Abstract
The Tabby markings of the domestic cat are unique coat patterns for which no causative candidate gene has been inferred from other mammals. In this study, a genome scan was performed on a large pedigree of cats that segregated for Tabby coat markings, specifically for the Abyssinian (Ta-) and blotched (tbtb) phenotypes. There was linkage between the Tabby locus and eight markers on cat chromosome B1. The most significant linkage was between marker FCA700 and Tabby (Z = 7.56, θ = 0.03). Two additional markers in the region supported linkage, although not with significant LOD scores. Pairwise analysis of the markers supported the published genetic map of the cat, although additional meioses are required to refine the region. The linked markers cover a 17-cM region and flank an evolutionary breakpoint, suggesting that the Tabby gene has a homologue on either human chromosome 4 or 8. Alternatively, Tabby could be a unique locus in cats.
Keywords: Abyssinian, blotched, cat, classic, Felis catus, mackerel, Tabby
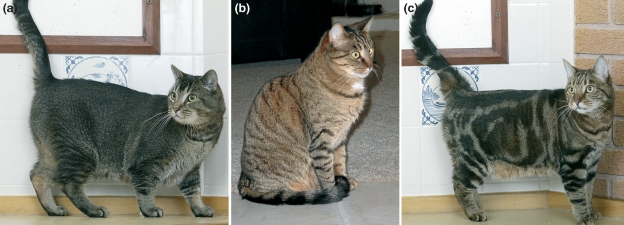
Coat colour striping patterns, commonly known as Tabby markings, are frequent in domestic cats, but a candidate is not available from other species. Traditionally, three autosomal alleles have been suggested for Tabby: Abyssinian (Ta; a.k.a. ticked), mackerel (Tm; a.k.a. striped) and blotched (tb; a.k.a. classic). Cats with traditional patterns resulting from Tabby alleles are presented in Fig. 1. The allelic series Ta > Tm > tb has been suggested; however, this series is disputed because the Tabby coat patterns that form a distinctive spotted pattern cannot be explained with a single, allelic model. However, traditional patterns are represented by three alleles: Ta, which produces few markings and possible stripes on the head, legs and tail but not on the torso; Tm, which produces stripes on the head, legs, tail and torso; and tb, which produces stripes on the head, legs and tail but circular patterns on the torso. Cats that are homozygous for the Abyssinian allele (TaTa) may have less barring on the legs and can be distinguished from heterozygotes. Hence, the Ta allele can be considered co-dominant to Tm and tb when considering the leg barring, but this demarcation has variable expression and is difficult to distinguish.
Figure 1.
Tabby patterns in the domestic shorthair cats: (a) Abysinnian, Ta-, (b) mackerel, TmTm or Ttb, and (c) blotched, tbtb. The Abysinnian pattern is also known as ticked, the mackerel pattern as striped and the blotched pattern as classic. Homozygous TaTa individuals have the same pattern on the torso but may have less barring on the legs and tail. The Tmtbmackerel pattern may appear as the broken mackerel or spotted mackerel patterns; however, these phenotypes have not segregated in pedigrees, and the spotted pattern may be affected by modifiers.
In the mouse, loss-of-function alleles for the sex-linked ectodysplasin-A (Eda) gene were originally described as ‘Tabby’, but this phenotype affects hair structure and is completely unrelated to the Tabby phenotypic patterns in cats (Ferguson et al. 1997; Srivastava et al. 1997). Therefore, a full-genome scan across the cat autosomes was performed to identify the locus for Tabby markings. An extended pedigree of 64 cats segregating for Tabby markings (Fig. 2) from the WALTHAM Centre for Pet Nutrition (Melton Mowbray, Leics, UK) was used for linkage analysis. Three marking patterns were used to demarcate the Tabby phenotypes: Abyssinian, mackerel and blotched (Fig. 1; Lomax & Robinson 1988). Buccal cells were non-invasively obtained by swabbing the internal cheek of each cat with a cytological brush. DNA was extracted from buccal cells from the cats representing the pedigree as previously described (Oberbauer et al. 2003).
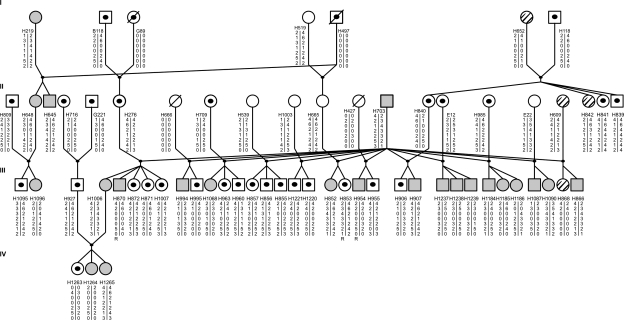
Figure 2.
Pedigree segregating for domestic cat Tabby patterns and linked markers from cat chromosome B1. Circles represent females, squares represent males, filled symbols indicate phenotypically Tabby Abyssinian cats, symbols with a center dot are Tabby blotched patterned cats and striped symbols indicate Tabby mackerel pattern cats. Unknown patterns are presented as open symbols. Unknown cats had solid, non-agouti colours, thus Tabby patterns could not be distinguished. Symbols with a diagonal slash represent cats that were not available for the analyses. Numbers under the symbols represent the laboratory sample numbers. Genotypes for the seven linked markers (FCA023, FCA809, FCA811, FCA812, FCA700, FCA254 and FCA813) are represented below the cat identification numbers respectively and are presented as haplotypes. No genotype data are represented by ‘0’. Cat H118 was duplicated to break an inbreeding loop and is also represented as B118. Cats that are recombinants between at least one marker in the haplotype and the Tabby phenotype are indicated with an ‘R’.
Approximately 150 feline-derived microsatellites were selected from the feline linkage (Menotti-Raymond et al. 1999, 2003) and radiation hybrid (RH) (Murphy et al. 2000) maps to conduct a genome scan with an average marker spacing of 75 cR. Genotyping for the markers (Young et al. 2005) and two-point linkage between the microsatellite genotypes and the Tabby phenotype (Lyons et al. 2005) was conducted using the linkage software program (Lathrop et al. 1984). Only torso patterns were considered for the phenotypic determination of Tabby Abyssinian to avoid problems when determining homozygotes and heterozygotes for this allele. To represent the dominant allelic series, the Tabby alleles were coded as binary factors for the linkage analysis: 111 representing Ta, 011 representing Tm and 001 representing the tb allele. Frequencies of the phenotype and of the co-dominant marker alleles were estimated by directly counting unrelated parents in the pedigree. One individual (H118) was duplicated to remove an inbreeding loop.
Pairwise analyses for markers on cat chromosome B1 and the Tabby phenotype are presented in Table 1. Linkage was suggested between eight markers on feline chromosome B1 and Tabby with LOD scores >3.0. Seven additional markers on cat chromosome B1 were genotyped to refine the linked region and the recombination map for this chromosome (Table 1). The most significant linkage was between marker FCA700 and Tabby (Z = 7.56, θ = 0.03). The small number of meioses in the pedigree restricts accurate multipoint mapping of the region; however, the Tabby locus likely resides between markers FCA559 and FCA254, which cover an estimated 113.6-cR region on the RH map of cat chromosome B1 and approximately 15.9 Mb or 17 cM on the linkage map (Menotti-Raymond et al. 2003). The other 55 markers genotyped across the pedigree were not linked to Tabby (data not shown).
Table 1.
LOD scores for pairwise comparisons between Tabby and chromosome B1 microsatellite markers.
| Marker | Feline B1 position (cR)1 | HSA chromosome2 | Max LOD (Z)3 | Recombination rate (θ) at LOD (Z) |
|---|---|---|---|---|
| FCA577 | 156.7 | No RH | NS | – |
| FCA519 | 248.7 | 8 | 1.42 | 0.191 |
| FCA559 | 416.9 | 8 | 3.17 | 0.099 |
| FCA339 | 566.1 | 8 | – | – |
| FCA023 | 597.1 | No RH | 5.38 | 0.038 |
| FCA809 | 608.7 | 8 | 5.34 | 0.081 |
| FCA810 | 618.1 | No RH | 0.48 | 0.000 |
| FCA811 | 630.2 | No RH | 3.69 | 0.048 |
| FCA812 | 648.1 | No RH | 3.76 | 0.053 |
| FCA700 | 663.0 | 4 | 7.56 | 0.030 |
| FCA254 | 679.7 | No RH | 5.09 | 0.106 |
| FCA813 | 701.2 | No RH | 3.68 | 0.05 |
| FCA550 | 731.8 | No RH | NS | – |
| FCA814 | 774.0 | No RH | NS | – |
| FCA815 | 783.6 | No RH | – | – |
| FCA513 | 793.6 | No RH | 2.64 | 0.144 |
| FCA816 | 826.1 | 4 | NS | – |
Radiation hybrid (RH) positions were previously published (Menotti-Raymond et al. 2003).
Human chromosome that has homologous sequence to the feline sequence. Markers without a chromosome position have not been assigned on the human radiation hybrid map (no RH).
NS, non-significant LOD score.
Positions (in cR) of the markers on cat chromosome B1 and the homologous sequences in humans are shown in Table 1. The comparative map between the cat and human indicates that a gene controlling the Tabby locus is on feline chromosome B1 near a breakpoint of evolutionarily conserved segments that are homologous to human chromosomes 4 and 8.
Domestic cats have several tabby coat patterns, including Abyssinian, mackerel, broken mackerel, spotted and blotched. The analysed pedigree was limited primarily to the Abyssinian and blotched phenotypes. One mating between phenotypically Abyssinian and mackerel cats produced one cat of each phenotype and one mating between phenotypically blotched and mackerel cats produced two blotched and one mackerel cat. The segregation of these patterns is consistent with the hypothesis of a tri-allelic locus with a dominant series of alleles; however, the number of meioses from mackerel patterned cats was limited in our pedigree. Leg barring was not used to distinguish Abyssinian heterozygotes, and the mackerel patterns were not delineated between mackerel and broken mackerel in the analysis. Thus, controlled matings are required to clarify these classifications. In wild felids, such as lions and pumas, spotted Tabby patterns are evident in cubs but are lacking in the TabbyAbyssinian patterns of the adults. Along with the presentation of other coat pattern markings such as the spotted patterns and broken mackerel patterns, alternative hypotheses for the dominance of alleles and the presence of potential modifiers are still possible. The identified 17-cM region on cat chromosome B1 should be explored for candidate genes and causative mutations for feline Tabby patterns.
Acknowledgments
Funding for this project was provided to L. A. Lyons from NIH-NCRR RR016094, the Winn Feline Foundation, and the George and Phyllis Miller Feline Health Fund, Center for Companion Animal Health, School of Veterinary Medicine, University of California, Davis. We acknowledge the National Cancer Institute for allocation of computing time and staff support at the Advanced Biomedical Computing Center of the Frederick Cancer Research and Development Center. We appreciate additional technical support and the use of instrumentation at the Veterinary Genetics Laboratory, University of California, Davis.
References
- Ferguson BM, Brockdorff N, Formstone E, et al. Cloning of Tabby, the murine homolog of the human EDA gene: evidence for a membrane-associated protein with a short collagenous domain. Human Molecular Genetics. 1997;6:1589–94. doi: 10.1093/hmg/6.9.1589. [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, et al. Strategies for multilocus linkage analysis in humans. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:3443–6. doi: 10.1073/pnas.81.11.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomax T, Robinson R. Tabby pattern alleles of the domestic cat. Journal of Heredity. 1988;79:21–3. doi: 10.1093/oxfordjournals.jhered.a110438. [DOI] [PubMed] [Google Scholar]
- Lyons LA, Imes DL, Rah HC, et al. Tyrosinase mutations associated with Siamese and Burmese patterns in the domestic cat (Felis catus) Animal Genetics. 2005;36:119–26. doi: 10.1111/j.1365-2052.2005.01253.x. [DOI] [PubMed] [Google Scholar]
- Menotti-Raymond M, David VA, Lyons LA, et al. A genetic linkage map of microsatellites in the domestic cat (Felis catus) Genomics. 1999;57:9–23. doi: 10.1006/geno.1999.5743. [DOI] [PubMed] [Google Scholar]
- Menotti-Raymond M, David VA, Roelke ME, et al. Second-generation integrated genetic linkage/radiation hybrid maps of the domestic cat (Felis catus) Journal of Heredity. 2003;94:95–106. doi: 10.1093/jhered/esg008. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Sun S, Chen Z, et al. A radiation hybrid map of the cat genome: implications for comparative mapping. Genome Research. 2000;10:691–702. doi: 10.1101/gr.10.5.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberbauer AM, Grossman DI, Eggleston ML, et al. Alternatives to blood as a source of DNA for large-scale scanning studies of canine genome linkages. Veterinary Research Communication. 2003;27:27–38. doi: 10.1023/a:1022006606796. [DOI] [PubMed] [Google Scholar]
- Srivastava AK, Pispa J, Hartung AJ, et al. The Tabby phenotype is caused by mutation in a mouse homologue of the EDA gene that reveals novel mouse and human exons and encodes a protein (ectodysplasin-A) with collagenous domains. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:13069–74. doi: 10.1073/pnas.94.24.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AE, Biller DS, Hergesell EJ, et al. Feline polycystic kidney disease is linked to the PKDI region. Mammalian Genome. 2005;16:59–65. doi: 10.1007/s00335-004-2412-2. [DOI] [PubMed] [Google Scholar]