Abstract
It is believed that replication capacity is an important determinant of human immunodeficiency virus type 1 (HIV-1) pathogenicity and transmissibility. To explore this, we conducted a comprehensive analysis of the replication properties of nine drug-resistant and nine drug-susceptible viral isolates derived from patients with primary HIV-1 infection. Viral isolates were tested for single-cycle infectivity in the GHOST cell line. The infectivity of isolates carrying resistance-associated mutations was significantly higher than that of drug-susceptible isolates. Additionally, the growth kinetics of these isolates were determined in CD4+ T lymphocytes. Drug-resistant isolates replicated as well as drug-susceptible viruses. Insertion of the resistance-conferring regions into an NL4-3-based molecular background resulted in chimeras that displayed a modest but significant reduction in replication capacity compared to the drug-susceptible chimeric viruses. Of note, two multidrug-resistant isolates and one protease inhibitor-resistant isolate displayed higher rates of infectivity and growth kinetics than the other drug-resistant or drug-susceptible isolates. These distinct replicative features, however, were not seen in the corresponding chimeras, indicating that changes within the C-terminal region of Gag as well as within the protease and reverse transcriptase genes contribute to but are not sufficient for the level of compensatory adaptation observed. These findings suggest that some drug-resistant viruses isolated during primary infection possess unique adaptive changes that allow for both high viral replication capacity and resistance to one or more classes of antiretroviral drugs. Further studies are needed to elucidate the precise regions that are essential for these characteristics.
The clinical benefits of antiretroviral treatment are limited by the selection of drug-resistant human immunodeficiency virus type 1 (HIV-1) strains during therapy (38). A recent U.S. survey reported that up to 70% of patients with detectable plasma viremia harbor drug-resistant viral variants (D. Richman, S. Bozzette, S. Morton, S. Chien, T. Wrin, K. Dawson, and N. Hellmann, Abstr. 41st Intersci. Conf. Antimicrob. Agents Chemother., abstr. LB-17, 2001). In contrast, the observed frequency of drug resistance in newly infected individuals is 5 to 10 times lower (range of 6 to 20% in countries with broad access to treatment [5, 18, 25, 26, 41, 42, 46]). The discrepancy between the prevalence of drug-resistant variants in the treated HIV-1-positive population and the observed transmission rate may in part be attributed to the impaired replication capacity (RC) of the drug-resistant variants (4). Several studies indicate that drug-resistant variants in the setting of chronic disease generally display replication deficiencies. First, drug-susceptible variants in vivo rapidly outgrow the drug-resistant viral quasispecies in the absence of selective pressure (i.e., discontinuation of treatment) (20, 21). Second, a number of in vitro studies have demonstrated that primary resistance-associated substitutions in protease (PR) and reverse transcriptase (RT) genes reduce the overall performance of the mutated viral enzymes (1, 2, 8, 10, 15, 34). It has also been shown that the impairment resulting from these defects depends on both the position of the mutation and the genetic viral background (13, 17). Ongoing viral replication in the presence of antiretroviral drugs can select for variants carrying additional compensatory substitutions that partially rescue the drug resistance-associated replication defects. These adaptive changes can be found in PR and RT (6), p6gag (35), and regions distal to (16) or within (9, 29, 30, 40, 48) the Gag cleavage sites.
HIV-1 transmission represents a selective evolutionary bottleneck. Although it remains unclear whether a single viral species or viral quasispecies are initially transmitted by sexual contact (24, 28) and to what extent gender influences viral heterogeneity (27), the viral population present during early primary infection in males is likely to be homogeneous, whether it is drug susceptible or drug resistant. Detection of drug-resistant variants in patients with acute primary infection implies that these viruses possess replication characteristics that allow them to ultimately establish themselves as the dominant viral population in a drug-free environment. Indeed, in the rhesus macaque model, viruses (e.g., simian immunodeficiency virus or simian/human immunodeficiency virus) with higher RC were found to be more efficiently transmitted by the vaginal route (31). In humans, the risk of transmission was associated with the level of plasma viremia (19), although no direct relation between variation in viral RC and plasma viremia has been established.
Due to the inherent difficulties in identifying individuals during early primary HIV-1 infection who harbor drug-resistant viruses, our present knowledge of the replicative capacity of these viruses is limited. This is especially true for viruses with resistance to two or three drug classes (multidrug resistance [MDR]) that have been transmitted to a new host. A better understanding of the properties of these viruses is, however, of great clinical relevance, since it will allow us to define their mode of spread and to design appropriate therapeutic strategies. Here we have evaluated the replication characteristics (i.e., single-cycle infectivity, growth kinetics, and Gag protein processing) of 18 viral isolates with or without drug resistance. Viral isolates were generated from samples obtained 10 to 79 days (median, 21 days) after onset of symptoms. In addition, we have studied two pairs of epidemiologically related, transmitted drug-resistant isolates in order to analyze the replication characteristics associated with sequential transmission of drug-resistant variants.
MATERIALS AND METHODS
Patient population.
Primary viral isolates were cultured from 18 patients who were identified as having been newly infected with HIV-1. The median time between onset of symptoms compatible with acute retroviral syndrome and sample collection was 21 days (range, 10 to 79 days; for two patients [MDR3 and WT8], documentation was not available). The clinical findings for newly infected individuals with drug-susceptible HIV-1 were comparable to the findings for patients infected with drug-resistant viruses (plasma viremia, P = 0.16; absolute CD4+ cell numbers, P = 0.55; and CD4+/CD8+ T-lymphocyte ratio, P = 0.29 [Table 1 ]). Transmission occurred sexually in all subjects (all were homosexual males except WT2, who was a heterosexual female). Pretreatment genotypic and phenotypic drug resistance data were available for all patients. Data for a subset of these patients have been reported previously (5, 42). Written informed consent was obtained from all subjects, and human experimentation guidelines of The Rockefeller University Institutional Review Board were followed.
TABLE 1.
Baseline characteristics of the 18 newly infected individuals from whom primary viral isolates were derived and resistance-associated mutations in the PR and RT genes of the primary isolates
| Virusa | Time (days)b | WBc | HIV-1 RNA (log10/ml) | CD4+ T cells (cells/mm3) | CD4+/CD8+ ratio | Genotyped
|
||
|---|---|---|---|---|---|---|---|---|
| PI | NRTI | NNRTI | ||||||
| MDR1 | 31 | ± | 6.2 | 263 | 0.18 | L10V, K20I, M36I A71V, G73S, L90M | M41L, T69D, K70R, V118I, T215F | V179D, Y188L |
| MDR2 | 79 | + | 5.6 | 395 | 0.48 | K20I, M36I, M46I, A71I, G73S, L90M | M41L, E44D, T69D, V118I, L210W, T215Y | —e |
| MDR3 | NAf | + | 3.5 | 420 | 0.75 | K20I, M36I, L90M | — | K103N, Y181C |
| R1+ | 19 | + | 6.1 | 475 | 0.43 | — | — | K103N |
| R2+ | 23 | ± | 6.4 | 528 | 0.59 | — | — | K103N |
| R3 | 10 | − | 5.2 | 211 | 0.30 | — | M184V | — |
| P1 | 16 | − | 6.1 | 635 | 0.73 | A71V, V82A | — | — |
| P2* | 30 | ± | 6.4 | 505 | 0.39 | A71V, L90M | — | — |
| P3* | 78 | + | 5.2 | 334 | 0.86 | A71V, L90M | — | — |
| WT1 | 10 | − | 7.4 | 221 | 0.97 | — | — | — |
| WT2 | 12 | + | 7.6 | 379 | 0.39 | K20R, M36I | — | — |
| WT3 | 28 | − | 5.8 | 423 | 0.80 | A71T | V118I | — |
| WT4 | 37 | + | 6.0 | 1281 | 0.30 | L10V | T215S | — |
| WT5 | 13 | + | 6.7 | 277 | 0.19 | L10I | — | — |
| WT6 | 10 | − | 7.2 | 210 | 0.29 | — | — | — |
| WT7 | 13 | − | 6.5 | 197 | 0.39 | — | — | — |
| WT8 | NA | + | 4.9 | 494 | 0.37 | M36I | — | V179D |
| WT9 | 55 | + | 5.5 | 510 | 0.28 | — | — | V179D |
| Rg | 36 | 5.6 | 418 | 0.52 | ||||
| WTh | 22 | 6.4 | 443 | 0.44 | ||||
* and +, viral isolates that are phylogenetically related.
Duration between onset of acute retroviral syndrome and sample collection.
WB, Western blot. +, positive; −, negative; ±, indeterminate.
Drug resistance-associated mutations were categorized based on the most recent guidelines (International AIDS Society-USA website updated November-December 2002 (www.iasusa.org) (20).
—, absence of drug resistance-associated mutations.
NA, not available.
R, mean values for all patients with drug-resistant viruses.
WT, mean values for all patients with drug susceptible viruses.
Cells.
Human osteosarcoma cells that stably express CD4 and the chemokine receptor CXCR4 or CCR5 and also carry the green fluorescent protein (GFP) gene under the transcriptional control of the HIV-2 long terminal repeat [GHOST(3) HIV indicator cells] were obtained from V. N. KewalRamani and D. R. Littman through the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program (32). Cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 100 μg of penicillin-streptomycin per ml, 2.3 mg of HEPES per ml, 1 μg of puromycin per ml, 500 μg of G418 per ml, and 100 μg of hygromycin per ml.
Human peripheral blood mononuclear cells (PBMC) were obtained by Ficoll density centrifugation. Enrichment for CD4+ cells was subsequently performed by depletion of CD8+ cells with M450 CD8 Dynabeads (Dynal AS, Oslo, Norway) according to the manufacturer's recommendations. Cells were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum, 100 μg of penicillin-streptomycin per ml, 2.3 mg of HEPES per ml, and 20U of recombinant interleukin-2 per ml.
Primary viral isolates.
Viral stocks were obtained by cocultivation of patient PBMC with phytohemagglutinin (PHA)-stimulated HIV-1-negative donor PBMC. Cultures were monitored for p24 antigen production by using a commercially available enzyme-linked immunosorbent assay (ELISA) (HIV-1 p24 antigen assay; Coulter, Miami, Fla.). Culture supernatants were filtered (0.2-μm-pore-size filter) and stored at −70°C until further use. Viral stocks of drug-resistant isolates were analyzed by sequencing to confirm that the genotype of the viral population matched the sequence (PR and the first 245 amino acids of RT [42]) originally obtained from plasma.
Determination of chemokine receptor usage.
Coreceptor usage was determined with the GHOST(3)-CXCR4 and GHOST(3)-CCR5 cell lines (44); 2 × 105 cells of each cell line were infected in parallel with 2 ng of p24 equivalent of virus, and the proportion of GFP-positive cells was assessed 48 h after infection by fluorescence-activated cell sorting.
Assessment of infectivity.
The infectivity of primary viral isolates was assessed in a single-cycle infectivity assay with GHOST(3)-CCR5 cells as described by Bleiber et al. (3) with the following modifications. A total of 4 × 104 cells (preincubated with 20 μg of Polybrene per ml for 1 h) were infected in duplicate with 2 ng of p24 antigen equivalent of virus by using the spinoculation technique (3 h at 168 × g and 22°C). A positive control (NL/JRFL [47]) and negative control were included in each experiment to assess interassay variation and cell autofluorescence. The proportion of GFP-expressing cells determined by fluorescence-activated cell sorting at 24 h postinfection was used as a measure of infectivity.
Assessment of growth kinetics.
HIV-1-negative CD4+ enriched cells were stimulated for 48 to 72 h with 1 μg of PHA per ml, and 106 cells were infected overnight with 2 ng of p24 antigen equivalent of each viral isolate, washed, and resuspended in complete medium supplemented with recombinant interleukin-2. All infections were performed in duplicate on cells from the same donor and then repeated with PBMC from a second donor. Aliquots of virus supernatant were collected every 3 to 4 days and monitored for p24 antigen production over 21 days with a commercially available ELISA (Coulter).
The individual growth kinetics for each virus were addressed mathematically by using the logistic growth law (33), which is based on a prey-predator model of population dynamics. This law states that the rate of growth depends on the number of virions (R) and becomes lower as this number of virions reaches some maximum, designated Rmax. The equation for logistic growth is dR/dt = gR[1 − (R/Rmax)], where R is the number of virions, g is the growth rate, and Rmax is the carrying capacity or maximum population size. For this reason, this equation is also known as the saturation law. Using nonlinear least squares to fit the number of virions (proportional to the concentration of p24, expressed in picograms per milliliter) to this equation, we estimated the parameter growth (day−1) for each virus. In this equation the parameter growth is indicative of how fast the number of virions increases in the earlier dynamics of viral replication, and exhaustion of target cells can therefore be neglected. Since we have two independent growth kinetics determined in duplicate on different PBMC donors for each viral isolate, the growth rate of each virus is given as the mean of the two estimates. Early data points below 80 pg of p24 antigen per ml were treated as censored data. The 95% confidence interval of the mean estimates was obtained from the cumulative distribution of the estimates computed by bootstrapping the residuals between the uncensored data and the theoretical values for both replicates. The doubling time (days), defined as the time required to double the number of virions per day, is given by ln(2)/g.
RCs of chimeric viruses encoding PR-RT regions.
RC was assessed by using an assay modification of PhenoSense (ViroLogic). Briefly, a 1.5-kb region spanning the p7-p1-p6 cleavage sites in gag, the entire PR gene, and the first 305 residues from RT was amplified from each filtered viral supernatant and inserted into an HIV-1 vector containing a luciferase expression cassette in the env position (45). (It should be noted that this assay requires reverse transcription and integration for the reporter signal to be generated.) Recombinant single-cycle viruses were generated upon cotransfection with a murine leukemia virus (4070A) envelope expression plasmid. Luciferase activity in infected cells was normalized for transfection efficiency between cultures and was used as a quantitative measure of infectivity (45). Adjusted RC rates were reported and reflect the percentage of the mean RC of a panel of wild-type HIV-1 isolates (ViroLogic) (unpublished data). These same chimeric viruses were used to perform phenotypic drug resistance testing (PhenoSense; ViroLogic) (36).
Virion-associated protein processing.
Filtered viral supernatants were concentrated by ultracentrifugation (3 h, 23,586 × g, 4°C). Similar amounts of viral particles, as assessed by p24 ELISA (Coulter), were separated on a sodium dodecyl sulfate-4 to 12% polyacrylamide gel (NuPage; Invitrogen, Carlsbad, Calif.), transferred to polyvinylidene difluoride membranes, and probed with mouse anti-Gag monoclonal antibodies (Coulter clone KC57). Signals were visualized by chemiluminescence (SuperSignal West; Pierce, Rockford, Ill.). The relative Gag protein distribution (p55/p24) was quantified with NIH Image software.
Sequencing analysis of the gag gene.
The gag gene was sequenced bidirectionally in order to assess amino acid substitutions in the cleavage sites as well as insertions and deletions. Viral RNA was extracted from filtered viral supernatants (QIAamp viral RNA kit; Qiagen, Valencia, Calif.), and the region encoding Gag and PR was amplified by using nested RT-PCR (SuperScript one-step RT-PCR system; Invitrogen) with outer primers fph3 [5′-AG(A/C)TCTCTCGACGCAGGACTCGGCTTG, positions 679 to 705 (HXB2)] and R7 (5′-CTGGGAAGTTCAATTAGGAATACC, positions 2810 to 2833) and inner primers Bssh1 (5′-TGCTGAAGCGCGCACGGCAAGAGGC, positions 704 to 728) and p2 (5′-CTTTTGGGCCATCCATTCCTGGC, positions 2588 to 2610). Purified PCR products were directly sequenced on an ABI sequencer (Applied Biosystems, Foster City, Calif.). Sequences were manually edited and aligned by using the DNASTAR software package and compared to the subtype B consensus gag sequence (Los Alamos HIV sequence database [http://hiv-web.lanl.gov]). To infer linkages among the 18 gag, PR, and RT sequences, phylogenetic analyses were performed with PAUP version 4.0 beta.
Statistical analysis.
The Mann-Whitney test was used to compare the infectivity rates and replication kinetics of the viral variants. All P values are two sided, and a P value of <0.05 was considered significant. All statistical analyses were performed with the Prism software (version 3.0; GraphPad Software Inc.).
Nucleotide sequence accession numbers.
The gag, PR, and RT nucleotide sequences of the 18 viral isolates are available under accession numbers AY206647 to AY206664 (gag) and AY261757 to AY261774 (PR and RT) in GenBank.
RESULTS
Characteristics of primary viral isolates.
All 18 viral isolates were CCR5-tropic as determined by parallel infection of GHOST(3)-CXCR4 and GHOST(3)-CCR5 cells (data not shown).
The drug-resistant group comprised three MDR viral isolates (MDR1 to -3) encoding primary resistance-conferring mutations in PR and RT, three isolates (R1 to -3) with isolated mutations in RT, and three isolates (P1 to -3) with mutations exclusively located in PR (Table 1). The PR and RT sequences derived from the viral isolates with drug resistance-associated mutations were similar to those derived from the plasma compartment (data not shown). Furthermore, the plasma-derived viral population and the viral isolates displayed comparable levels of drug susceptibility (variation of less than 2.5-fold [data not shown]). The only exception was isolate MDR2, which displayed a higher level of resistance to zidovudine (ZDV) than was originally measured with plasma-derived cell-free viruses (plasma-derived virus, 50-fold reduced susceptibility to ZDV; isolate, 500-fold reduced susceptibility to ZDV [data not shown]). Taken together, these control experiments show that in vitro culture did not reduce the level of drug resistance.
In order to study the replication characteristics of related primary isolates encoding primary drug resistance-associated mutations, two pairs of epidemiologically linked viruses (P2-P3 [L90M in PR] and R1-R2 [K103N in RT] [Table 1]) were included in the drug-resistant group. In the case of P2-P3, secondary transmission occurred during primary infection, with the linkage being confirmed by the patients' histories. In the second case, phylogenetic analysis of the PR-RT regions of R1 and R2 suggested a previously unrecognized linkage, potentially through one or more intermediates since the dates of the respective primary infections were 2 years apart. These findings were subsequently confirmed by independent sequencing of the viral isolates in which the gag, PR, and RT sequences of the pairs P2-P3 and R1-R2 formed clades distinct from the rest of the data set, with bootstrap values of 100 and small intrasample pairwise distances (data not shown).
Four isolates in the drug-susceptible group displayed single amino acid substitutions in RT that, in the context of other resistance-associated mutations, can either contribute to decreased susceptibility towards RT inhibitors (V179D and V118I) or reflect reversion from ZDV resistance-associated mutation T215Y (e.g., 215S [12, 15]). None of these changes are considered to confer primary resistance when identified in isolation.
Single-cycle infectivity of viral isolates with or without drug resistance.
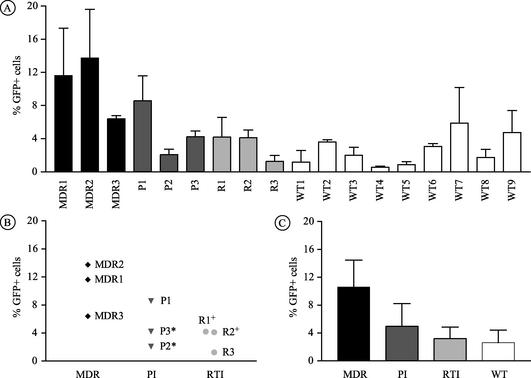
The 18 isolates were tested simultaneously in parallel infections performed in duplicate. The range of infectious titers, expressed as the percentage of GFP-positive cells 24 h after infection, represents the mean values from two or three independent experiments (mean for all isolates, 4.5; range for drug-resistant isolates, 1.2 to 13.7; range for drug-susceptible isolates, 0.6 to 5.9). The infectivity of the variants with drug resistance (mean, 6.4) was higher than that of the drug-susceptible variants (mean, 2.6; P = 0.032) (Fig. 1A). Detailed analysis of the group of drug-resistant isolates (Fig. 1B) showed that the infectivity rates of isolates MDR1, MDR2, and P1 differed significantly from those of the other drug-resistant viruses (P = 0.024). In the case of the linked viruses P2 and P3, a twofold increase of infectivity was observed (2.1 for P2 versus 4.2 for P3). The two phylogenetically related nonnucleoside RT inhibitor (NNRTI)-resistant isolates yielded similar infectivity rates (4.2 for R1 versus 4.1 for R2). The lamivudine-resistant isolate R3 displayed the lowest infectivity of all drug-resistant isolates.
FIG. 1.
Infectivities of the 18 viral isolates as assessed with GHOST reporter cells. The rate of infectivity is expressed as the percentage of GFP-positive cells at 24 h postinfection. The results shown represent the means ± standard deviations from two or three independent experiments. (A) Infectivity rates of the individual viral isolates; (B) infectivity rates of the nine drug-resistant viruses with respect to the drug class affected; (C) comparison of the infectivity rates for each drug resistance class with that of the group of wild-type (WT) (drug-susceptible) isolates. * and +, viral isolates that are phylogenetically related.
Kinetics of replication of viral isolates with or without drug resistance.
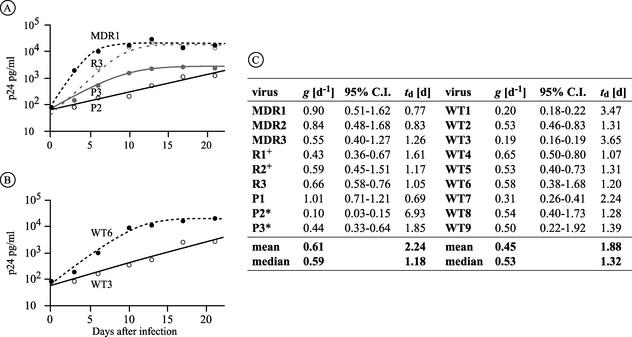
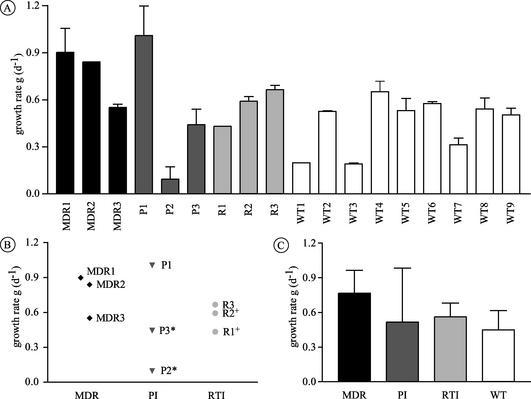
We next assessed the ability of the viral isolates to undergo multiple rounds of infection in stimulated CD4+ T lymphocytes. The viral growth kinetics were analyzed using a mathematical equation that allowed a precise curve fitting over a wide range of individual growth rates (see Fig. 3). The growth estimates were well reproduced with cells from the same donor and were further confirmed by repetition of the experiment with CD4+ T lymphocytes from a different donor. Drug-resistant isolates displayed growth rates (mean, 0.61) similar to those of drug-susceptible isolates (mean, 0.45) (P = 0.136) (Fig. 2A). The growth kinetics of isolates MDR1, MDR2, and P1 (Fig. 2B), however, were significantly higher than those of the wild-type viruses (P = 0.009) or the other drug-resistant isolates (P = 0.024). Analysis of the replication kinetics of the related isolates revealed that isolate P3 replicated more efficiently than P2 (approximately fourfold), whereas R1 and R2 had similar kinetics (Fig. 2B and 3). The lamivudine-resistant isolate R3 yielded growth rates similar to those of the wild-type viruses. In summary, all isolates were able to yield productive infections in mitogen-stimulated T lymphocytes, and drug-resistant isolates replicated as efficiently and in some case more efficiently than wild-type viruses.
FIG. 3.
Replication kinetics of viral isolates as determined by using a mathematical equation. (A and B) Representative curve fits for drug-resistant (A) and drug-susceptible (B) isolates over a 21-day period. Isolates were selected to visually represent the observed range of growth kinetics. Note that P2 and P3 are epidemiologically related. (C) Mean values for the growth rates, 95% confidence intervals (C.I.), and doubling times from two independent infections performed with PBMC from different donors. *and +, viral isolates that are phylogenetically related.
FIG. 2.
Replication kinetics of the 18 viral isolates as assessed by parallel infection of PHA-stimulated CD4+ T-lymphocytes. The growth rates (g) shown represent the means ± standard deviations from two different infections, each performed in duplicate. (A) Growth rates of the individual viral isolates; (B) growth rates of the nine drug-resistant viruses with respect to the drug class affected; (C) comparison of the growth rates for each drug resistance class with that of the group of wild-type (WT) (drug-susceptible) isolates. * and +, viral isolates that are phylogenetically related.
Virion-associated Gag processing of isolates with or without drug resistance.
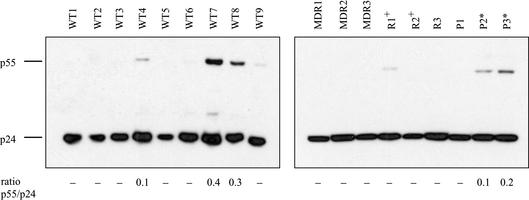
Western blotting of concentrated viruses was performed in order to demonstrate that the MDR and PR inhibitor (PI)-resistant isolates studied here do not display sizable Gag processing defects compared to RTI-resistant or wild-type isolates. Indeed, isolates MDR1, MDR2, MDR3, and P1 revealed no sizable processing defects despite the presence of primary mutations in PR (M46I, L90M, or V82A). The related isolates P2 and P3 (PR L90M) both yielded a small portion of unprocessed p55 (p55/p24 ratios of 0.1 and 0.2, for P2 and P3, respectively) (Fig. 4). The most sizable processing defects within this panel of isolates were observed in two of the drug-susceptible viruses (p55/p24 ratios of 0.4 and 0.3 for WT7 and WT8, respectively).
FIG. 4.
Virion-associated Gag processing of the viral isolates. Viral supernatants were concentrated by ultracentrifugation. Equivalent amounts of p24 antigen (2 ng) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes, and probed with the same mouse anti-Gag monoclonal antibody used for p24 quantification (KC57; Coulter). The ratio of uncleaved p55 to mature p24 was determined, using NIH Image software. WT, wild type (drug-susceptible isolates). * and +, viral isolates that are phylogenetically related.
Thus, none of the 18 isolates displayed substantial processing defects as defined by Gag processing intermediates. Moreover, all six MDR and PI-resistant isolates were able to efficiently process Gag, with isolates P2 and P3 displaying minor defects.
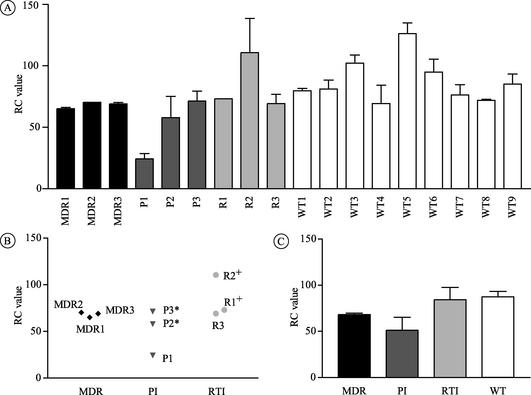
RCs of recombinant viruses encoding the drug resistance-associated regions.
Chimeric viruses encoding the two C-terminal Gag cleavage sites, PR, and the first 305 residues of RT were constructed in order to delineate the impact of compensatory changes located beyond the drug resistance-encoding regions themselves. Chimeric viruses encoding regions with drug resistance-associated mutations yielded lower RC values than the drug-susceptible recombinants (68 versus 87%; P = 0.011) (Fig. 5A). Analysis of the group of drug-resistant chimeras showed a narrow range of RC rates (58 to 73%), except for P1 (24%) and R2 (110%) (Fig. 5B). In contrast to the data generated by using viral isolates, the recombinant viruses MDR1, MDR2, and P1 displayed RC values similar to those of the other drug-resistant chimeras (P = 0.26) (compare Fig. 1B and 5B). Although the related isolates P2 and P3 differed with respect to infectivity and replication kinetics (Fig. 1 and 2), the PR-RT sequences of the two viruses were found to be identical. It is therefore not surprising that the RC values of the corresponding chimeric viruses P2 and P3 were comparable (Fig. 5).
FIG. 5.
RCs of the 18 chimeric viruses that encode the drug resistance-conferring regions (PR-RT). RC values were assessed by using a variation of the PhenoSense assay (ViroLogic). The results shown represent the means ± standard deviations from two different experiments. (A) RC rates of the individual viral isolates; (B) RC rates of the nine drug-resistant viruses with respect to the drug class affected; (C) comparison of the RC rates for each drug resistance class with that of the group of wild-type (WT) (drug-susceptible) isolates.* and +, viral isolates that are phylogenetically related.
In summary, insertion of the PR-RT regions of the drug-resistant isolates into an NL4-3-based molecular background resulted in chimeras that displayed a modest but significant reduction in RC.
Patterns of amino acid substitutions in Gag of isolates with or without drug resistance.
It has been shown that adaptive changes in different regions of Gag can contribute to restored replication of drug-resistant HIV-1 (9, 29, 48). For the purpose of differentiation between viral isolates and recombinant viruses, the Gag substitutions are reported based upon their location, with the C-terminal Gag region being included in the chimeric viruses (Table 2).
TABLE 2.
Sequences of the five cleavage sites in Gag from 18 viral isolates
| Gaga | Cleavage site sequenceb
|
||||
|---|---|---|---|---|---|
| p17/p24 | p24/p2 | p2/p7 | p7/p1 | p1/p6 | |
| conB | VSQNY/PIVQN | KARVL/AEAMS | SATIM/MQRGN | ERQAN/FLGKI | RPGNF/LQSRP |
| MDR1 | -----/----- | ---I-/----- | P-A--/----- | ----/------ | -----/--N-T |
| MDR2 | I----/----- | -----/----- | --A--/----- | ---V-/----- | -----/--N-- |
| MDR3 | --R--/----- | -----/----- | ---V-/--KS- | -----/----- | -----/--R-- |
| R1+ | -----/----- | -----/----- | NPA--/----- | -----/----- | -----/----- |
| R2+ | -----/----- | -----/----- | NPV--/----- | -----/----- | -----/----- |
| R3 | -----/----- | -----/----- | -----/--K-- | -----/----- | -----/P---T |
| P1 | ----L/----- | -----/----- | SSA--/----- | -----/---R- | -----/----- |
| P2* | D----/----- | -----/----- | ---V-/--K-- | -----/----L | -----/----- |
| P3* | -----/----- | -----/----- | ---V-/--K-- | -----/----L | -----/----- |
| WT1 | A----/----- | -----/----- | -GAA-/----- | -----/----- | -----/----- |
| WT2 | --R--/----- | -----/----- | PNNV-/----- | -----/----- | -----/P---- |
| WT3 | -----/----- | -----/----- | ---V-/----- | -----/----- | -----/--N-- |
| WT4 | -----/----- | -----/----- | --A--/----- | -----/----- | -----/----- |
| WT5 | -----/----- | -----/----- | --NI-/----- | -----/----- | -----/----- |
| WT6 | -----/----- | ---I-/----- | -----/----- | -----/----- | -----/----- |
| WT7 | S----/----- | -----/----- | --NV-/----- | -----/----- | -----/----- |
| WT8 | -----/---R- | -----/----- | A-N--/--K-- | -----/----- | -----/--N-- |
| WT9 | -----/----- | ---I-/----- | PT---/----- | -G---/----- | -----/----- |
MDR and PI-resistant isolates are in boldface. conB, subtype B Gag consensus sequence. * and +, viral isolates that are phylogenetically related.
Dashes indicate homology to the consensus B reference sequence.
(i) Substitutions in the C-terminal Gag cleavage sites and in the p6 region.
It has been reported that specific substitutions within the p7/p1 and p1/p6 Gag cleavage sites occur more frequently in patients failing PI-based regimens and that these changes can compensate for the reduced cleavage efficacy of drug-resistant PR (9, 40). All MDR and PI-resistant isolates studied encoded changes in at least one of the two C-terminal cleavage sites (Table 2), but only isolate MDR2 harbored a substitution clearly associated with restored infectivity in the context of PI resistance (A431V in p7/p1).
It has been proposed that the duplication of the PTAP motif in the p6 region of Gag can lead to increased infectivity and contribute to NRTI resistance (35). We observed insertions of three to nine residues in p6 involving partial as well as complete duplication of this motif in two MDR isolates (MDR2, PAE SFP SAP; MDR3, PAP) and in two of the drug-susceptible isolates (WT3, SAP; WT4, PEP TAP). Three of the four viruses also encoded mutations in RT that directly mediate resistance to NRTIs (e.g., T215Y/F in the MDR viruses) or are consistent with previous exposure to NRTIs (T215S in WT4).
(ii) Substitutions in the N-terminal cleavage sites of Gag.
Mutations within the p17/p24, p24/p2, and p2/p7 cleavage sites have previously been described to be present in both PI-treated and naive patients (9). It is therefore likely that the changes we observed within these cleavage sites of both drug-susceptible and drug-resistant viruses represent naturally occurring polymorphisms rather than relevant compensatory changes (Table 2).
Two substitutions in the p17/p24 sites of isolates P1 and P2 may, however, have an influence on viral replication efficiency. Isolate P1 displayed high infectivity and growth rates (Fig. 1 and 2), but the corresponding chimera showed low RC values (Fig. 5). In this isolate a tyrosine (Y)→leucine (L) substitution at the C-terminal position of p17 was observed (Table 2). The Gag amino acid sequences from the related PI-resistant isolates P2 and P3 differed in only one residue. At position P5 of the p17/p24 cleavage site, P2 encoded aspartic acid (D) instead of valine (V), which was seen in P3 as well as in the consensus B reference sequence (Table 2).
In conclusion, only one of the PI-resistant and MDR viruses encoded a known compensatory Gag cleavage site substitution. The differences in replication efficiency observed between the primary isolates and recombinant viruses encoding PR-RT and the C-terminal portion of Gag are therefore likely to be in part reflective of other compensatory mechanisms.
DISCUSSION
This study represents a comprehensive consideration of the replicative characteristics of 18 viral isolates derived from patients with primary HIV-1 infection, 9 of which encode some primary drug resistance-associated mutation in PR, RT, or both. The data described here suggest that drug-resistant HIV-1 isolates can bear adaptive mutations that allow for wild-type-level replicative function, thereby overcoming the potential defects associated with the genetic changes necessary for drug resistance.
Drug-resistant isolates displayed growth kinetics similar to those of drug-susceptible isolates in CD4+ T-lymphocyte cell culture. Furthermore, the single-cycle infectivity rates of these viruses were on average higher than those of drug-susceptible viruses. Additionally, the range of infectivity and growth rate values for the drug-resistant isolates was broader than that for the drug-susceptible isolates, with the latter group representing a more homogeneous set (Fig. 1 and 2).
The creation of chimeric viruses proved to be a useful tool for determining the contribution of certain viral regions towards the replicative fitness of the drug-resistant isolates (39, 40, 45). Specifically, the introduction of the C-terminal region of Gag and the PR-RT regions of the resistant viruses into an NL4-3 molecular background resulted in chimeras with lower RC values than the drug-susceptible recombinants. Moreover, the majority of chimeric viruses with drug resistance had very similar RC values within a relatively narrow range (58 to 73%).
Interestingly, three drug-resistant isolates (MDR1, MDR2, and P1) were characterized by a significantly higher level of infectivity and better growth kinetics than the remaining drug-resistant viruses and all of the drug-susceptible viruses (Fig. 1 and 2). Indeed, the aforementioned differences between isolates and chimeras were most dramatic for these cases. Because the inserted regions here included the C-terminal region of Gag as well as PR and RT, we may surmise that mutations in these regions do not account for the high replication efficiency displayed by these isolates. Specifically, we can conclude that the well-characterized compensatory substitutions in the p7/p1 cleavage site and the PTAP duplication in p6 (e.g., MDR2), while they may have contributed to the restoration of fitness, were not sufficient for the high replication levels of these two MDR viruses. While further studies are needed to elucidate the viral regions essential for the observed phenotypes of MDR1 and MDR2 (e.g., the N-terminal portion of Gag or the C-terminal part of Pol or Env), we may speculate that in the case of isolate P1 the Y→L substitution in the p17/p24 cleavage site contributed to efficient replication (Table 2). Although substitutions within this site were documented to take place under PI treatment (29), this specific substitution is not known to be of compensatory relevance. Recent reports to the contrary (37) demonstrate that wild-type PR cleaves p17/p24 less efficiently if Y is replaced by L in position P1 of the cleavage site. However, it is conceivable that the altered substrate specificity of drug-resistant PR (10) results in better viral maturation and consequently restores viral replication to levels observed in isolate P1. Lastly, we need to take into consideration the possibility that other regions of the viral genome of isolate P1 contribute to the observed phenotype.
Also of interest was the difference in replication characteristics observed among the related resistant isolates (Fig. 1B and 2B). Secondary transmission of the PI-resistant isolates occurred during primary infection. Isolate P3 showed a modest, but reproducible, twofold increase in infectivity relative to isolate P2. This difference was seen to be amplified (approximately fourfold) over multiple rounds of replication in PBMC culture in comparison to the single-cycle assay performed with the GHOST cell line. The chimeric viruses encoding the PR-RT sequences of these related viral isolates displayed similar RCs, however, suggesting that the PR functions in P2 and P3 are equivalent. The reversion to the consensus B p17/p24 cleavage site amino acid sequence in P3, therefore, may contribute to the increased replication efficiency of this isolate. Alternatively, the differences in replication efficiencies of P2 and P3 may reflect the differences in the duration of infection (P2, 30 days; P3, 70 days), since it has been proposed that replication efficiency increases over the time of infection (23). If the latter is true, the relative adaptive changes could have taken place in regions such as env, which are both highly variable and subject to the selective pressure of the immune system (14).
The growth rates of the linked NNRTI-resistant viruses, on the other hand, remained stable after sequential transmission. The replication rates of these isolates were comparable to those of P3. The preservation of replication kinetics by transmitted NNRTI-resistant viruses observed here is not surprising, since the relevant mutations (e.g., K103N) are known to confer little RT impairment (11).
Some have observed the persistence of transmitted drug-resistant variants within a host over time (e.g., 4 to 6 months) (3, 7), which might suggest intrinsic fitness. These results are in agreement with reports that some MDR isolates replicate to high levels both in vitro (3) and in vivo (22). We must consider, however, the likely homogeneity of the original inocula in these cases. In the setting of an emergent drug-resistant viral variant in chronic disease, for example, it is seen that discontinuation of antiretroviral treatment results in a rapid overgrowth by wild-type viruses. This suggests that these drug-resistant viruses are “fit” only in the presence of their respective drug pressures. The persistence of transmitted drug-resistant virus within a new host, therefore, may simply reflect an absence of competition.
Furthermore, in vitro observation may misrepresent the in vivo case. Indeed, a high level of replication in PBMC does not necessarily imply similar kinetics in other cell populations. Some PI-resistant viruses, for example, replicate well in PBMC but show profound replication defects in thymocytes (43). Thus, it may be that the lower virological set point observed in patients infected with MDR viruses does not reflect a poor viral replication potential in general; rather, these viruses may simply be selectively incapable of replicating efficiently in certain cell populations (e.g., thymocytes) and therefore are less generally pathogenic to the host.
It is important, however, to emphasize that our data are based on isolates derived from PBMC collected within 10 to 79 days (median, 21 days) after the onset of symptoms. The virus populations were sampled before being subjected to a number of immunological and host cell-associated constraints (24), but we cannot exclude the possibility that the viral characteristics analyzed in this study reflect but a snapshot of the viral replicative potential before the development of a mature HIV-1-specific immune response.
In conclusion, these findings indicate that the biological characteristics of newly transmitted viruses are similar, irrespective of the presence or absence of drug resistance-associated mutations. The specific compensatory mechanisms used to overcome defects imposed by primary drug resistance-associated mutations in PR and RT are likely to be complex and probably reflect an interplay between different viral regions. Elucidating how transmitted drug-resistant viruses regain infective potency can lead to the identification of new therapeutic targets which may someday allow us to prevent the emergence of such otherwise intractable variants.
Acknowledgments
We are indebted to the clinical team of the Aaron Diamond AIDS Research Center for their ongoing efforts in identifying newly infected individuals. We also thank David D. Ho for his consistent support and Paul Bieniasz for providing reagents and helpful discussions. Additionally, we thank Deborah M. Gurner, Lubbertus C. F. Mulder, and Lisa A. Chakrabarti for critical reading of the manuscript and Gregor M. Cardué for assistance with graphics.
This work was supported by Rockefeller University General Clinical Grant M01-RR00102 and NIH grants AI47033 and AI47033. V.S. was supported by a Research Fellowship from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Archer, R. H., C. Dykes, P. Gerondelis, A. Lloyd, P. Fay, R. C. Reichman, R. A. Bambara, and L. M. Demeter. 2000. Mutants of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase resistant to nonnucleoside reverse transcriptase inhibitors demonstrate altered rates of RNase H cleavage that correlate with HIV-1 replication fitness in cell culture. J. Virol. 74:8390-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Back, N. K., and B. Berkhout. 1997. Limiting deoxynucleoside triphosphate concentrations emphasize the processivity defect of lamivudine-resistant variants of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 41:2484-2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bleiber, G., M. Munoz, A. Ciuffi, P. Meylan, and A. Telenti. 2001. Individual contributions of mutant protease and reverse transcriptase to viral infectivity, replication, and protein maturation of antiretroviral drug-resistant human immunodeficiency virus type 1. J. Virol. 75:3291-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blower, S. M., A. N. Aschenbach, H. B. Gershengorn, and J. O. Kahn. 2001. Predicting the unpredictable: transmission of drug-resistant HIV. Nat. Med. 7:1016-1020. [DOI] [PubMed] [Google Scholar]
- 5.Boden, D., A. Hurley, L. Zhang, Y. Cao, Y. Guo, E. Jones, J. Tsay, J. Ip, C. Farthing, K. Limoli, N. Parkin, and M. Markowitz. 1999. HIV-1 drug resistance in newly infected individuals. JAMA 282:1135-1141. [DOI] [PubMed] [Google Scholar]
- 6.Borman, A. M., S. Paulous, and F. Clavel. 1996. Resistance of human immunodeficiency virus type 1 to protease inhibitors: selection of resistance mutations in the presence and absence of the drug. J. Gen. Virol. 77:419-426. [DOI] [PubMed] [Google Scholar]
- 7.Brenner, B. G., J. P. Routy, M. Petrella, D. Moisi, M. Oliveira, M. Detorio, B. Spira, V. Essabag, B. Conway, R. Lalonde, R. P. Sekaly, and M. A. Wainberg. 2002. Persistence and fitness of multidrug-resistant human immunodeficiency virus type 1 acquired in primary infection. J. Virol. 76:1753-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caliendo, A. M., A. Savara, D. An, K. DeVore, J. C. Kaplan, and R. T. D'Aquila. 1996. Effects of zidovudine-selected human immunodeficiency virus type 1 reverse transcriptase amino acid substitutions on processive DNA synthesis and viral replication. J. Virol. 70:2146-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cote, H. C., Z. L. Brumme, and P. R. Harrigan. 2001. Human immunodeficiency virus type 1 protease cleavage site mutations associated with protease inhibitor cross-resistance selected by indinavir, ritonavir, and/or saquinavir. J. Virol. 75:589-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dauber, D. S., R. Ziermann, N. Parkin, D. J. Maly, S. Mahrus, J. L. Harris, J. A. Ellman, C. Petropoulos, and C. S. Craik. 2002. Altered substrate specificity of drug-resistant human immunodeficiency virus type 1 protease. J. Virol. 76:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deeks, S. G., T. Wrin, T. Liegler, R. Hoh, M. Hayden, J. D. Barbour, N. S. Hellmann, C. J. Petropoulos, J. M. McCune, M. K. Hellerstein, and R. M. Grant. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344:472-480. [DOI] [PubMed] [Google Scholar]
- 12.de Ronde, A., M. van Dooren, L. van Der Hoek, D. Bouwhuis, E. de Rooij, B. van Gemen, R. de Boer, and J. Goudsmit. 2001. Establishment of new transmissible and drug-sensitive human immunodeficiency virus type 1 wild types due to transmission of nucleoside analogue-resistant virus. J. Virol. 75:595-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dykes, C., K. Fox, A. Lloyd, M. Chiulli, E. Morse, and L. M. Demeter. 2001. Impact of clinical reverse transcriptase sequences on the replication capacity of HIV-1 drug-resistant mutants. Virology 285:193-203. [DOI] [PubMed] [Google Scholar]
- 14.Ferbas, J., E. S. Daar, K. Grovit-Ferbas, W. J. Lech, R. Detels, J. V. Giorgi, and A. H. Kaplan. 1996. Rapid evolution of human immunodeficiency virus strains with increased replicative capacity during the seronegative window of primary infection. J. Virol. 70:7285-7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Lerma, J. G., S. Nidtha, K. Blumoff, H. Weinstock, and W. Heneine. 2001. Increased ability for selection of zidovudine resistance in a distinct class of wild-type HIV-1 from drug-naive persons. Proc. Natl. Acad. Sci. USA 98:13907-13912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gatanaga, H., Y. Suzuki, H. Tsang, K. Yoshimura, M. F. Kavlick, K. Nagashima, R. J. Gorelick, S. Mardy, C. Tang, M. F. Summers, and H. Mitsuya. 2002. Amino acid substitutions in Gag protein at non-cleavage sites are indispensable for the development of a high multitude of HIV-1 resistance against protease inhibitors. J. Biol. Chem. 277:5952-5961. [DOI] [PubMed] [Google Scholar]
- 17.Gotte, M., and M. A. Wainberg. 2000. Biochemical mechanisms involved in overcoming HIV resistance to nucleoside inhibitors of reverse transcriptase. Drug Resist. Update 3:30-38. [DOI] [PubMed] [Google Scholar]
- 18.Grant, R. M., F. M. Hecht, M. Warmerdam, L. Liu, T. Liegler, C. J. Petropoulos, N. S. Hellmann, M. Chesney, M. P. Busch, and J. O. Kahn. 2002. Time trends in primary HIV-1 drug resistance among recently infected persons. JAMA 288:181-188. [DOI] [PubMed] [Google Scholar]
- 19.Gray, R. H., M. J. Wawer, R. Brookmeyer, N. K. Sewankambo, D. Serwadda, F. Wabwire-Mangen, T. Lutalo, X. Li, T. vanCott, and T. C. Quinn. 2001. Probability of HIV-1 transmission per coital act in monogamous, heterosexual, HIV-1-discordant couples in Rakai, Uganda. Lancet 357:1149-1153. [DOI] [PubMed] [Google Scholar]
- 20.Hance, A. J., V. Lemiale, J. Izopet, D. Lecossier, V. Joly, P. Massip, F. Mammano, D. Descamps, F. Brun-Vezinet, and F. Clavel. 2001. Changes in human immunodeficiency virus type 1 populations after treatment interruption in patients failing antiretroviral therapy. J. Virol. 75:6410-6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izopet, J., P. Massip, C. Souyris, K. Sandres, B. Puissant, M. Obadia, C. Pasquier, E. Bonnet, B. Marchou, and J. Puel. 2000. Shift in HIV resistance genotype after treatment interruption and short-term antiviral effect following a new salvage regimen. AIDS 14:2247-2255. [DOI] [PubMed] [Google Scholar]
- 22.Kaufmann, D., M. Munoz, G. Bleiber, S. Fleury, B. Lotti, R. Martinez, W. Pichler, P. Meylan, and A. Telenti. 2000. Virological and immunological characteristics of HIV treatment failure. AIDS 14:1767-1774. [DOI] [PubMed] [Google Scholar]
- 23.Lawson, V. A., R. Oelrichs, C. Guillon, A. A. Imrie, D. A. Cooper, N. J. Deacon, and D. A. McPhee. 2002. Adaptive changes after human immunodeficiency virus type 1 transmission. AIDS Res. Hum. Retroviruses 18:545-556. [DOI] [PubMed] [Google Scholar]
- 24.Learn, G. H., D. Muthui, S. J. Brodie, T. Zhu, K. Diem, J. I. Mullins, and L. Corey. 1959. 2002. Virus population homogenization following acute human immunodeficiency virus type 1 infection. J. Virol. 76:11953-11959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Little, S. J., E. S. Daar, R. T. D'Aquila, P. H. Keiser, E. Connick, J. M. Whitcomb, N. S. Hellmann, C. J. Petropoulos, L. Sutton, J. A. Pitt, E. S. Rosenberg, R. A. Koup, B. D. Walker, and D. D. Richman. 1999. Reduced antiretroviral drug susceptibility among patients with primary HIV infection. JAMA 282:1142-1149. [DOI] [PubMed] [Google Scholar]
- 26.Little, S. J., S. Holte, J. P. Routy, E. S. Daar, M. Markowitz, A. C. Collier, R. A. Koup, J. W. Mellors, E. Connick, B. Conway, M. Kilby, L. Wang, J. M. Whitcomb, N. S. Hellmann, and D. D. Richman. 2002. Antiretroviral-drug resistance among patients recently infected with HIV. N. Engl. J. Med. 347:385-394. [DOI] [PubMed] [Google Scholar]
- 27.Long, E. M., H. L. Martin, Jr., J. K. Kreiss, S. M. Rainwater, L. Lavreys, D. J. Jackson, J. Rakwar, K. Mandaliya, and J. Overbaugh. 2000. Gender differences in HIV-1 diversity at time of infection. Nat. Med. 6:71-75. [DOI] [PubMed] [Google Scholar]
- 28.Lukashov, V. V., and J. Goudsmit. 1997. Founder virus population related to route of virus transmission: a determinant of intrahost human immunodeficiency virus type 1 evolution? J. Virol. 71:2023-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mammano, F., C. Petit, and F. Clavel. 1998. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and Gag coevolution in protease inhibitor-treated patients. J. Virol. 72:7632-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mammano, F., V. Trouplin, V. Zennou, and F. Clavel. 2000. Retracing the evolutionary pathways of human immunodeficiency virus type 1 resistance to protease inhibitors: virus fitness in the absence and in the presence of drug. J. Virol. 74:8524-8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, C. J., M. Marthas, J. Greenier, D. Lu, P. J. Dailey, and Y. Lu. 1998. In vivo replication capacity rather than in vitro macrophage tropism predicts efficiency of vaginal transmission of simian immunodeficiency virus or simian/human immunodeficiency virus in rhesus macaques. J. Virol. 72:3248-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morner, A., A. Bjorndal, J. Albert, V. N. Kewalramani, D. R. Littman, R. Inoue, R. Thorstensson, E. M. Fenyo, and E. Bjorling. 1999. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 73:2343-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murray, J. 1989. Continuous population models for single species. Mathematical biology. Springer Verlag, Berlin, Germany.
- 34.Olsen, D. B., M. W. Stahlhut, C. A. Rutkowski, H. B. Schock, A. L. vanOlden, and L. C. Kuo. 1999. Non-active site changes elicit broad-based cross-resistance of the HIV-1 protease to inhibitors. J. Biol. Chem. 274:23699-23701. [DOI] [PubMed] [Google Scholar]
- 35.Peters, S., M. Munoz, S. Yerly, V. Sanchez-Merino, C. Lopez-Galindez, L. Perrin, B. Larder, D. Cmarko, S. Fakan, P. Meylan, and A. Telenti. 2001. Resistance to nucleoside analog reverse transcriptase inhibitors mediated by human immunodeficiency virus type 1 p6 protein. J. Virol. 75:9644-9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettit, S. C., G. J. Henderson, C. A. Schiffer, and R. Swanstrom. 2002. Replacement of the P1 amino acid of human immunodeficiency virus type 1 Gag processing sites can inhibit or enhance the rate of cleavage by the viral protease. J. Virol. 76:10226-10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Precious, H. M., H. F. Gunthard, J. K. Wong, R. T. D'Aquila, V. A. Johnson, D. R. Kuritzkes, D. D. Richman, and A. J. Leigh Brown. 2000. Multiple sites in HIV-1 reverse transcriptase associated with virological response to combination therapy. AIDS 14:31-36. [DOI] [PubMed] [Google Scholar]
- 39.Resch, W., R. Ziermann, N. Parkin, A. Gamarnik, and R. Swanstrom. 2002. Nelfinavir-resistant, amprenavir-hypersusceptible strains of human immunodeficiency virus type 1 carrying an N88S mutation in protease have reduced infectivity, reduced replication capacity, and reduced fitness and process the Gag polyprotein precursor aberrantly. J. Virol. 76:8659-8666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson, L. H., R. E. Myers, B. W. Snowden, M. Tisdale, and E. D. Blair. 2000. HIV type 1 protease cleavage site mutations and viral fitness: implications for drug susceptibility phenotyping assays. AIDS Res. Hum. Retroviruses 16:1149-1156. [DOI] [PubMed] [Google Scholar]
- 41.Salomon, H., M. A. Wainberg, B. Brenner, Y. Quan, D. Rouleau, P. Cote, R. LeBlanc, E. Lefebvre, B. Spira, C. Tsoukas, R. P. Sekaly, B. Conway, D. Mayers, J. P. Routy, et al. 2000. Prevalence of HIV-1 resistant to antiretroviral drugs in 81 individuals newly infected by sexual contact or injecting drug use. AIDS 14:F17-F23. [DOI] [PubMed] [Google Scholar]
- 42.Simon, V., J. Vanderhoeven, A. Hurley, B. Ramratnam, M. Louie, K. Dawson, N. Parkin, D. Boden, and M. Markowitz. 2002. Evolving patterns of HIV-1 resistance to antiretroviral agents in newly infected individuals. AIDS 16:1511-1519. [DOI] [PubMed] [Google Scholar]
- 43.Stoddart, C. A., T. J. Liegler, F. Mammano, V. D. Linquist-Stepps, M. S. Hayden, S. G. Deeks, R. M. Grant, F. Clavel, and J. M. McCune. 2001. Impaired replication of protease inhibitor-resistant HIV-1 in human thymus. Nat. Med. 7:712-718. [DOI] [PubMed] [Google Scholar]
- 44.Vodros, D., C. Tscherning-Casper, L. Navea, D. Schols, E. De Clercq, and E. M. Fenyo. 2001. Quantitative evaluation of HIV-1 coreceptor use in the GHOST3 cell assay. Virology 291:1-11. [DOI] [PubMed] [Google Scholar]
- 45.Wrin, T., A. Gamarnik, N. Whitehurst, J. Beauchaine, J. M. Whitcomb, N. S. Hellmann, and C. J. Petropoulos. 2001. Natural variation of replication capacity measurements in drug-naïve/susceptible HIV-1. Antiviral Ther. 6:20. [Google Scholar]
- 46.Yerly, S., L. Kaiser, E. Race, J. P. Bru, F. Clavel, and L. Perrin. 1999. Transmission of antiretroviral-drug-resistant HIV-1 variants. Lancet 354:729-733. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, Y. J., T. Hatziioannou, T. Zang, D. Braaten, J. Luban, S. P. Goff, and P. D. Bieniasz. 2002. Envelope-dependent, cyclophilin-independent effects of glycosaminoglycans on human immunodeficiency virus type 1 attachment and infection. J. Virol. 76:6332-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, Y. M., H. Imamichi, T. Imamichi, H. C. Lane, J. Falloon, M. B. Vasudevachari, and N. P. Salzman. 1997. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J. Virol. 71:6662-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]