Abstract
Herpes simplex virus (HSV) DNA polymerase (Pol) mutations can confer resistance to all currently available antiherpetic drugs. However, discrimination between mutations responsible for drug resistance and those that are part of viral polymorphism can be difficult with current methodologies. A new system is reported for rapid generation of recombinant HSV type 1 (HSV-1) DNA Pol mutants based on transfection of a set of overlapping viral cosmids and plasmids. With this approach, twenty HSV-1 recombinants with single or dual mutations within the DNA pol gene were successfully generated and subsequently evaluated for their susceptibilities to acyclovir (ACV), foscarnet (FOS), cidofovir (CDV), and adefovir (ADV). Mutations within DNA Pol conserved regions II (A719T and S724N), VI (L778M, D780N, and L782I), and I (F891C) were shown to induce cross-resistance to ACV, FOS, and ADV, with two of these mutations (S724N and L778M) also conferring significant reduction in CDV susceptibility. Mutant F891C was associated with the highest levels of resistance towards ACV and FOS and was strongly impaired in its replication capacity. One mutation (D907V) lying outside of the conserved regions was also associated with this ACV-, FOS-, and ADV-resistant phenotype. Some mutations (K522E and Y577H) within the δ-region C were lethal, whereas others (P561S and V573M) induced no resistance to any of the drugs tested. Recombinants harboring mutations within conserved regions V (N961K) and VII (Y941H) were resistant to ACV but susceptible to FOS. Finally, mutations within conserved region III were associated with various susceptibility profiles. This new system allows a rapid and accurate evaluation of the functional role of various DNA Pol mutations, which should translate into improved management of drug-resistant HSV infections.
Two categories of antivirals are available for the management of herpes simplex virus (HSV) infections. The first class of agents includes molecules that, following viral and/or cellular phosphorylation, compete with natural deoxynucleoside triphosphates for incorporation into the elongating viral DNA chain. Acyclovir (ACV) and penciclovir with their respective prodrugs, valacyclovir and famciclovir, are agents representative of this class and are considered the standard therapy for HSV infections. Although not indicated for the treatment of HSV infections, cidofovir [(S)-1-(3-hydroxy-2-phosphonylmethoxypropyl)cytosine; CDV] and adefovir [9-(2-phosphonylmethoxyethyl)adenine; ADV] are acyclic nucleoside phosphonate derivatives that, once activated, are also alternative substrates for the viral DNA polymerase (Pol). The other class of HSV inhibitors consists of pyrophosphate analogues such as foscarnet (FOS). These agents are direct noncompetitive inhibitors of the viral DNA Pol. A single DNA Pol mutation could thus confer resistance to more than one antiviral depending on the specific drug binding site on the enzyme.
Resistance to ACV, the most widely used anti-HSV agent, is due mainly to mutations in the viral thymidine kinase (TK) gene which impair the initial drug phosphorylation (23). Although less frequently encountered, ACV- and/or FOS-resistant clinical isolates containing DNA Pol mutations have been reported following prolonged therapy with these agents (9, 15, 39, 41, 43). HSV DNA Pol mutations leading to CDV resistance have also been found in laboratory-derived strains (1, 2), although they have not yet been identified in clinical isolates from patients failing therapy. Moreover, our group has previously reported the emergence of HSV DNA Pol mutations associated with a reduction in both FOS and ADV susceptibility in patients receiving FOS therapy (6).
The putative catalytic domain of the HSV DNA Pol enzyme contains eight conserved regions designated I to VII and δ-region C (8, 27, 47). These regions share significant amino acid sequence homology with all other herpesvirus DNA Pol enzymes. Drug resistance mutations have been found mainly within such conserved regions of the HSV DNA pol gene, with regions II and III containing the greatest clusters of mutations (reviewed in reference 23). The δ-region C, which is common to cellular DNA Pols δ (47), is part of the 3′-5′ exonuclease editing domain of the enzyme. When not lethal, some mutations within this region have been shown to confer resistance to pyrophosphate analogues (22, 29, 33, 34). Only a few drug-resistant mutations have been described within the other conserved regions or outside such regions (1, 6, 18, 22, 27, 36, 43). Because of the reported viral polymorphism (11, 32), the role of any DNA Pol mutation must be carefully assessed. Marker transfer experiments, in which a specific mutation is transferred to a wild-type (wt) virus background by homologous recombination, have been used to generate recombinant HSV DNA Pol mutants (10, 27, 28, 35, 36, 39). In addition to being fastidious and inefficient, such methods rely on selection of recombinant mutants through drug pressure which may frequently lead to additional undesired mutations.
Cihlar et al. have elegantly demonstrated the possibility of using a system of overlapping cosmids and plasmids to generate recombinant human cytomegalovirus (HCMV) strains with specific DNA Pol mutations (12). Although not specifically designed for site-directed mutagenesis experiments with the DNA pol gene, a similar system has been reported for HSV-1 by Cunningham and Davison (16). Herein, we report a modification of the previous cosmid-based recombination system aimed at generating specific HSV-1 DNA Pol mutants. Such an approach was used to evaluate the drug susceptibility phenotypes and replicative capacities of several HSV-1 DNA Pol mutants.
MATERIALS AND METHODS
Cells and viruses.
Vero cells were maintained in Eagle's minimum essential medium supplemented with 10% fetal calf serum and antibiotics. HSV-1 laboratory strain 17 and all recombinant viruses were propagated in Vero cells.
DNA constructs.
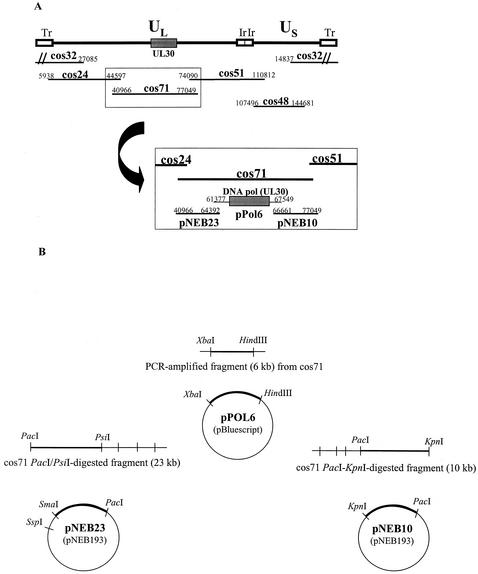
The set of five overlapping DNA cosmids containing the entire genome of strain 17 was kindly donated by Charles Cunningham, MRC Virology Unit, Glasgow, United Kingdom (16). The original viral fragment of cosmid 71, corresponding to nucleotides (nt) 40966 to 77049, was replaced by three viral plasmids (pNEB23, pPol6, and pNEB10) as shown in Fig. 1A. Briefly, a viral fragment corresponding to nt 61377 to 67549 and encompassing the entire HSV DNA pol gene (UL30; nt 62807 to 66514) was cloned into the pBluescript II vector (Stratagene) to generate pPol6. This DNA fragment was amplified from cosmid 71 by using primers 5′-CAATCTAGAGGTAAATTTCTCCCCGA-3′ (forward) and 5′-CAAAAGCTTTGGCCTGCGGGAACGAGTT-3′ (reverse), containing, respectively, an XbaI and a HindIII restriction site (underlined). The purified XbaI-HindIII-digested PCR product was then cloned into the XbaI-HindIII site of the pBluescript vector as illustrated in Fig. 1B, and the exact sequence of the catalytic domain of the DNA pol gene was verified. Plasmids pNEB23 and pNEB10 were generated by cloning DNA fragments (nt 40966 to 64392 and nt 66661 to 77049, respectively) from PacI-PsiI- and PacI-KpnI-digested cosmid 71 products into the PacI-SmaI and PacI-KpnI sites of pNEB193 (New England Biolabs) (Fig. 1B).
FIG. 1.
Schematic representation of viral cosmids and plasmids used to generate HSV-1 recombinants. (A) Inserts of the five cosmids (24, 32, 48, 51, and 71) covering the complete genome of the HSV-1 strain 17 (16), along with the three plasmids (pNEB23, pPol6, and pNEB10) designed to replace cos71. Tr, terminal repeat; UL, unique long; US, unique short; Ir, internal repeat. (B) Construction of the pNEB23, pPol6, and pNEB10 plasmids. The nucleotide positions of inserts in the HSV strain 17 genome are indicated.
Site-directed mutagenesis of the DNA pol gene.
Mutagenesis was performed by using the QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. Plasmid pPol6 was used as a template for all mutagenesis experiments. Each mutation was introduced by using a specific pair of mutagenic primers ranging in length from 28 to 40 bp. The correct nucleotide sequence of the catalytic domain of the DNA pol gene from each mutated plasmid was verified by sequence analysis and compared to that from reference strain 17.
Generation of DNA Pol mutants.
Inserts from cosmids 24, 32, 48, and 51 were released by digestion with PacI (16). Inserts from plasmids pPol6, pNEB23, and pNEB10 were released by digestion with XbaI-HindIII, PacI-SspI, and PacI-KpnI, respectively. All enzymes were inactivated, and the seven overlapping DNA fragments were cotransfected into Vero cells in 6-well plates by using Lipofectamine 2000 (Life Technologies) according to the manufacturer's instructions. Viral cytopathic effects were usually observed within a 3-day incubation period. The catalytic domain of the DNA pol gene from each recombinant was amplified and verified by DNA sequencing.
Structural analysis of viral genomes.
Extracellular virions were collected from 18 ml of culture medium by centrifugation at 100,000 × g for 120 min. The virus pellets were resuspended in 0.5 ml of TNE-sodium dodecyl sulfate buffer (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 25 mM EDTA, 0.5% sodium dodecyl sulfate) and incubated overnight with 0.2 mg of proteinase K at 56°C with moderate agitation. DNA was extracted twice with phenol-chloroform-isoamyl alcohol (25:24:1) and once with chloroform-isoamyl alcohol (24:1), precipitated with ethanol, and resuspended in H2O. The genomic structure of recombinant HSV-1 strains was determined by digestion of purified extracellular recombinant virus with KpnI and separation on a 0.5% agarose gel. Viral DNA was then transferred onto Hybond-N+ membranes (Amersham) and probed with the various cosmids labeled by the ECL direct nucleic acid labeling system (Amersham) according to the manufacturer's instructions.
Antiviral susceptibility assays.
Susceptibility to ACV, FOS, CDV (gift from Gilead), and ADV (gift from Gilead) was determined by a plaque reduction assay with Vero cells (41). The mean 50% inhibitory concentration (IC50) for each mutant virus was determined by testing in duplicate each of two independently generated recombinants. A twofold difference in IC50s between the mutant virus and the wt recombinant virus was considered significant, as previously reported (12). In the case of ACV, this difference is about the same as the one between the established cutoff value for resistance (2 μg/ml) and the mean IC50 for susceptible clinical isolates (40, 41).
In vitro single-cycle replication experiments.
The replication capacities of selected DNA Pol mutants were compared to that of the wt recombinant virus by assessing the yield of extracellular virus production after infection of Vero cells at a high multiplicity of infection (MOI) (19, 28). On the day of infection, cells were infected with each recombinant at a MOI of 5, and then 200 μl of supernatant was collected at specific times postinfection, ranging from 2 to 144 h. Supernatants were stored at −80°C before titration in 24-well plates.
RESULTS
Efficiency of DNA recombination between cosmid and plasmid inserts.
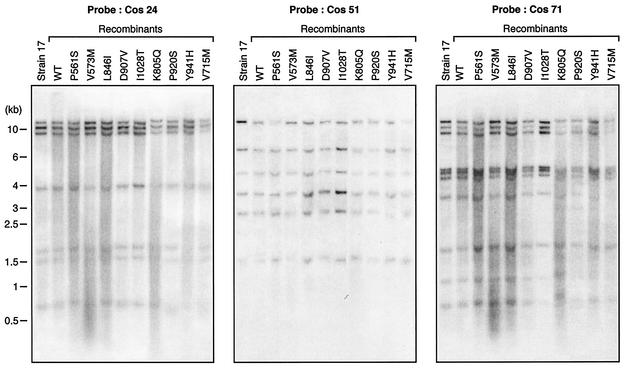
Cotransfection into Vero cells of digested cosmids and plasmids generated infectious recombinant viruses, with typical cytopathic effects usually observed within 3 days. The genomic structures of the recombinant viruses were verified by performing Southern blot analysis of KpnI-digested purified genomes with independent cosmids as hybridization probes. Figure 2 shows representative results with three different probes. Overall, Southern blot analysis of the recombinants revealed no structural differences between the genomes of the recombinant viruses and that of the parental HSV-1 strain 17.
FIG. 2.
Southern blot analysis of some recombinant HSV-1 wt and mutant viruses along with the parental laboratory strain HSV 17 by using some of the viral cosmids (cos 24, 51, and 71) as probes. Numbers on the left indicate molecular sizes in kilobase pairs.
Susceptibility of HSV recombinants to antivirals.
A total of 20 DNA Pol mutants were evaluated in this study by generating two independent recombinants for each specific mutation. Those mutations were selected on the basis of their presence within previously reported clinical HSV isolates (6, 27, 43) or within laboratory-derived HSV strains (1, 22, 28, 29, 32, 34, 36). Furthermore, a few HSV-1 DNA Pol mutants were generated based on the presence of similar mutations within the HCMV DNA pol gene (4, 5, 12, 31, 44). Each recombinant was tested at least twice for its susceptibility to four antivirals (ACV, FOS, CDV, and ADV) by using the plaque reduction assay (Table 1). On all occasions, the IC50s of all antivirals were similar for the two generated recombinant viruses harboring the same mutation.
TABLE 1.
Genotypic and phenotypic characterization of recombinant HSV-1a with regard to antivirals
| Conserved region(s) | Nucleotide change(s) | Amino acid change(s) | Reference(s)d | IC50 (μg/ml) of ACV for:
|
SDf | Ratiog | IC50 (μg/ml) of FOS | SD | Ratio | IC50 (μg/ml) of CDV | SD | Ratio | IC50 (μg/ml) of ADV | SD | Ratio | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mutant 1 | Mutant 2 | |||||||||||||||
| wt | wt | 0.0057 | 0.0064 | ±0.0012 | 1.00 | 15.0275 | ±1.61 | 1.00 | 0.1605 | ±0.0377 | 1.00 | 0.8475 | ±0.05 | 1.00 | ||
| δ-C | A1654G | K552Eb | 12, 44 (K513Ec) | |||||||||||||
| δ-C | C1681T | P561S | 12, 30, 46 (P522Sc) | 0.0054 | 0.0054 | ±0.0007 | 0.85 | 13.6725 | ±1.91 | 0.91 | 0.2383 | ±0.0414 | 1.48 | 0.1800 | ±0.07 | 0.21 |
| δ-C (exoIII) | G1717A | V573M | 1 | 0.0122 | 0.0120 | ±0.0030 | 1.90 | 13.0340 | ±3.95 | 0.87 | 0.1861 | ±0.0530 | 1.16 | 1.1108 | ±0.42 | 1.31 |
| δ-C (exoIII) | T1729C | Y577Hb | 28, 29 | |||||||||||||
| II | G2099T | R700M | 1 | 0.0044 | 0.0062 | ±0.0021 | 0.98 | 2.0350 | ±0.52 | 0.14 | 0.5886 | ±0.0385 | 3.67 | 0.4449 | ±0.01 | 0.52 |
| II | G2143A | V715Mc | 4, 5, 12, 46 | 0.0198 | 0.0182 | ±0.0045 | 2.86 | 12.1932 | ±2.76 | 0.81 | 0.0561 | ±0.0151 | 0.35 | 1.5076 | ±0.50 | 1.78 |
| II | G2155A | A719T | 6, 43 (A724Te) | 0.0373 | 0.0377 | ±0.0043 | 5.94 | 32.6985 | ±2.57 | 2.18 | 0.2965 | ±0.0345 | 1.85 | 3.1958 | ±0.64 | 3.77 |
| II | G2171A | S724N | 1, 22, 34, 6, 43 (S729Ne) | 0.0633 | 0.0627 | ±0.0253 | 9.87 | 40.4150 | ±2.96 | 2.69 | 0.3284 | ±0.0235 | 2.05 | 4.6750 | ±1.31 | 5.52 |
| VI | C2332A | L778M | 6, 43 (D783Me) | 0.0300 | 0.0351 | ±0.0106 | 5.53 | 30.5800 | ±6.52 | 2.03 | 0.3281 | ±0.0561 | 2.04 | 2.7312 | ±1.70 | 3.22 |
| VI | G2338A | D780N | 6, 43 (D785Ne) | 0.0445 | 0.0388 | ±0.0097 | 6.10 | 32.6100 | ±5.06 | 2.17 | 0.1937 | ±0.0387 | 1.21 | 2.3979 | ±0.31 | 2.83 |
| VI | C2344A | L782I | 4, 12, 46 (V781Ic) | 0.0373 | 0.0408 | ±0.0086 | 6.43 | 34.3525 | ±4.60 | 2.29 | 0.2825 | ±0.0658 | 1.76 | 3.1875 | ±0.40 | 3.76 |
| III | A2413C | K805Qc | 12, 44 | 0.0032 | 0.0041 | ±0.0012 | 0.64 | 5.0978 | ±0.25 | 0.34 | 0.2914 | ±0.0660 | 1.82 | 0.2523 | ±0.06 | 0.30 |
| III | C2462T | T821M | 32 | 0.0294 | 0.0397 | ±0.0136 | 6.25 | 22.1939 | ±2.90 | 1.48 | 0.3392 | ±0.0364 | 2.11 | 2.3464 | ±0.53 | 2.77 |
| III | C2536A | L846I | 6, 43 (L850Ie) | 0.0094 | 0.0096 | ±0.0013 | 1.52 | 10.5263 | ±2.26 | 0.70 | 0.2015 | ±0.0332 | 1.26 | 0.3913 | ±0.11 | 0.46 |
| I | T2672G | F891C | 26, 36 | 0.2222 | 0.2222 | ±0.0196 | 34.99 | 58.7450 | ±15.15 | 3.91 | 0.2795 | ±0.0542 | 1.74 | 5.9300 | ±0.08 | 7.00 |
| I-VII | A2720T | D907V | 6, 43 (D912Ve) | 0.0280 | 0.0240 | ±0.0070 | 3.78 | 34.1425 | ±4.76 | 2.27 | 0.2555 | ±0.0594 | 1.59 | 2.1440 | ±0.68 | 2.53 |
| I-VII | C2758T | P920S | 6, 43 | 0.0051 | 0.0067 | ±0.0023 | 1.05 | 10.9750 | ±2.88 | 0.73 | 0.1037 | ±0.0130 | 0.65 | 0.8275 | ±0.09 | 0.98 |
| VII | T2821C | Y941H | 27 | 0.0555 | 0.0596 | ±0.0228 | 9.39 | 20.7158 | ±3.77 | 1.38 | 0.0629 | ±0.0164 | 0.39 | 4.8400 | ±1.31 | 5.71 |
| V | C2883G | N961K | 22 | 0.0369 | 0.0339 | ±0.008 | 5.34 | 12.4200 | ±2.12 | 0.83 | 0.2261 | ±0.1141 | 1.41 | 0.9250 | ±0.27 | 1.09 |
| After V | C3019A | L1007M | 1 | 0.0118 | 0.0117 | ±0.0012 | 1.84 | 8.8320 | ±1.20 | 0.59 | 0.1955 | ±0.0305 | 1.22 | 0.3050 | ±0.15 | 0.36 |
| After V | T3083C | I1028T | 1 | 0.0112 | 0.0111 | ±0.0023 | 1.74 | 8.6550 | ±2.15 | 0.58 | 0.3658 | ±0.0070 | 2.28 | 0.5373 | ±0.16 | 0.63 |
| II and I-VII | G2171A and C2758T | S724N and P920S | 6, 43 | 0.0811 | 0.0737 | ±0.0142 | 11.61 | 38.0000 | ±2.44 | 2.53 | 0.3819 | ±0.1218 | 2.38 | 6.5873 | ±1.59 | 7.77 |
Two independently generated recombinant viruses were tested for each mutation. IC50s are averages from at least two determinations for each of the recombinant viruses. Bold numbers indicate significant (i.e., at least twofold) changes in HSV drug susceptibility.
This mutation could not be generated.
Mutation originally described in the HCMV DNA pol (UL54) gene.
Mutations shown in parentheses are those specifically described by the references.
Mutation originally described in the HSV-2 DNA pol gene.
SD, standard deviation.
Ratios of IC50s for HSV recombinant mutants to that for parental wt recombinant.
Effects of specific mutations within conserved regions of the viral DNA pol gene.
When not lethal, some of the previously reported HSV mutations in δ-region C have been associated with resistance to pyrophosphate analogues (22, 29, 33), whereas other mutations within the same region of the HCMV DNA pol gene have been associated with resistance to ganciclovir (GCV) and CDV (12, 20, 30, 44). In this study, we were able to evaluate two recombinants with mutations (P561S and V573M) within this region, whereas mutants K552E and Y577H could not be generated. The former mutants were susceptible to all antivirals, although a slight increase in IC50s of ACV compared to that for the wt was observed for mutant V573M (mean increase, 1.90-fold). Recombinant P561S demonstrated a marked increase in ADV susceptibility compared to that of the wt virus (mean decrease in IC50, 4.76-fold).
Mutations located in conserved region II of the HSV DNA pol gene are frequently associated with resistance to ACV and pyrophosphate analogues (1, 22, 34, 42). Two (A719T and S724N) of the four mutations located within this region conferred similar phenotypes and were also associated with ADV resistance. In addition, a significant decrease in CDV susceptibility was noted for these mutants. Mutation V715M led to significant and almost significant (1.8-fold) decreases in levels of susceptibility to ACV and ADV, respectively, whereas it was associated with susceptibility to FOS and hypersusceptibility to CDV (mean decrease in IC50, 2.86-fold). The mutant R700M remained susceptible to ACV and ADV and even hypersusceptible to FOS (mean decrease in IC50, 7.14-fold), although it exhibited a significant level of resistance to CDV (mean increase in IC50, 3.67-fold).
Only a few mutations in conserved region VI of the DNA pol gene of HSV (6, 43) or HCMV (4, 12, 44, 46) have been described, and all were identified in clinical isolates. We generated three corresponding HSV-1 DNA Pol mutants (L778M, D780N, and L782I) and found that all three recombinants induced resistance to ACV, FOS, and ADV. Moreover, mutants L778M and L782I showed some reduction in CDV susceptibilities (mean increases in IC50, 2.04- and 1.76-fold, respectively). The three recombinants containing mutations within conserved region III were associated with variable levels of resistance or hypersusceptibility towards the different antiviral agents. One mutant (T821M) had a phenotype of resistance to ACV, CDV, and ADV, whereas the other two (K805Q and L846I) were susceptible or hypersusceptible to the various drugs.
Region I is one of the most conserved regions among α-like DNA Pols. Mutation F891C within this region was associated with the highest levels of resistance towards ACV and FOS (mean increases in IC50, 34.99- and 3.91-fold, respectively), with significant levels of resistance to ADV (mean increase in IC50, 7.00-fold). In contrast to previous results (27, 39), the mutant Y941H (region VII) was not associated with a significant increase in IC50s of FOS, although a marked decrease in ACV susceptibility (mean increase in IC50, 9.39-fold) was noted as previously reported. Furthermore, this amino acid substitution was associated with a significant level of resistance to ADV but induced hypersusceptibility to CDV. Mutation N961K within conserved region V conferred resistance only to ACV, as previously reported for laboratory-derived HSV-1 strains (22).
Effects of mutations outside conserved regions of the viral DNA pol gene.
Only a few mutations located between conserved regions of the HSV DNA pol gene in drug-resistant viruses have been reported (1, 6, 22, 43). Mutation D907V was identified in a clinical HSV-2 isolate that also contained a TK mutation (43). Our results confirm the role of this DNA Pol mutation in conferring resistance to ACV, FOS, and ADV. Mutation P920S was also found in combination with mutation S724N (region II) in three clinical HSV-1 isolates from a human immunodeficiency virus-infected patient (6, 43). The role of these two mutations was studied by generating three different recombinant viruses. IC50s for the mutant P920S were similar to those reported for the wt recombinant, while mutants S724N and S724N/P920S were associated with comparable multidrug resistance phenotypes. Two mutations in the C-terminal part of the HSV DNA pol gene have been described in laboratory-derived strains selected with HPMP (S-1-3-hydroxy-2-phosphonylmethoxypropyl) derivatives (1). When generated in our system, mutation L1007M conferred an almost significant increase in IC50s of ACV (mean increase in IC50, 1.84-fold) but conferred susceptibility to CDV, in contrast to previous data. On the other hand, mutant I1028T exhibited the same phenotype as previously reported, i.e., resistance to CDV only (1).
Replicative capacities of selected recombinant viruses.
The DNA pol gene encodes one of the pivotal proteins involved in viral replication. Mutations within this gene can thus alter the replicative capacities of herpesviruses (5, 13, 24, 28, 33, 36). For instance, we were unable, despite many attempts, to generate two recombinants with mutations (K552E and Y577H) located within δ-region C. The codon corresponding to Y577 is part of the exoIII motif involved in the 3′-5′ exonuclease activity of the enzyme (28, 29, 33). Although codon 552 is not part of the critical exo motifs, the Lys at the corresponding position in the protein is highly conserved among all herpesviruses.
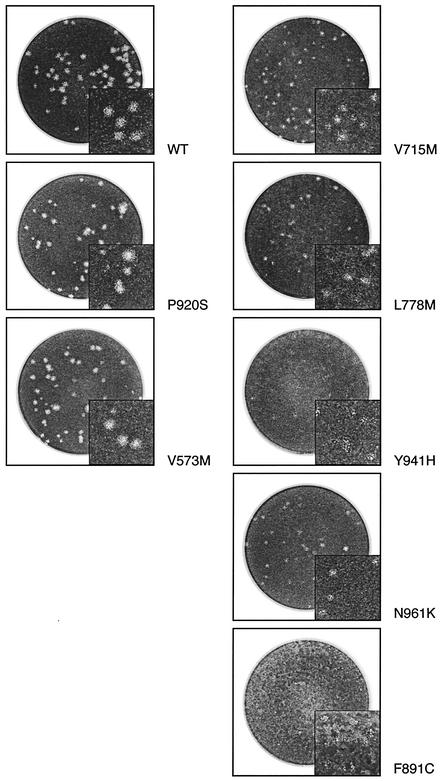
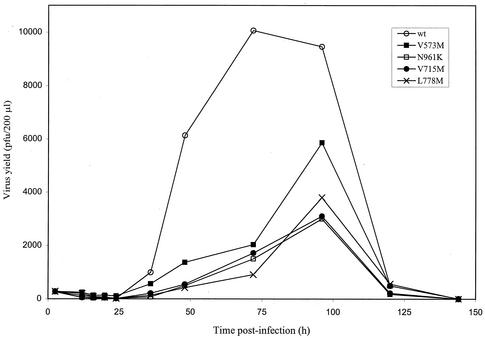
The indicated mutants were selected for evaluation of their replication kinetics based on several factors, such as (i) an obvious growth delay and/or smaller plaques when evaluated in cell culture (V715M, L778M, and N961K) (Fig. 3), (ii) a specific location within the 3′-5′ exonuclease motif (V573M), and (iii) the absence of a phenotype of resistance to all tested drugs (P920S). Mutants F891C and Y941H were so severely impaired in their ability to propagate in cell culture that sufficient titers could not be obtained to perform replication kinetic experiments (Fig. 3). The extracellular yields of the other recombinant mutants were determined over a period of 144 h postinfection and compared to that of the wt (Fig. 4). At most time points where measurement of virion production was possible (i.e., between 24 and 120 h postinfection), the yields of extracellular viruses were reduced from 5- to 15-fold for mutants V573M, V715M, L778M, and N961K compared to that of the wt. The susceptible mutant P920S had replication kinetics similar to those of the wt (data not shown).
FIG. 3.
Plaque morphologies of some recombinant HSV-1 wt and mutant viruses in 24-well plates containing Vero cells. At 96 h postinfection, cells were fixed and stained. Note that viral inoculums were not necessarily the same for each recombinant virus.
FIG. 4.
Influence of DNA Pol mutations on HSV replication capacities in single-step growth kinetics. On the day of infection, confluent Vero cells in 24-well plates were infected with either wt virus or different recombinant mutants (V573M, N961K, V715M, and L778M) at a MOI of 5. Extracellular viruses were collected at different times (up to 144 h postinfection) and titrated. Curves are representative of results from two independent experiments.
DISCUSSION
Although HSV resistance to ACV is mostly due to alterations within the TK gene (23), mutations within the viral DNA pol gene can be more problematic as the latter gene encodes the protein that is the target of all antiherpetic agents. In this study, we designed a novel approach to elucidate the role of specific DNA Pol mutations by using a set of overlapping viral plasmids and cosmids. Twenty DNA Pol recombinant mutants were generated and tested for their drug susceptibility patterns. Our results have revealed that alterations in a specific conserved region are not necessarily associated with uniform drug resistance patterns. Furthermore, we showed the importance of the central part of the enzyme's catalytic domain (regions II and VI) as a multidrug recognition site.
The main advantage of our method is the lack of a drug selection step, since only homogenous progeny of mutant virus is generated from the overlapping viral fragments. Although we could not exclude the emergence of mutations at the different recombination sites, we believe that such events were unlikely due to the following observations: (i) IC50s for both independently generated recombinants containing the same DNA Pol mutation were virtually identical, and (ii) no major genomic rearrangements occurred during recombination, as assessed by Southern blot hybridization. The HSV DNA Pol is a multifunctional enzyme that shows several similarities with α-like DNA Pols (8). Although no direct crystallographic data are available to place specific HSV DNA Pol mutations in a three-dimensional context, the structural significance of some mutations may be deduced based on similarity with Pol regions from other members of this family (21, 26, 45).
Our results demonstrate that mutations in all conserved regions of the enzyme could be associated with ACV resistance. Although the chosen resistance definition for our recombinants was somewhat arbitrary (twofold increase in IC50 over that for the wt), we were able to corroborate most previously reported ACV resistance phenotypes associated with DNA Pol mutations in laboratory-derived (1, 22, 32, 34, 36) and clinical (27, 39) HSV isolates. Of interest, mutation F891C, within conserved region I, was shown to confer the highest level of resistance to ACV in our study. It has been previously suggested that this mutation could alter the conformation of this highly conserved region so that the Pol is less able to incorporate ACV triphosphate (26). Of note, we found a significant level of ACV resistance for DNA Pol mutants previously reported to be susceptible based on an ACV resistance cutoff of 2 μg/ml (6, 43). This fact illustrates the difficulty of relying on a specific IC50 for defining resistance in the absence of a pretherapy isolate. Our system was also useful for sorting out the role of dual DNA Pol and/or TK mutations previously reported in two clinical isolates (6, 43). In the first case, we were able to confirm the role of mutation D907V located outside conserved regions in the phenotype of resistance to ACV. This finding, combined with previous confirmation of the role of a TK mutation in the same clinical isolate (7), establishes the additive effect of mutations in two genes with respect to the final ACV resistance phenotype (6, 42). In the second case, our data now support a role for the S724N (region II) mutation but not for a concomitant DNA Pol mutation (P920S) located outside a conserved region in the ACV resistance phenotype.
Mutations associated with FOS resistance were found to be located mostly within conserved regions II, VI, and I. Mutation D907V, which is located in a nonconserved region, was also associated with resistance to FOS. While some mutations that are part of conserved region III have been associated with resistance to pyrophosphate analogues (6, 22, 25, 43), the three mutants tested in this study remained susceptible to FOS. Our results confirm the role of most DNA Pol mutations (A719T, S724N, L778M, D780N, and D907V) present in previously reported FOS-resistant clinical isolates (6, 43). The mutation L846I (region III) generated in this study was not associated with FOS resistance. This mutation was located next to the previously reported corresponding HSV-1 mutation (L845I) (6, 43). Thus, the role of newly observed mutations in drug resistance must be adequately confirmed even when the mutations are located close to a known mutation associated with a specific drug phenotype. Most notably, our recombinant Y941H mutant was susceptible to FOS despite the fact that this exact mutation was found in a FOS-resistant HSV-1 clinical isolate (27, 39). It is possible that additional DNA Pol mutations (not part of the sequenced region) could have been present in the previous isolate. Alternatively, differences in the genetic backgrounds of the two HSV-1 strains could also possibly explain the discrepant phenotypes. Of note, the acute HSV infection associated with this clinical DNA Pol mutant was successfully managed with intravenous FOS therapy (39).
Our results expand on the limited information regarding HSV resistance to CDV. We found that a few DNA Pol mutants with mutations in regions II, VI, and III exhibited decreased susceptibility towards this antiviral. We were able to confirm a role in the CDV resistance phenotype for some (R700M and I1028T) but not all (V573M and L1007M) mutations previously reported in laboratory-derived strains selected with HPMP derivatives (1). As the authors of the previous study did not sequence the entire DNA Pols of those laboratory-derived strains, it is possible that additional mutations could be responsible for the CDV-resistant phenotype. Finally, we confirmed the role of the unique mutant (L778M) reported to be CDV resistant in our previous study (6). We also found the same phenotype for the mutant S724N, otherwise reported to be susceptible to CDV (6). However, the use of a few wt isolates (not obtained from the same patients) to compare CDV IC50s may explain such discrepancy. Importantly, we did not find a correlation between ACV and CDV resistance in HSV mutants as reported between GCV and CDV resistance in HCMV DNA Pol mutants, suggesting that the binding sites of ACV and GCV may differ (3, 14) or that those sites may vary according to the virus.
ADV resistance was associated with several DNA Pol mutations, mainly in regions II, VI, I, and VII, as previously reported for laboratory-derived (1) and clinical (6) HSV isolates. Moreover, consistent with the results of the previous HSV studies (1, 6) and with those of studies of HCMV recombinants (12), an almost perfect cross-resistance pattern between FOS and ADV was seen with our HSV recombinants. Furthermore, all of those FOS- and ADV-resistant recombinants were also resistant to ACV in our study, with two of these (S724N and L778M) also exhibiting a notable decrease in CDV susceptibility. These results suggest the possibility that a unique DNA Pol mutation can confer resistance to all available antiherpetic agents. Importantly, it was previously reported that such mutations can be selected in vivo in patients who sequentially receive ACV and FOS therapy only (6, 43). However, there were two notable exceptions (mutants T821M and Y941H) which did not share this typical FOS and ADV resistance profile. This unusual phenotype has also been reported occasionally for other clinical DNA Pol mutants (L850I and N815S) (6; G. Andrei, P. Fiten, P. Goubau, H. Van Landuyt, B. Gordts, D. Selleslag, E. De Clercq, and R. Snoeck, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. V-921, p. 436, 2002), suggesting that subtle differences do exist in the interactions of the two drugs with the enzyme.
The viral Pol is an essential enzyme, part of a complex of seven proteins involved in the viral DNA replication cycle. As such, it is not unexpected that some DNA Pol mutations could affect the efficiency of viral growth (5, 13, 24, 28, 33, 36). Indeed, we were unable to generate two recombinants with mutations (K552E and Y577H) located in δ-region C. Hwang et al. (28, 29) have found a drastic increase in the mutational rate of recombinant Y577H (exoIII motif) generated in a complementary cell line expressing a wt HSV-1 DNA Pol (28). Also, a K513N HCMV DNA Pol mutant (with a substitution corresponding to the K552 amino acid in HSV-1) had severely impaired replication kinetics in vitro (13). Along the same line, our V715M HSV-1 mutant had decreased replication capacity, as reported for the corresponding HCMV mutant (5). Interestingly, we were successful in generating mutant V573M, which involves a nonconserved residue in the exoIII motif. However, this mutant had some impairment in replication as previously reported and was also shown to be less neurovirulent in mice (1). The recombinant with mutation F891C, which lies two amino acids away from the invariant YGDTDS motif of conserved region I (36), could also be generated in our study, albeit with severe impairment in replication capacities. Previous investigators have shown that most mutations in this highly conserved motif inactivate Pol elongation activity, with a few notable exceptions, such as the S889A mutation (17, 18).
We were able to generate three HSV-1 recombinants (P561S, V715M, and K805Q) containing amino acid changes previously reported in drug-resistant HCMV strains (4, 5, 12, 30, 44). Only one of these HSV-1 mutants (K805Q) had the same drug resistance phenotype as that described for HCMV (12). Some differences in drug susceptibilities were found for the other two recombinants. Thus, despite structural similarities among α-like DNA Pols, functional differences regarding interactions with antivirals can be present among distinct viral members (38). Such differences in drug interactions could be explained by slight variance in the conserved regions of the different viruses combined with distinct conformations created by the diversity of nonconserved domains (37). Indeed, interaction of an antiviral with the Pol requires the folding of several noncontiguous regions within the protein (26, 37). Further experiments, such as structural and conformational analyses of the different DNA Pol proteins, are therefore needed to clarify these observations.
In summary, we showed that the structure-function relationship of the HSV DNA Pol catalytic domain was more complex than what could be assumed based on HCMV recombinant studies (12). Also, our results confirm that resistance to a single drug such as ACV can be mapped to numerous conserved regions as well as nonconserved regions of the DNA Pol, which will limit rapid genotyping analysis based on discriminative PCR or restriction fragment length polymorphism analysis. Importantly, we found that some HSV DNA Pol mutations (including ones previously reported in clinical isolates) are associated with a phenotype of resistance to all currently available antiherpetic agents, emphasizing the need to develop new drugs with different mechanisms of action. The development of efficient methods like the one reported here to generate recombinant viruses should facilitate the understanding of drug resistance mechanisms and the assessment of various viral gene functions.
Acknowledgments
We thank Dominic Gagné for technical support and helpful discussions.
This work was supported by a grant (no. MT-13924) from the Canadian Institutes of Health Research to G.B. J.B.-S. holds a student fellowship award from the Canadian Institutes of Health Research.
REFERENCES
- 1.Andrei, G., R. Snoeck, E. De Clercq, R. Esnouf, P. Fiten, and G. Opdenakker. 2000. Resistance of herpes simplex virus type 1 against different phosphonylmethoxyalkyl derivatives of purines and pyrimidines due to specific mutations in the viral DNA polymerase gene. J. Gen. Virol. 81:639-648. [DOI] [PubMed] [Google Scholar]
- 2.Andrei, G., R. Snoeck, and E. De Clercq. 1997. Differential susceptibility of several drug-resistant strains of herpes simplex virus type 2 to various antiviral compounds. Antivir. Chem. Chemother. 8:457-461.
- 3.Andrei, G., R. Snoeck, J. Neyts, M. L. Sandvold, F. Myhren, and E. De Clercq. 2000. Antiviral activity of ganciclovir elaidic acid ester against herpesviruses. Antivir. Res. 45:157-167. [DOI] [PubMed] [Google Scholar]
- 4.Baldanti, F., A. Sarasini, E. Silini, M. Barbi, A. Lazzarin, K. K. Biron, and G. Gerna. 1995. Four dually resistant human cytomegalovirus strains from AIDS patients: single mutations in UL97 and UL54 open reading frames are responsible for ganciclovir- and foscarnet-specific resistance, respectively. Scand. J. Infect. Dis. Suppl. 99:103-104. [PubMed] [Google Scholar]
- 5.Baldanti, F., M. R. Underwood, S. C. Stanat, K. K. Biron, S. Chou, A. Sarasini, E. Silini, and G. Gerna. 1996. Single amino acid changes in the DNA polymerase confer foscarnet resistance and slow-growth phenotype, while mutations in the UL97-encoded phosphotransferase confer ganciclovir resistance in three double-resistant human cytomegalovirus strains recovered from patients with AIDS. J. Virol. 70:1390-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bestman-Smith, J., and G. Boivin. 2002. Herpes simplex virus isolates with reduced adefovir susceptibility selected in vivo by foscarnet therapy. J. Med. Virol. 67:88-91. [DOI] [PubMed] [Google Scholar]
- 7.Bestman-Smith, J., I. Schmit, B. Papadopoulou, and G. Boivin. 2001. Highly reliable heterologous system for evaluating resistance of clinical herpes simplex virus isolates to nucleoside analogues. J. Virol. 75:3105-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanco, L., A. Bernad, M. A. Blasco, and M. Salas. 1991. A general structure for DNA-dependent DNA polymerases. Gene 100:27-38. [DOI] [PubMed] [Google Scholar]
- 9.Chibo, D., A. Mijch, R. Doherty, and C. Birch. 2002. Novel mutations in the thymidine kinase and DNA polymerase genes of acyclovir and foscarnet resistant herpes simplex viruses infecting an immunocompromised patient. J. Clin. Virol. 25:165-170. [DOI] [PubMed] [Google Scholar]
- 10.Chiou, H. C., D. Kumura, A. Hu, K. M. Kerns, and D. M. Coen. 1995. Penciclovir-resistance mutations in the herpes simplex virus DNA polymerase gene. Antivir. Chem. Chemother. 6:281-288. [Google Scholar]
- 11.Chou, S., N. S. Lurain, A. Weinberg, G. Y. Cai, P. L. Sharma, and C. S. Crumpacker. 1999. Interstrain variation in the human cytomegalovirus DNA polymerase sequence and its effect on genotypic diagnosis of antiviral drug resistance. Antimicrob. Agents Chemother. 43:1500-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cihlar, T., M. Fuller, and J. Cherrington. 1998. Characterization of drug resistance-associated mutations in the human cytomegalovirus DNA polymerase gene by using recombinant mutant viruses generated from overlapping DNA fragments. J. Virol. 72:5927-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cihlar, T., M. D. Fuller, A. S. Mulato, and J. M. Cherrington. 1998. A point mutation in the human cytomegalovirus DNA polymerase gene selected in vitro by cidofovir confers a slow replication phenotype in cell culture. Virology 248:382-393. [DOI] [PubMed] [Google Scholar]
- 14.Coen, D. M. 1996. Nucleosides and foscarnet mechanisms, p. 81-102. In D. D. Richman (ed.), Antiviral drug resistance. John Wiley & Sons Ltd., New York, N.Y.
- 15.Collins, P., B. A. Larder, N. M. Oliver, S. Kemp, I. W. Smith, and G. Darby. 1989. Characterization of a DNA polymerase mutant of herpes simplex virus from a severely immunocompromised patient receiving acyclovir. J. Gen. Virol. 70:375-382. [DOI] [PubMed] [Google Scholar]
- 16.Cunningham, C., and A. J. Davison. 1993. A cosmid-based system for constructing mutants of herpes simplex virus type 1. Virology 197:116-124. [DOI] [PubMed] [Google Scholar]
- 17.Dorsky, D. I., and C. S. Crumpacker. 1990. Site-specific mutagenesis of a highly conserved region of the herpes simplex virus type 1 DNA polymerase gene. J. Virol. 64:1394-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorsky, D. I., and C. Plourde. 1993. Resistance to antiviral inhibitors caused by the mutation S889A in the highly-conserved 885-GDTDS motif of the herpes simplex virus type 1 DNA polymerase. Virology 195:831-835. [DOI] [PubMed] [Google Scholar]
- 19.Duffy, K. E., M. R. Quail, T. T. Nguyen, R. J. Wittrock, J. O. Bartus, W. M. Halsey, J. J. Leary, T. H. Bacon, and R. T. Sarisky. 7 May 2002, posting date. Assessing the contribution of the herpes simplex virus DNA polymerase to spontaneous mutations. BMC Infect. Dis. 2:7. [Online.] http://www .biomedcentral.com/bmcinfectdis. [DOI] [PMC free article] [PubMed]
- 20.Eckle, T., L. Prix, G. Jahn, T. Klingebiel, R. Handgretinger, B. Selle, and K. Hamprecht. 2000. Drug-resistant human cytomegalovirus infection in children after allogeneic stem cell transplantation may have different clinical outcomes. Blood 96:3286-3289. [PubMed] [Google Scholar]
- 21.Franklin, M. C., J. Wang, and T. A. Steitz. 2001. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell 105:657-667. [DOI] [PubMed] [Google Scholar]
- 22.Gibbs, J. S., H. C. Chiou, K. F. Bastow, Y. C. Cheng, and D. M. Coen. 1988. Identification of amino acids in herpes simplex virus DNA polymerase involved in substrate and drug recognition. Proc. Natl. Acad. Sci. USA 85:6672-6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert, C., J. Bestman-Smith, and G. Boivin. 2002. Resistance of herpesviruses to antiviral drugs: clinical impacts and molecular mechanisms. Drug Resist. Updates 5:88-114. [DOI] [PubMed] [Google Scholar]
- 24.Hall, J. D., K. L. Orth, K. L. Sander, B. M. Swihart, and R. A. Senese. 1995. Mutations within conserved motifs in the 3′-5′ exonuclease domain of herpes simplex virus DNA polymerase. J. Gen. Virol. 76:2999-3008. [DOI] [PubMed] [Google Scholar]
- 25.Hall, J. D., Y. S. Wang, J. Pierpont, M. S. Berlin, S. E. Rundlett, and S. Woodward. 1989. Aphidicolin resistance in herpes simplex virus type I reveals features of the DNA polymerase dNTP binding site. Nucleic Acids Res. 17:9231-9244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, L., K. K. Ishii, H. Zuccola, A. M. Gehring, C. B. Hwang, J. Hogle, and D. M. Coen. 1999. The enzymological basis for resistance of herpesvirus DNA polymerase mutants to acyclovir: relationship to the structure of alpha-like DNA polymerases. Proc. Natl. Acad. Sci. USA 96:447-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang, C. B., K. L. Ruffner, and D. M. Coen. 1992. A point mutation within a distinct conserved region of the herpes simplex virus DNA polymerase gene confers drug resistance. J. Virol. 66:1774-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hwang, Y. T., B. Y. Liu, D. M. Coen, and C. B. C. Hwang. 1997. Effects of mutations in the Exo III motif of the herpes simplex virus DNA polymerase gene on enzyme activities, viral replication, and replication fidelity. J. Virol. 71:7791-7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang, Y. T., J. F. Smith, L. Gao, and C. B. Hwang. 1998. Mutations in the Exo III motif of the herpes simplex virus DNA polymerase gene can confer altered drug sensitivities. Virology 246:298-305. [DOI] [PubMed] [Google Scholar]
- 30.Jabs, D. A., B. K. Martin, M. S. Forman, J. P. Dunn, J. L. Davis, D. V. Weinberg, K. K. Biron, and F. Baldanti. 2001. Mutations conferring ganciclovir resistance in a cohort of patients with acquired immunodeficiency syndrome and cytomegalovirus retinitis. J. Infect. Dis. 183:333-337. [DOI] [PubMed] [Google Scholar]
- 31.Jabs, D. A., C. L. Meinert, and J. P. Lalezari. 1997. Cidofovir for cytomegalovirus retinitis—response. Ann. Intern. Med. 127:490-491. [DOI] [PubMed] [Google Scholar]
- 32.Knopf, C. W., and K. Weisshart. 1988. The herpes simplex virus DNA polymerase: analysis of the functional domains. Biochim. Biophys. Acta 951:298-314. [DOI] [PubMed] [Google Scholar]
- 33.Kuhn, F. J., and C. W. Knopf. 1996. Herpes simplex virus type 1 DNA polymerase. Mutational analysis of the 3′-5′-exonuclease domain. J. Biol. Chem. 271:29245-29254. [DOI] [PubMed] [Google Scholar]
- 34.Larder, B. A., S. D. Kemp, and G. Darby. 1987. Related functional domains in virus DNA polymerases. EMBO J. 6:169-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larder, B. A., J. J. Lisle, and G. Darby. 1986. Restoration of wild-type pathogenicity to an attenuated DNA polymerase mutant of herpes simplex virus type 1. J. Gen. Virol. 67:2501-2506. [DOI] [PubMed] [Google Scholar]
- 36.Marcy, A. I., C. B. Hwang, K. L. Ruffner, and D. M. Coen. 1990. Engineered herpes simplex virus DNA polymerase point mutants: the most highly conserved region shared among α-like DNA polymerases is involved in substrate recognition. J. Virol. 64:5883-5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matthews, J. T., B. J. Terry, and A. K. Field. 1993. The structure and function of the HSV DNA replication proteins: defining novel antiviral targets. Antivir. Res. 20:89-114. [DOI] [PubMed] [Google Scholar]
- 38.Reha-Krantz, L. J., R. L. Nonay, and S. Stocki. 1993. Bacteriophage T4 DNA polymerase mutations that confer sensitivity to the PPi analog phosphonoacetic acid. J. Virol. 67:60-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sacks, S. L., R. J. Wanklin, D. E. Reece, K. A. Hicks, K. L. Tyler, and D. M. Coen. 1989. Progressive esophagitis from acyclovir-resistant herpes simplex. Clinical roles for DNA polymerase mutants and viral heterogeneity? Ann. Intern. Med. 111:893-899. [DOI] [PubMed] [Google Scholar]
- 40.Safrin, S., C. Crumpacker, P. Chatis, R. Davis, R. Hafner, J. Rush, H. A. Kessler, B. Landry, and J. Mills. 1991. A controlled trial comparing foscarnet with vidarabine for acyclovir-resistant mucocutaneous herpes simplex in the acquired immunodeficiency syndrome. N. Engl. J. Med. 325:551-555. [DOI] [PubMed] [Google Scholar]
- 41.Safrin, S., S. Kemmerly, B. Plotkin, T. Smith, N. Weissbach, D. De Veranez, L. D. Phan, and D. Cohn. 1994. Foscarnet-resistant herpes simplex virus infection in patients with AIDS. J. Infect. Dis. 169:193-196. [DOI] [PubMed] [Google Scholar]
- 42.Saijo, M., Y. Yasuda, H. Yabe, S. Kato, T. Suzutani, E. De Clercq, M. Niikura, A. Maeda, I. Kurane, and S. Morikawa. 2002. Bone marrow transplantation in a child with Wiskott-Aldrich syndrome latently infected with acyclovir-resistant (ACVr) herpes simplex virus type 1: emergence of foscarnet-resistant virus originating from the ACVr virus. J. Med. Virol. 68:99-104. [DOI] [PubMed] [Google Scholar]
- 43.Schmit, I., and G. Boivin. 1999. Characterization of the DNA polymerase and thymidine kinase genes of herpes simplex virus isolates from AIDS patients in whom acyclovir and foscarnet therapy sequentially failed. J. Infect. Dis. 180:487-490. [DOI] [PubMed] [Google Scholar]
- 44.Smith, I. L., J. M. Cherrington, R. E. Jiles, M. D. Fuller, W. R. Freeman, and S. A. Spector. 1997. High-level resistance of cytomegalovirus to ganciclovir is associated with alterations in both the UL97 and DNA polymerase genes. J. Infect. Dis. 176:69-77. [DOI] [PubMed] [Google Scholar]
- 45.Wang, J., A. K. Sattar, C. C. Wang, J. D. Karam, W. H. Konigsberg, and T. A. Steitz. 1997. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell 89:1087-1099. [DOI] [PubMed] [Google Scholar]
- 46.Weinberg, A., D. A. Jabs, S. Chou, B. K. Martin, N. S. Lurain, M. S. Forman, and C. Crumpacker. 2003. Mutations conferring foscarnet resistance in a cohort of patients with acquired immunodeficiency syndrome and cytomegalovirus retinitis. J. Infect. Dis. 187:777-784. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, J., D. W. Chung, C. K. Tan, K. M. Downey, E. W. Davie, and A. G. So. 1991. Primary structure of the catalytic subunit of calf thymus DNA polymerase delta: sequence similarities with other DNA polymerases. Biochemistry 30:11742-11750. [DOI] [PubMed] [Google Scholar]