitor of amber/W editing in wild-type genotype III nonreplicating RNA. HDAg-S also efficiently inhibited editing in the double mutant M24/25, but in this case HDAg-L was a less efficient inhibitor than HDAg-S (Fig. 4, bottom).
FIG. 4.
Sequences and structures around SL2 are required for the differential inhibitory effects of HDAg-S and HDAg-L on genotype III amber/W editing. Huh-7 cells were cotransfected with the 500 ng of the nonreplicating genotype III antigenomic RNA expression construct pHDV-III-NR containing the indicated stem-loop deletion mutations and indicated amounts of genotype III HDAg-S or HDAg-L expression constructs. Editing was analyzed as in Fig. 1. Symbols: ▪, HDAg-S; □, HDAg-L. Also indicated on the right are schematic diagrams of the effects of the mutations on the predicted structure required for editing. Elements of schematic diagrams are as in Fig. 3; deleted sequences are shown by a dotted line. The mutations analyzed were as follows: ΔSL1-M24, top panel; ΔSL2-M25, middle panel; and ΔSL1,ΔSL2-M24/25, bottom panel.
Regulation of Amber/W site editing in replicating HDV genotype III RNA.
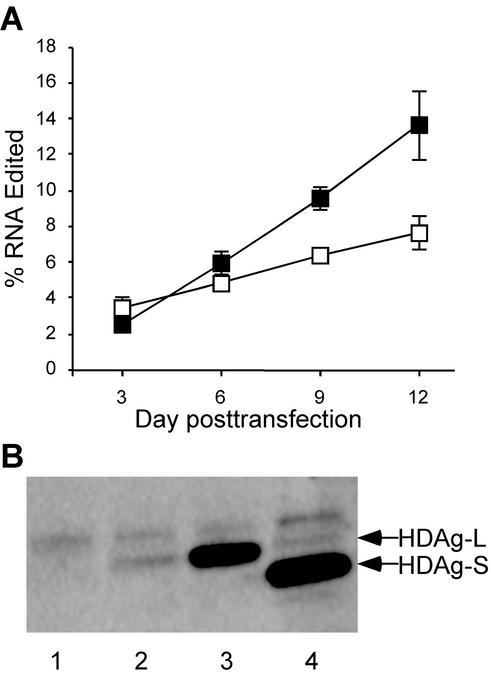
The results presented in Fig. 1, 2, and 4 indicate that HDAg-L can potently inhibit editing in nonreplicating HDV genotype III RNAs. To examine whether HDAg-L can affect amber/W editing in replicating HDV genotype III RNA, we generated the cell line Huh-AgSIII, which stably expresses genotype III HDAg-S under the control of the cytomegalovirus promoter. These cells were then transfected with replication-competent expression construct pHDV-III(−) or pHDV-III(−)Ag(−). The former construct produces wild-type genomic HDV genotype III RNA; the latter produces an RNA that differs by a single nucleotide insertion that destroys the HDAg-S reading frame (6); we refer to this RNA as Ag(−) to indicate that it does not express HDAg. The Ag(−) RNA replicates in cells that express HDAg, such as Huh-AgSIII; although this RNA is edited at the amber/W site, no HDAg-L is produced because the open reading frame for HDAg has been disrupted. Replicating wild-type RNA is edited and produces both HDAg-S and HDAg-L. Comparison of the kinetics of editing accumulation for the wild-type and Ag(−) RNAs shows that editing accumulated at a greater rate in the Ag(−) RNA than in wild-type RNA (Fig. 5A). By day 9, when HDAg-L was readily detectable in cells transfected with the wild-type expression construct pHDV-III, editing was nearly 50% greater in Ag(−) RNA than in wild-type RNA; by 12 days posttransfection editing was nearly twofold greater in Ag(−) RNA than in wild-type RNA (Fig. 5A). This result is consistent with the interpretation that amber/W editing is regulated during HDV genotype III replication by the level of HDAg-L.
FIG. 5.
(A) The kinetics of editing accumulation are inhibited by HDAg-L. Huh-AgSIII cells were transfected with either the wild-type genotype III RNA expression construct pHDV-III(−) (□) or pHDV-III(−)Ag(−) (▪). Total cellular RNA was harvested on days 3, 6, 9, and 12 posttransfection and analyzed for editing at the amber/W site by RT-PCR-StyI digestion, as described previously (36). The values shown are the average of editing in three independent samples from one experiment and are representative of other experiments that gave similar results. Standard deviations are indicated by vertical lines. (B) SDS-PAGE-immunoblot analysis of HDAg-S and HDAg-L. Lanes 1 to 3, detection of HDAg-L in samples harvested from cells 2 days posttransfection with 500 ng of pHDV-III-NR and the following amounts of pHDAg-L-III: lane 1, 0.5 ng; lane 2, 5 ng; and lane 3, 50 ng. Lane 4, sample harvested 9 days posttransfection with the wild-type replicating construct pHDV-III(−), which produces both HDAg-S and HDAg-L. Equivalent amounts of protein were loaded in all lanes. HDAg-L and HDAg-S bands are indicated by arrows. The band just above the HDAg-L band is present in lysates from untransfected cells and is likely due to nonspecific interaction with components of the horseradish peroxidase-conjugated antibody detection system.
We further explored the relationship between the observed inhibition of HDV genotype III amber/W site editing by HDAg-L and the possible regulation of editing during HDV genotype III RNA replication by comparing the relative amounts of HDAg and HDV RNA in cells transfected with the replicating construct pHDV-III(−), on the one hand, with those cotransfected with the HDAg-L expression construct pHDAg-L-III and the nonreplicating RNA expression construct pHDV-III-NR, on the other hand. We found that the amount of HDAg-L that gave slightly greater than 50% inhibition of editing on the nonreplicating RNA was similar to the level of HDAg-L in cells 9 days posttransfection with the replicating HDV genotype III expression construct pHDV-III(−) (Fig. 5B). Comparison of editing levels in Ag(−) and wild-type RNAs (Fig. 5A) indicated that there was a readily detectable difference at 9 days posttransfection, a finding consistent with an inhibitory effect of HDAg-L at this time. Further, analysis of RNA levels by blot hybridization showed that the amounts of nonreplicating RNA 2 days posttransfection were similar to the level of replicating antigenomic RNA on day 9. Thus, not only were the levels of HDAg-L similar but the HDAg-L/antigenomic RNA ratios were also similar.
Genotype I HDAg-S and HDAg-L also exhibit differential inhibition of HDV genotype III RNA editing.
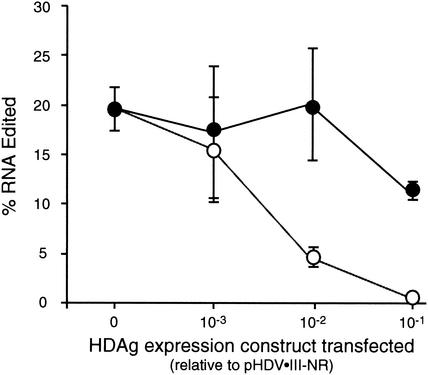
Previous studies have indicated that HDAg-S from genotypes I and III exhibit genotype-specific support of HDV RNA replication; i.e., they can be functionally distinct (6). Moreover, the 19 C-terminal amino acids of genotype I HDAg-L differ substantially from the 20 C-terminal amino acids of genotype III HDAg-L; indeed, these C-terminal sequences may be defining features of the HDV genotypes (5, 41, 46). To determine whether the different editing-inhibitory patterns of genotype III HDAg-S and HDAg-L were specific for genotype III HDAg, we examined the abilities of genotype I HDAg-S and HDAg-L to inhibit amber/W editing on genotype III RNA (Fig. 6). For this analysis we used the genotype III M8/9 “stem-flip” mutant because nonreplicating RNA containing this mutation is edited at significantly higher levels than the wild-type RNA but retains the differential inhibitory affects of HDAg-S and HDAg-L (Fig. 1 and 2). We observed that genotype I HDAg-L strongly inhibited editing of the genotype III RNA amber/W site, whereas type I HDAg-S inhibited editing only weakly (Fig. 6). This result is similar to that observed for the inhibition of genotype III amber/W editing by genotype III HDAg-S and HDAg-L (Fig. 1 and 2). Thus, the differential abilities of HDAg-S and HDAg-L to inhibit editing of genotype III HDV RNA amber/W editing is a general property of genotype I and III HDAg and may reflect a difference between HDAg-S and HDAg-L that applies to all three HDV genotypes.
FIG. 6.
Inhibition of genotype III amber/W editing by genotype I HDAg-S and HDAg-L. Huh-7 cells were transfected with the nonreplicating genotype III antigenomic RNA expression construct pHDV-III-NR-M8/9 and indicated amounts of the genotype I HDAg expression constructs pHDAg-S-I or pHDAg-L-I (36). Editing was analyzed as in Fig. 1. Symbols: •, HDAg-S expression; ○, HDAg-L expression.
DISCUSSION
RNA editing via adenosine deamination has been increasingly recognized as an important posttranscriptional control mechanism, allowing for the production of multiple protein variants from the same gene (1). Indeed, editing at the HDV amber/W site has been shown to play a central role in the HDV replication cycle (3, 16, 17). On host RNAs that are substrates for editing, editing levels can vary developmentally and in a tissue-specific manner (19), but aside from autoregulation of ADAR2 expression via editing (31, 38), the regulatory mechanisms are not well understood. HDV must control both the rate and the extent of editing because HDAg-L, which is produced as a result of editing, is necessary for virion production but inhibits viral RNA replication. HDV does not appear to regulate editing by affecting ADAR1 expression because ADAR1 levels are unaffected by HDV replication (45).
Here we show that, for HDV genotype III, HDAg-L can strongly inhibit amber/W site editing in nonreplicating RNAs in transfected cells. Remarkably, this inhibition occurs at HDAg-L expression levels 100-fold lower than required for similar inhibition by genotype III HDAg-S. This pattern is consistent with the interpretation that amber/W site editing is controlled during HDV genotype III replication by a negative-feedback mechanism. This possibility is supported by the observation that amber/W editing accumulates more rapidly in replicating RNAs that produce no HDAg-L than in HDAg-L-producing RNAs (Fig. 5). By 9 days posttransfection, a time at which HDAg-L production becomes readily detectable, replicating RNAs that produce HDAg-L are edited at lower levels than replicating RNAs that produce no HDAg-L (Fig. 5). It is important to note that the inhibitory effect of HDAg-L on editing in nonreplicating RNAs was not likely due to overexpression of HDAg-L because the inhibition occurred at protein and RNA levels similar to those observed in cells 9 days posttransfection with replication-competent constructs.
The mechanistic basis for the differential activities of HDAg-S and HDAg-L is not clear. The most likely mechanism by which HDAg-L inhibits genotype III amber/W editing is by binding HDV RNA near the editing site and preventing access of the deaminase. HDAg has been shown to bind HDV RNA specifically (10-12, 22, 23, 33), and RNA binding is required for ADAR1 activity (15, 20, 25, 30). In this regard, we previously showed that deletion of the RNA-binding region of genotype I HDAg eliminated its ability to inhibit amber/W site editing in a nonreplicating genotype I reporter RNA (36). Moreover, mutational analysis indicated that deletion of the SL2 region eliminated the difference between the activities of HDAg-S and HDAg-L (Fig. 4); the role of this region as a potential binding site for HDAg that could interfere with ADAR1 activity is consistent with studies indicating that ADAR1 binds base-paired segments on the 3′ side of the adenosine to be edited (15, 24, 34, 47).
Possibly, HDAg-L and HDAg-S interact differently with structures in the vicinity of SL2; these differences could include binding affinity or even altered geometries that could affect the ability of HDAg to interfere with ADAR1 binding near the editing site. The improved inhibitory ability of HDAg-S for the M25 mutation (Fig. 4), which affects the RNA structure by removing SL2 sequences, may be explained by improved HDAg-S binding to RNA lacking SL2 and neighboring structures. The lack of an effect of the SL1 deletion mutant M24 on HDAg-S inhibition does not necessarily indicate that this deletion has no effect on HDAg-S binding, because ADAR1 might not need to bind to the SL1 region, which is 5′ of the amber/W editing site, in order to edit the site. Alternatively, the differences between HDAg-S and HDAg-L may be related to other factors, such as protein-protein interactions. Whatever the nature of the difference, it is interesting that, despite significant sequence variations between genotypes I and III, genotype I HDAg-S and HDAg-L exhibit inhibitory effects similar to their genotype III counterparts (Fig. 6), an indication that the differential inhibitory effect is a general property of HDAg-S and HDAg-L. Further studies will be required to determine (i) which specific structures are actually formed by the SL2 region and (ii) the extent to which SL2, structural elements formed by the other bases (nt 601 to 612) in the deleted region, or other as-yet-unknown structures are responsible for the reduced inhibitory activity of HDAg-S.
The high editing activities observed for the M24, M25, and M24/25 mutants (Fig. 3) indicate that the sequences and structures deleted in these mutants are not essential for editing. Although we showed previously that the branched double-hairpin structure is required for genotype III amber/W editing, these results show that the essential elements of this structure do not include SL1 and SL2. Rather, as for the genotype I amber/W site, the base pairs formed immediately around the amber/W site are the critical elements. Perhaps the primary role of SL2 is to provide a mechanistic basis for negative-feedback regulation, because deletion of SL2 and neighboring sequences eliminates the difference between the editing-inhibitory activities of HDAg-S and HDAg-L. In this event, the primary role of SL1 could be to stabilize the overall structure required for editing by providing base-pairing partners for bases whose base-pairing partners from the unbranched rod were rearranged within SL2 or within the amber/W base-pairing region.
It seems likely that the mechanisms by which HDV RNA regulates amber/W site editing will turn out to be yet another example of functional differences between HDV genotypes. We previously reported that genotype I HDAg, which binds HDV RNA, can strongly inhibit genotype I HDV RNA editing (36), but we have not observed differential inhibition by HDAg-S and HDAg-L (Q. F. Cheng, G. C. Jayan, and J. L. Casey, unpublished data). In this regard, it has been reported that genotype I HDAg-L does not inhibit HDV RNA replication late in the replication cycle (27). It remains to be seen whether the same holds true for genotype III. Conceivably, different regulatory functions for editing could be related to different effects on RNA replication.
It is interesting that the different abilities of genotype I and genotype III HDAg-S to inhibit editing are correlated with the relative levels of editing in replicating and nonreplicating RNAs for these two genotypes. For genotype I, in the absence of HDAg-S, nonreplicating reporter RNAs are efficiently edited (30 to 40%) within 2 to 3 days posttransfection (7, 16, 36), whereas replicating RNAs, which produce HDAg-S, are edited at much lower rates: editing is not detectable at 2 days posttransfection and is no more than 6% on day 4 (16). In contrast, for HDV genotype III, both nonreplicating and replicating RNAs exhibit similar low levels of editing. Editing levels 2 to 3 days posttransfection are typically ca. 5% for nonreplicating genotype III RNAs (see Fig. 1 and 3) and are similar for replicating RNAs (17; Cheng et al., unpublished). Because of the deleterious effects of premature excessive editing and HDAg-L production (16), both genotypes must limit the extent of editing that occurs early in the replication cycle. We suggest that for genotype I editing is limited by HDAg-S, which is a potent inhibitor of editing on nonreplicating RNAs (36), and that for genotype III editing is limited instead by the requirement to form the double-hairpin structure, and further suppression of editing by HDAg-S is not required. Clearly, a more thorough understanding of the mechanisms by which editing is controlled and regulated during replication of HDV genotypes I and III will require further study.
The results presented here lend further support to the idea that distinctions between the HDV genotypes are functional as well as genetic (6). The extent to which these differences may be responsible for various levels of disease associated with these genotypes remains to be determined. Further comparative analysis of HDV functions among the genotypes is likely to uncover more important differences among them. In the process, we are also likely to uncover new attributes that are shared, such as the finding reported here that HDAg-S and HDAg-L appear to interact differently with different RNA structures.
Acknowledgments
This work was supported by grant R01-AI42324 from the National Institutes of Health.
REFERENCES
- 1.Bass, B. L. 2002. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71:817-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergmann, K. F., and J. L. Gerin. 1986. Antigens of hepatitis delta virus in the liver and serum of humans and animals. J. Infect. Dis. 154:702-706. [DOI] [PubMed] [Google Scholar]
- 3.Casey, J. L. 2002. RNA editing in hepatitis delta virus genotype III requires a branched double-hairpin RNA structure. J. Virol. 76:7385-7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casey, J. L., K. F. Bergmann, T. L. Brown, and J. L. Gerin. 1992. Structural requirements for RNA editing in hepatitis delta virus: evidence for a uridine-to-cytidine editing mechanism. Proc. Natl. Acad. Sci. USA 89:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casey, J. L., T. L. Brown, E. J. Colan, F. S. Wignall, and J. L. Gerin. 1993. A genotype of hepatitis D virus that occurs in northern South America. Proc. Natl. Acad. Sci. USA 90:9016-9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casey, J. L., and J. L. Gerin. 1998. Genotype-specific complementation of hepatitis delta virus RNA replication by hepatitis delta antigen. J. Virol. 72:2806-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casey, J. L., and J. L. Gerin. 1995. Hepatitis D virus RNA editing: specific modification of adenosine in the antigenomic RNA. J. Virol. 69:7593-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casey, J. L., G. A. Niro, R. E. Engle, A. Vega, H. Gomez, M. McCarthy, D. M. Watts, K. C. Hyams, and J. L. Gerin. 1996. Hepatitis B virus (HBV)/hepatitis D virus (HDV) coinfection in outbreaks of acute hepatitis in the Peruvian Amazon basin: the roles of HDV genotype III and HBV genotype F. J. Infect. Dis. 174:920-926. [DOI] [PubMed] [Google Scholar]
- 9.Chang, F. L., P. J. Chen, S. J. Tu, C. J. Wang, and D. S. Chen. 1991. The large form of hepatitis delta antigen is crucial for assembly of hepatitis delta virus. Proc. Natl. Acad. Sci. USA 88:8490-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, M. F., S. C. Baker, L. H. Soe, T. Kamahora, J. G. Keck, S. Makino, S. Govindarajan, and M. M. Lai. 1988. Human hepatitis delta antigen is a nuclear phosphoprotein with RNA-binding activity. J. Virol. 62:2403-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, M. F., C. Y. Sun, C. J. Chen, and S. C. Chang. 1993. Functional motifs of delta antigen essential for RNA binding and replication of hepatitis delta virus. J. Virol. 67:2529-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chao, M., S. Y. Hsieh, and J. Taylor. 1991. The antigen of hepatitis delta virus: examination of in vitro RNA-binding specificity. J. Virol. 65:4057-4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chao, M., S. Y. Hsieh, and J. Taylor. 1990. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J. Virol. 64:5066-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glenn, J. S., and J. M. White. 1991. Trans-dominant inhibition of human hepatitis delta virus genome replication. J. Virol. 65:2357-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herbert, A., and A. Rich. 2001. The role of binding domains for dsRNA and Z-DNA in the in vivo editing of minimal substrates by ADAR1. Proc. Natl. Acad. Sci. USA 98:12132-12137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayan, G. C., and J. L. Casey. 2002. Increased RNA Editing and inhibition of hepatitis delta virus replication by high-level expression of ADAR1 and ADAR2. J. Virol. 76:3819-3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jayan, G. C., and J. L. Casey. 2002. Inhibition of hepatitis delta virus RNA editing by short inhibitory RNA-mediated knockdown of Adar1 but not Adar2 expression. J. Virol. 76:12399-12404. [DOI] [PMC free article] [PubMed]
- 18.Kuo, M. Y., M. Chao, and J. Taylor. 1989. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 63:1945-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai, F., C. X. Chen, V. M. Lee, and K. Nishikura. 1997. Dramatic increase of the RNA editing for glutamate receptor subunits during terminal differentiation of clonal human neurons. J. Neurochem. 69:43-52. [DOI] [PubMed] [Google Scholar]
- 20.Lai, F., R. Drakas, and K. Nishikura. 1995. Mutagenic analysis of double-stranded RNA adenosine deaminase, a candidate enzyme for RNA editing of glutamate-gated ion channel transcripts. J. Biol. Chem. 270:17098-17105. [DOI] [PubMed] [Google Scholar]
- 21.Lazinski, D. W., and J. M. Taylor. 1994. Expression of hepatitis delta virus RNA deletions: cis and trans requirements for self-cleavage, ligation, and RNA packaging. J. Virol. 68:2879-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazinski, D. W., and J. M. Taylor. 1993. Relating structure to function in the hepatitis delta virus antigen. J. Virol. 67:2672-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, C. Z., J. H. Lin, M. Chao, K. McKnight, and M. M. Lai. 1993. RNA-binding activity of hepatitis delta antigen involves two arginine-rich motifs and is required for hepatitis delta virus RNA replication. J. Virol. 67:2221-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lehmann, K. A., and B. L. Bass. 1999. The importance of internal loops within RNA substrates of ADAR1. J. Mol. Biol. 291:1-13. [DOI] [PubMed] [Google Scholar]
- 25.Liu, Y., C. X. George, J. B. Patterson, and C. E. Samuel. 1997. Functionally distinct double-stranded RNA-binding domains associated with alternative splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase. J. Biol. Chem. 272:4419-4428. [DOI] [PubMed] [Google Scholar]
- 26.Luo, G. X., M. Chao, S. Y. Hsieh, C. Sureau, K. Nishikura, and J. Taylor. 1990. A specific base transition occurs on replicating hepatitis delta virus RNA. J. Virol. 64:1021-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macnaughton, T. B., and M. M. Lai. 2002. Large hepatitis delta antigen is not a suppressor of hepatitis delta virus RNA synthesis once RNA replication is established. J. Virol. 76:9910-9919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakano, T., C. N. Shapiro, S. C. Hadler, J. L. Casey, M. Mizokami, E. Orito, and B. H. Robertson. 2001. Characterization of hepatitis D virus genotype III among Yucpa Indians in Venezuela. J. Gen. Virol. 82:2183-2189. [DOI] [PubMed] [Google Scholar]
- 29.Naumann, H., S. Schaefer, C. F. Yoshida, A. M. Gaspar, R. Repp, and W. H. Gerlich. 1993. Identification of a new hepatitis B virus (HBV) genotype from Brazil that expresses HBV surface antigen subtype adw4. J. Gen. Virol. 74:1627-1632. [DOI] [PubMed] [Google Scholar]
- 30.Ohman, M., A. M. Kallman, and B. L. Bass. 2000. In vitro analysis of the binding of ADAR2 to the pre-mRNA encoding the GluR-B R/G site. RNA 6:687-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palladino, M. J., L. P. Keegan, M. A. O'Connell, and R. A. Reenan. 2000. dADAR, a Drosophila double-stranded RNA-specific adenosine deaminase is highly developmentally regulated and is itself a target for RNA editing. RNA 6:1004-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pohl, C., B. M. Baroudy, K. F. Bergmann, P. J. Cote, R. H. Purcell, J. Hoofnagle, and J. L. Gerin. 1987. A human monoclonal antibody that recognizes viral polypeptides and in vitro translation products of the genome of the hepatitis D virus. J. Infect. Dis. 156:622-629. [DOI] [PubMed] [Google Scholar]
- 33.Poisson, F., P. Roingeard, A. Baillou, F. Dubois, F. Bonelli, R. A. Calogero, and A. Goudeau. 1993. Characterization of RNA-binding domains of hepatitis delta antigen. J. Gen. Virol. 74:2473-2478. [DOI] [PubMed] [Google Scholar]
- 34.Polson, A. G., and B. L. Bass. 1994. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 13:5701-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polson, A. G., B. L. Bass, and J. L. Casey. 1996. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature 380:454-456. [DOI] [PubMed] [Google Scholar]
- 36.Polson, A. G., H. L. Ley III, B. L. Bass, and J. L. Casey. 1998. Hepatitis delta virus RNA editing is highly specific for the amber/W site and is suppressed by hepatitis delta antigen. Mol. Cell. Biol. 18:1919-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rizzetto, M. 1983. The delta agent. Hepatology 3:729-737. [DOI] [PubMed] [Google Scholar]
- 38.Rueter, S. M., T. R. Dawson, and R. B. Emeson. 1999. Regulation of alternative splicing by RNA editing. Nature 399:75-80. [DOI] [PubMed] [Google Scholar]
- 39.Ryu, W. S., M. Bayer, and J. Taylor. 1992. Assembly of hepatitis delta virus particles. J. Virol. 66:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato, S., S. K. Wong, and D. W. Lazinski. 2001. Hepatitis delta virus minimal substrates competent for editing by ADAR1 and ADAR2. J. Virol. 75:8547-8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shakil, A. O., S. Hadziyannis, J. H. Hoofnagle, A. M. Di Bisceglie, J. L. Gerin, and J. L. Casey. 1997. Geographic distribution and genetic variability of hepatitis delta virus genotype I. Virology 234:160-167. [DOI] [PubMed] [Google Scholar]
- 42.Wang, J. G., J. Cullen, and S. M. Lemon. 1992. Immunoblot analysis demonstrates that the large and small forms of hepatitis delta virus antigen have different C-terminal amino acid sequences. J. Gen. Virol. 73:183-188. [DOI] [PubMed] [Google Scholar]
- 43.Wang, K. S., Q. L. Choo, A. J. Weiner, J. H. Ou, R. C. Najarian, R. M. Thayer, G. T. Mullenbach, K. J. Denniston, J. L. Gerin, and M. Houghton. 1986. Structure, sequence and expression of the hepatitis delta viral genome. Nature 323:508-514. [DOI] [PubMed] [Google Scholar]
- 44.Weiner, A. J., Q. L. Choo, K. S. Wang, S. Govindarajan, A. G. Redeker, J. L. Gerin, and M. Houghton. 1988. A single antigenomic open reading frame of the hepatitis delta virus encodes the epitope(s) of both hepatitis delta antigen polypeptides p24 delta and p27 delta. J. Virol. 62:594-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wong, S. K., and D. W. Lazinski. 2002. Replicating hepatitis delta virus RNA is edited in the nucleus by the small form of ADAR1. Proc. Natl. Acad. Sci. USA 99:15118-15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu, J. C., T. Y. Chiang, and I. J. Sheen. 1998. Characterization and phylogenetic analysis of a novel hepatitis D virus strain discovered by restriction fragment length polymorphism analysis. J. Gen. Virol. 79:1105-1113. [DOI] [PubMed] [Google Scholar]
- 47.Yang, J. H., P. Sklar, R. Axel, and T. Maniatis. 1997. Purification and characterization of a human RNA adenosine deaminase for glutamate receptor B pre-mRNA editing. Proc. Natl. Acad. Sci. USA 94:4354-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and biotechnology. Kluwer Academic Publishers, Dordrecht, The Netherlands.