Abstract
We previously established a high-efficiency, retrovirus-mediated expression cloning method. Using this system, we now have developed an expression cloning method (FL-REX; fluorescence localization-based retrovirus-mediated expression cloning) in which cDNAs can be isolated based on the subcellular localization of their protein products. Complementary DNAs generated from mRNA using random hexamers were fused to the cDNA of green fluorescent protein (GFP) in the pMX retrovirus vector. The resulting cDNA-GFP fusion library was transfected into retrovirus-packaging cells, and the derived retroviruses were used to infect NIH 3T3 cells. Infected cells then were screened to identify cDNAs of interest through the subcellular localization of the GFP-fusion products. Using FL-REX, we have identified 25 cDNAs, most of which showed reasonable subcellular localization as GFP-fusion proteins, indicating that FL-REX is useful for identification of proteins that show specific intracellular localization.
Retrovirus-mediated expression cloning (1–3) is applicable to many types of experiments because retrovirally transduced cDNAs are stably expressed in the cells, which enables a variety of selection methods in the screening. In fact, we and others have identified a variety of novel genes through functional assays by using retrovirus-mediated expression cloning (4–7). We also applied the screening method to identify constitutive active forms of a cytokine receptor, MPL (8), and a transcription factor, STAT5 (9, 10), in combination with PCR-driven random mutagenesis. In addition, we recently have succeeded in establishing an efficient signal sequence trap method based on retrovirus-mediated cDNA library screening (SST-REX; ref. 11) and have cloned novel cDNAs for secreted and membrane proteins.
In our retrovirus-mediated expression cloning, combination of an efficient transient packaging cell line, BOSC23 (12), and a retrovirus vector, pMX (13), which is designed for the construction of cDNA expression libraries, is used. The pMX vector has multicloning sites suitable for cDNA library construction and avoids a drug-resistance gene. It has an extended packaging signal derived from the MFG vector (14) and generates higher titers of retroviruses compared with the conventional retrovirus vectors such as LXSN (15) and pBabe-puro (16). In our hands, the titer of the retrovirus usually exceeds 3 × 106 plaque-forming units (pfu)/ml and sometimes reaches 1 × 107 pfu/ml, sufficient to cover the large complexities of cDNA libraries (1, 13).
Recently, green fluorescent protein (GFP) has become popular as a useful intracellular localization marker (17–19). When GFP is fused to a certain protein, we can directly observe the physiological localization of the protein and the movement of the protein in response to various stimuli, which suggest its functions. With this in mind, we now have developed a method to clone cDNAs based on the localization of cDNA products by using a GFP-fused cDNA library and a retrovirus-mediated expression cloning system. This system is named FL-REX, which stands for fluorescence localization-based retrovirus-mediated expression cloning. Using FL-REX, we have cloned and characterized 25 cDNAs from fetal mouse liver cells that showed various GFP-staining patterns including nuclear, nuclear speckle, nuclear spotty, nucleolar, cytoplasmic reticular, cytoplasmic speckle, and perinuclear staining.
Materials and Methods
Construction of the Retrovirus Expression Vector for FL-REX, pMX-FL.
The enhanced GFP (EGFP) cDNA was cut from pEGFP-N1 (CLONTECH) by SalI and NotI and inserted into the SalI site downstream of multicloning sites of the pMX vector by using a SalI adapter (Invitrogen).
Construction of the cDNA Library for FL-REX.
Poly(A)+ RNA was purified from mouse day 12 fetal liver cells by using FastTrack (Invitrogen). cDNA was synthesized from the poly(A)+ RNA by random hexamers by using a cDNA synthesis kit (GIBCO/BRL) according to the manufacturer's instructions. The resulting cDNAs were size-fractionated through agarose gel electrophoresis, and cDNA fragments longer than 1 kbp were extracted from the gel by using Qiaex II (Qiagen, Chatsworth, CA). The cDNA fragments then were inserted into BstXI sites of pMX-FL by using BstXI adapters (Invitrogen). The ligated DNA was ethanol-precipitated and then electroporated into DH10B-competent cells (Electromax DH10B; GIBCO/BRL) by using E-coli Pulser (Bio-Rad) and a 0.1-cm cuvette (Gene Pulser Cuvette; Bio-Rad) at 1.8 kV. Plasmid DNA was purified by using JETstar (Genomed, Research Triangle Park, NC) after 200 ml of culture of the transformed bacteria for 16 h.
Preparation of High-Titer Retroviruses and Infection of NIH 3T3.
A murine fibroblastic cell line, NIH 3T3, was cultured in DMEM containing 10% FCS. High-titer retroviruses representing the cDNA-EGFP library were produced by using packaging cell line BOSC23, and NIH 3T3 cells were infected as described (1). One day later, the cells were spread into 10-cm-diameter culture dishes (104/dish). When the cells became confluent, the clones with specific phenotypes via fluorescent microscope (Olympus, Tokyo) were subjected to subcloning by using cylinders, syringes, or tips.
Isolation of cDNA Fragments from NIH 3T3 Clones Showing Specific GFP-Staining Patterns.
Genomic DNAs extracted from NIH 3T3 clones with specific GFP staining were subjected to PCR to recover the integrated cDNAs by using vector primers (GGTGGACCATCCTCTAGACT and CCCTTTTTCTGGAGACTAAAT). The PCR was run for 30 cycles (30 s at 98°C for denaturation, 30 s at 58°C for annealing, and 1 min at 72°C for extension) by using a GeneAmp PCR System 2400 (Perkin–Elmer) and LA Taq polymerase (Takara Shuzo, Kyoto). The resulting PCR fragments were sequenced by using the Taq DyeTerminator Cycle Sequencing Kit (Applied Biosystems) and were analyzed by an automatic sequencer (310 Genetic Analyzer; Applied Biosystems).
For the rescue of integrated retroviruses, a helper virus construct, SV-Ψ−-E-MLV (20), harboring a packaging signal-deficient murine leukemia virus genome was transfected to NIH 3T3 cells by using Lipofectamine (GIBCO/BRL), and the culture supernatant was saved 2 days later as a virus stock, which then was used to infect uninfected NIH 3T3 cells, as described (1). The NIH 3T3 colony showing the phenotype of interest was isolated and subjected for further characterization as described above.
Results
Construction and Screening of the cDNA Library.
The EGFP cDNA was inserted into the SalI site of the pMX vector to construct pMX-FL as described in the experimental protocol. The two BstXI sites upstream of the inserted GFP enabled the insertion of cDNA fragments, creating cDNA-GFP fusion libraries (Fig. 1). One microgram of poly(A)+ RNA extracted from day 12 fetal mouse liver cells was used to construct an FL-REX library as described in the experimental protocol. The resulting library contained 8.2 × 106 independent clones, with the size of cDNA inserts averaging 1,370 bp. The library was converted to retroviruses by using BOSC23 cells, and 5 × 105 NIH 3T3 cells were infected with 2 ml of the 4-fold-diluted retroviral supernatant to achieve 30% infection efficiency as determined by the efficiency of the control pMX-GFP vector on NIH 3T3 cells in a simultaneous experiment.
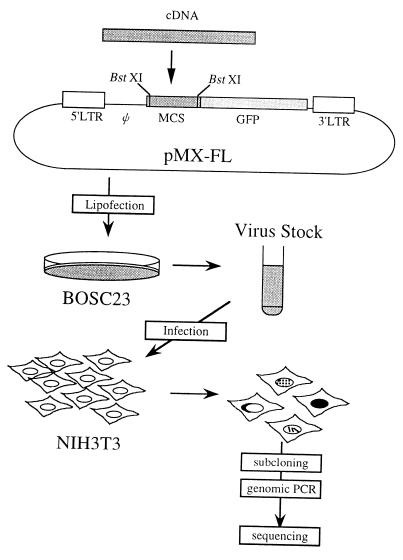
Figure 1.
A rationale of FL-REX. A retrovirus vector pMX-FL was designed for the construction of the cDNA-GFP fusion library; cDNAs can be inserted into BstXI sites for the fusion with the GFP cDNA. After converting the library to retroviruses by using Bosc23-packaging cells, the derived retroviruses were used to infect NIH 3T3 cells, and the clones of interest were subjected for further analysis as described in the experimental protocol. LTR, long terminal repeat; Ψ, retrovirus-packaging signal; MCS, multicloning site.
After a 24-h incubation with the retroviruses, these cells were spread into 30 10-cm-diameter dishes. When the dishes became confluent, a total of 500–1,000 colonies demonstrated characteristic staining patterns under fluorescent microscope. Among them, as a pilot experiment, we subcloned 25 colonies, including 20 with a nuclear staining pattern and five clones with a cytoplasmic staining pattern (Table 1). These NIH 3T3 clones were expanded, and genomic DNA was isolated from each clone. Integrated cDNAs were recovered by PCR by using the vector primers, sequenced, and then expressed in NIH 3T3 cells as a GFP-fused form to confirm the subcellular localization. To identify which band was responsible for the cellular localization of GFP in five clones that were found to harbor multiple integrations, a helper virus construct was used; the expression vector SV-Ψ−-E-MLV (20), harboring a packaging signal-deficient murine leukemia virus genome, was introduced into the NIH 3T3 clones via lipofection, and 2 days later the produced retroviruses were used to infect NIH 3T3 cells. Subsequently, the GFP-positive clone that had the original phenotype was isolated and subjected for the recovery of the integrated cDNA. In this secondary screening, the titers of the retroviruses obtained by the helper virus rescue were much lower, which resulted in a single integration in cells in the secondary infection.
Table 1.
Isolated cDNAs using FL-REX
| Type of localization | Clone no. | Gene | GenBank accession no.* | Detected length† | Full-length‡ |
|---|---|---|---|---|---|
| Cytoplasmic | |||||
| Fibrous | 1 | Vimentin | M24849 | 460 | 469 |
| Spot | 2 | Homologue of human R27216-1 (72%/97%)§ | (AC005306) | 152 | [>297] |
| Reticular | 3 | Stearoyl-CoA desaturase | M26270 | 335 | 357 |
| 4 | Homologue of human CIT987SK-A-319E8 (86%/100%)§ | (AC004020) | 233 | [238] | |
| Speckle | 5 | Novel (related to keratin) | 1,400 bp | ||
| Nuclear | |||||
| Homogenous | 6 | Histone H2A.Z | U70494 | 111 | 129 |
| 7 | Transaldorase | U67611 | 335 | 338 | |
| 8 | Homologue of human PHAPI2a (79%/91%)§ | (Y07569) | 247 | [252] | |
| 9 | Homologue of c. elegans C02F5.4 (47%/55%)§ | (P34281) | 295 | [261] | |
| 10 | Homologue of human ZNF207 (94%/99%)§ | (AF046001) | 343 | 495 | |
| 11, 12 | Pyrimidine binding protein | X52101 | 279, 329 | 528 | |
| 13 | Homologue of human MOZ (72%/86%)§ | (U47742) | 334 | [2,004] | |
| 14 | hnRNP G | AF031568 | 225 | 389 | |
| 15 | Homologue of human DEK (99%/100%)§ | (X64229) | 43 | [376] | |
| Spot | 16, 17 | hnRNP C1/C2 | AF095257 | 276, 286 | 313 |
| Speckle | 18 | SET | AB015613 | 242 | [278] |
| 19, 20 | PML | U33626 | 463, 540 | 809 | |
| 21 | Novel (under investigation) | AB033168 | 378 | 1,386 | |
| Nucleolar | |||||
| 22 | Homologue of human KIAA0670 (86%/99%)§ | (AB014570) | 291 | [>1,281] | |
| 23, 24, 25 | Nucleophosmin | M33212 | 143, 249, 256 | 293 |
Accession no. for GenBank nucleotide sequence databases. The numbers of human sequence are in parentheses.
† Number of amino acid residues fused to EGFP. The cDNA fragment of clone 22 is not fully investigated.
‡ Number of amino acid residues of the full-length protein. The lengths of human sequence are in square brackets. The genes for clone 2 and 22 have been reported only with partial coding.
§ Percent homology at the DNA level and percent similarity at the amino acid levels between the cloned cDNA fragments and probable human counterparts.
Analysis of Individual cDNA Clones.
Complementary DNAs cloned by FL-REX are listed in Table 1. The length of the cDNAs ranged from 130 to 1,700 bp, and the average was 1,000 bp. Photographs of the representative clones with characteristic staining patterns are presented in Fig. 2. With a few exceptions, most cDNA clones encoded proteins showing proper subcellular localization. Except for a cDNA for monocytic leukemia zinc finger protein MOZ (clone 13), which encodes a stretch from the 893rd to the 1,226th amino acids of the 2,004 full-length amino acids, and a cDNA for DEK (clone 15), which was an experimental background as described below, all of the known cDNAs cover the 5′ end of the coding region, and some of the cDNAs were nearly full-length (Table 1).
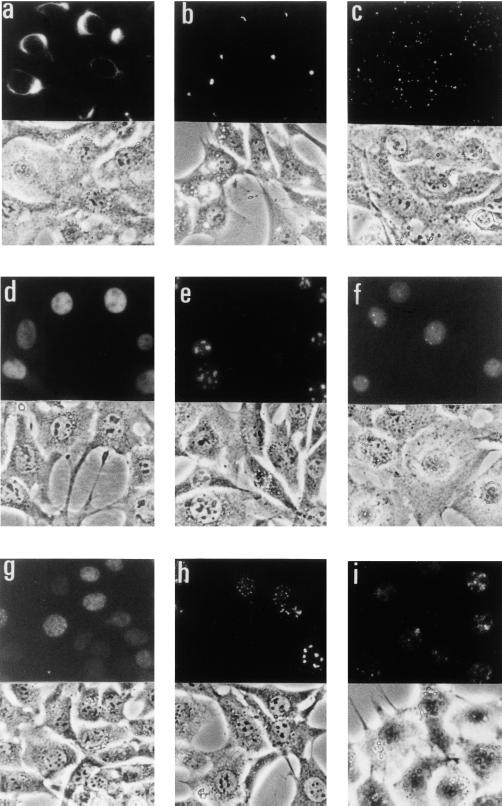
Figure 2.
Photographs of the clones with specific intercellular localization patterns. The upper part of each photo shows GFP fluorescence, and the lower part shows corresponding phase-contrast images. The lower part of b is the combination of both images. (a) Clone 1, vimentin (fibrous). (b) Clone 2, mouse homologue of human R27216–1 (perinuclear spotty). (c) Clone 5, a novel gene related to keratin family (cytoplasmic speckle). (d) Clone 10, mouse ZNF207 (nuclear). (e) Clone 24, nucleophosmin (nucleolar). (f) Clone 17, hnRNP C (nuclear spotty). (g) Clone 18, mouse SET (nuclear speckle). (h) Clone 20, mouse PML (nuclear speckle). (i) Clone 21, a novel gene (nuclear speckle).
The cells exhibiting specific cytoplasmic staining patterns were found to harbor reasonable cDNAs (Table 1). A cDNA for vimentin (clone 1) generated a fibrous pattern that is characteristic for cytoskeleton proteins (Fig. 2a). A cDNA for an uncharacterized protein R27216–1 (clone 2) gave rise to a unique perinuclear spotty pattern (Fig. 2b), and the psort ii program (http://psort.nibb.ac.jp:8800/form2.html) predicted that this protein localized in the cytoplasm. The reticular pattern characteristic for endoplasmic reticulum (ER) staining was obtained by cDNAs for Stearoyl-CoA desaturase (clone 3) and an uncharacterized protein, CIT987SK-A-319E8 (clone 4). The former is an ER protein, and the latter was predicted to exist in ER by psort ii. A characteristic cytoplasmic speckle pattern was obtained by a novel gene (clone 5) that is related to keratin (Fig. 2c).
Among the cDNAs showing a nuclear staining pattern (Fig. 2d), all but one seemed to encode nuclear proteins. Two were zinc finger transcription factors, mouse homologues of human ZNF207 (clone 10) and MOZ (clone 13). Three were known nuclear proteins: histone H2A.Z (clone 6), pyrimidine-binding protein (clones 11 and 12), and heterogeneous nuclear ribonucleoprotein (hnRNP) G (clone 14). The cDNA derived from clone 8 encoded a mouse homologue of human PHAPI2a, which is a nuclear protein related to DEK and SET. CO2F5.4 (clone 9) is an uncharacterized protein but has a nuclear localization signal (NLS) and was predicted to be a nuclear protein by psort ii. One of the clones with the nuclear staining pattern (clone 15) turned out to be “false-positive,” harboring an NLS-like sequence RRRRR upstream of GFP, which was not the actual reading frame of the cDNA, although it happened to encode a nuclear protein, DEK. A unique nuclear spotty pattern (Fig. 2f) was observed in two NIH 3T3 clones, both of which harbored cDNAs for hnRNP C (clones 16 and 17). Nucleolar staining patterns (Fig. 2e) were obtained by two proteins: nucleophosmin (clones 23–25) and a mouse homologue of human KIAA0670 (clone 22). The former is a nucleolar protein, and the latter is an uncharacterized protein harboring an NLS. The nuclear speckle patterns were observed in the cells transduced with cDNAs for a mouse homologue of SET (clone 18, Fig. 2g), a mouse homologue of PML (clones 19 and 20, Fig. 2h), and a novel protein (clone 21, Fig. 2i). The first two were transcription factors.
We also examined 10 NIH 3T3 cell clones showing nonspecific homogeneous staining patterns; however, we did not find any meaningful coding sequence fused to the GFP cDNA, indicating that these cells simply expressed the GFP protein itself, which homogeneously localizes in the cells.
Discussion
In this paper, we describe an expression cloning method, FL-REX, by which cDNAs can be isolated based on the subcellular localization of their protein products. To obtain the normal localization of a cDNA product by visualization with GFP, two conditions are required. First, the reading frames of the cDNA and GFP have to be matched. Second, the cDNA must be long enough to contain localization signals. Because some proteins may lose proper localization after truncation of the C-terminal portion and fusion to GFP, not all cDNAs would be cloned readily by using FL-REX. Also, a false-positive resulting from a misreading frame of a cDNA that makes an NLS-like sequence being fused “appropriately” with GFP has been recovered (Table 1, clone 15). Nevertheless, as shown in Table 1, most of the other clones identified by FL-REX exhibited physiologically correct cellular localization of cDNAs' products except for clone 7. This clone harbored an integration in which GFP was fused to a nearly full-length cDNA for transaldorase, which presumably is a cytoplasmic protein. Although nuclear localization of transaldorase-GFP fusion protein was confirmed in a reinfection experiment, it is not clear why it located in the nucleus. We also recovered cDNAs from 10 NIH 3T3 clones showing homogeneous staining patterns, which were found to express GFP itself. To avoid this type of background, it would be better to delete the initiation ATG of the GFP sequence in the pMX-FL vector. However, these backgrounds will not hamper the experiments in which specific staining patterns such as nuclear, perinuclear, or cytoplasmic speckle are searched for. One of our major purposes through FL-REX screening is identification of novel transcription factors. In this pilot experiment, we have obtained cDNAs for three zinc finger transcription factors, MOZ, PML, and ZNF207, and one novel cDNA giving rise to a speckle GFP pattern like PML, which may be a transcription factor.
Any infectable cells can be used as target cells, although NIH 3T3 cells offer the advantage of size for easy observation of subcellular structures. In addition, they proliferate quickly and are easy to clone. When the virus produced from the pMX vector and BOSC23 cells is infected directly to NIH 3T3 cells [10 ml viruses/3 × 106 cells; multiplicity of infection (moi) = 10–30], the infection efficiency is 100% and the average number of retrovirus integration is 5–10/cell (unpublished results). To avoid multiple integrations that complicate subsequent experiments, we limited the infection efficiency below 30% by using diluted retrovirus supernatant. In our experiments, 90% and 25% infection efficiency of the pMX-GFP vector in a simultaneous control experiment resulted in expression of cDNA-GFP fusion protein in 30% and 3% of the infected NIH 3T3 cells, respectively (data not shown). This result is reasonable, assessing from the probability of successful generation of fusion proteins that requires in-frame fusion of cDNA and GFP (one in three), the right orientation of the cDNA (one in two), and the cDNA to be truncated before its stop codon. In a typical experiment, we make 3 ml of the library viruses (107 virus particles) and dilute it to 30 ml to infect 1 × 107 NIH 3T3 cells (moi = 1). From this we can screen 3 × 105 GFP-positive clones (effective clones).
Two important advantages in using retrovirus-mediated expression screening in FL-REX are high efficiency of the screening and the restricted number of integrations. The latter point is particularly important in FL-REX; in the conventional expression cloning using transfection of simian virus 40-replication-origin-bearing plasmid DNA into COS cells, we cannot control the copy number of the plasmid in one cell. Recently, two similar methods have been reported. One used a GFP-fused plasmid library that was subdivided into many small pools and expressed in mammalian cells to identify cDNA based on the subcellular localization of GFP-fusion products, and a novel type of nuclear envelop protein was identified (21). The other used the yeast system in which the cDNA-encoded NLS is expected to carry a modified transcription factor (LexA) harboring the nuclear exporting signal (NES) of the HIV rev protein into the nucleus, which will result in transcriptional activation of a LexA-responsive leu2 reporter (22). The former system cannot control the copy number of plasmid vectors in one cell and, therefore, needs to use partitioned libraries, which is time-consuming. In the latter system, only nuclear proteins can be isolated when its NLS is stronger than the NES included in the modified transcription factor. Moreover, in both methods, cDNAs were generated by using oligo(dT) primers, and the fusion partner GFP or LexA cDNA was fused to 5′ of the cDNAs. In this way, stop codons will be included frequently between the fusion partner and library sequences, which makes screening inefficient. We believe that in this type of experiment it is much better to fuse randomly primed cDNAs to 5′ of the fusion partner as we did in FL-REX.
The FL-REX screening is applicable to a variety of experiments in addition to cloning of nuclear proteins, including transcription factors. Thus, FL-REX will be useful to identify proteins that dynamically function during mitosis or that can bind to a particular part of chromosomes such as telomeres and centromeres. It also enables identification of proteins that change subcellular localization in response to various stimuli including cytokine stimulation, irradiation, and heat shock.
Acknowledgments
We thank Dr. Yoshitsugu Yamada for continuous encouragement. This work was supported in part by the Ministry of Education, Science, Sports, and Culture of Japan. The Department of Hematopoietic Factors was supported in part by Chugai Pharmaceutical Ltd.
Abbreviations
- FL-REX
fluorescence localization-based retrovirus-mediated expression cloning
- GFP
green fluorescent protein
- EGFP
enhanced GFP
- NLS
nuclear localization signal
- hnRNP
heterogeneous nuclear ribonucleoprotein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB033168).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.060489597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.060489597
References
- 1.Kitamura T, Onishi M, Kinoshita S, Shibuya A, Miyajima A, Nolan G P. Proc Natl Acad Sci USA. 1995;92:9146–9150. doi: 10.1073/pnas.92.20.9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitehead I, Kirk H, Kay R. Mol Cell Biol. 1995;15:704–710. doi: 10.1128/mcb.15.2.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rayner J R, Gonda T J. Mol Cell Biol. 1994;14:880–887. doi: 10.1128/mcb.14.2.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng H K, Unutmaz H K, Kewal Ramani V N, Littman D R. Nature (London) 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 5.Hitoshi Y, Lorens J, Kitada S, Fisher J, LaBarge M, Ring H Z, Francke U, Reed J C, Kinoshita S, Nolan G P. Immunity. 1998;8:461–471. doi: 10.1016/s1074-7613(00)80551-8. [DOI] [PubMed] [Google Scholar]
- 6.Yamaoka S, Courtois G, Bessia C, Whiteside S T, Weil R, Agou F, Kirk H E, Kay R J, Israel A. Cell. 1998;93:1231–1240. doi: 10.1016/s0092-8674(00)81466-x. [DOI] [PubMed] [Google Scholar]
- 7.Yang Y L, Guo L, Xu S, Holland C A, Kitamura T, Hunter K, Cunningham J M. Nat Genet. 1999;21:216–219. doi: 10.1038/6005. [DOI] [PubMed] [Google Scholar]
- 8.Onishi M, Mui A L F, Morikawa Y, Cho L, Kinoshita S, Nolan G P, Gorman D M, Miyajima A, Kitamura T. Blood. 1996;88:1399–1406. [PubMed] [Google Scholar]
- 9.Onishi M, Nosaka T, Misawa K, Mui A L F, Gorman D, McMahon M, Miyajima A, Kitamura T. Mol Cell Biol. 1998;18:3871–3879. doi: 10.1128/mcb.18.7.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nosaka T, Kawashima T, Misawa K, Ikuta K, Mui A L F, Kitamura T. EMBO J. 1999;18:4754–4765. doi: 10.1093/emboj/18.17.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kojima T, Kitamura T. Nat Biotechnol. 1999;17:487–490. doi: 10.1038/8666. [DOI] [PubMed] [Google Scholar]
- 12.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onishi M, Kinoshita S, Morikawa Y, Shibuya A, Phillips J, Lanier L L, Gorman D M, Nolan G P, Miyajima A, Kitamura T. Exp Hematol. 1996;24:324–329. [PubMed] [Google Scholar]
- 14.Dranoff G, Jaffee E, Lazenby A, Golumbek P, Levitsky H, Brose K, Jackson V, Hamada H, Pardoll D, Mulligan R C. Proc Natl Acad Sci USA. 1993;90:3539–3543. doi: 10.1073/pnas.90.8.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller A D, Rosman G J. BioTechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 16.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Misteli T, Spector D L. Nat Biotechnol. 1997;15:961–964. doi: 10.1038/nbt1097-961. [DOI] [PubMed] [Google Scholar]
- 18.Kendall J M, Badminton M N. Trends Biotechnol. 1998;16:216–224. doi: 10.1016/s0167-7799(98)01184-6. [DOI] [PubMed] [Google Scholar]
- 19.Tsien R Y. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 20.Landau N R, Littman D R. J Virol. 1992;66:5110–5113. doi: 10.1128/jvi.66.8.5110-5113.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rolls M M, Stein P A, Talor S S, Ha E, McKeon F. J Cell Biol. 1999;146:29–43. doi: 10.1083/jcb.146.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ueki N, Oda T, Kondo M, Yano K, Noguchi T, Muramatsu M. Nat Biotechnol. 1998;16:1338–1342. doi: 10.1038/4315. [DOI] [PubMed] [Google Scholar]