Abstract
The replication of hepatitis B virus (HBV) can be regulated by a variety of factors, including hormones, growth factors, and cytokines. However, the molecular mechanisms of these regulations are largely unknown. Ras is a small GTPase that responds to many of these external stimuli. In this study, we investigated the possible effect of Ras on the replication of HBV. Our results indicated that activated Ras could suppress the replication of HBV in both Huh7 and HepG2 cells. This suppression was independent of the X protein and most likely occurred at the transcriptional level. Deletion-mapping analysis of the HBV core promoter and its upstream ENI and ENII enhancers revealed multiple elements responsive to activated Ras. This suppression of HBV replication by activated Ras was apparently mediated by the mitogen-activated protein (MAP) kinase pathway, as it was accompanied by activation of ERK1/2 and abolished by the MEK1/2 inhibitor U0126. Our results thus indicate that external stimuli may suppress HBV replication through the Ras-MAP kinase pathway.
Hepatitis B virus (HBV) is a small DNA virus with a high liver tropism (for a review, see reference 11). This virus contains a 3.2-kb circular, partially double-stranded DNA genome. Upon infection of hepatocytes, this genomic DNA is transported into the nucleus, where it is repaired to form a covalently closed circular DNA (cccDNA) molecule. This cccDNA then serves as the template for the transcription of HBV RNAs. The HBV genome contains four promoters named the pre-S1, S, core, and X promoters, which direct transcription of the pre-S1 protein mRNA and the pre-S2 protein, the major S protein mRNAs, the precore protein and the core protein mRNAs, and the X protein mRNA, respectively. Pre-S1, pre-S2, and major S proteins, which are collectively called surface antigens, are the viral envelope proteins. The precore protein is the precursor of the e antigen found in the serum of HBV patients, and the core protein is the viral capsid protein. The X protein is a transcriptional transactivator that can regulate cellular signaling pathways and modify the DNA binding activities of transcription factors (37). The core protein mRNA also encodes the viral DNA polymerase, which is also a reverse transcriptase. In addition to these four promoters, the HBV genome also contains two enhancer elements, termed ENI and ENII. Both enhancers display liver-specific activities and are recognized by liver-enriched transcription factors. ENII is located in the upstream regulatory region of the core promoter and is also known as the core upstream regulatory sequence (38).
After its translation, the core protein packages its own mRNA as well as the viral polymerase to form the core particle. The core mRNA, which is also known as the pregenomic RNA, is then reverse transcribed to form the circular, partially double-stranded HBV DNA genome. The core particle then interacts with the surface antigens on the membrane of the endoplasmic reticulum to form the progeny virions, which are then secreted from the infected cell (11).
The replication of HBV can be affected by a variety of factors, including hormones, growth factors, and cytokines. For example, glucagon interferes with the replication of the related duck HBV (14); transforming growth factor beta suppresses the replication of duck HBV; epidermal growth factor, transforming growth factor alpha, and hepatocyte growth factor stimulate the accumulation of duck HBV cccDNA in the nuclei of infected cells (5); and interferons, tumor necrosis factor alpha, and insulin suppress the replication of HBV (12, 30). In addition, the cell cycle has also been shown to affect HBV replication: the replication of HBV is enhanced in cells arrested in the G1 or G2 phase and suppressed if cells enter the S phase (29), and cccDNA accumulation appears to increase in the G1 phase (36). In spite of these observations, the molecular mechanisms by which external factors or the cell cycle affects HBV replication remain largely unknown.
Ras is a small GTP-binding protein in the cell that can be activated by many external stimuli, including epidermal growth factor and insulin (15, 27). The activated form of Ras, which binds to GTP, can associate with a number of downstream effectors to exert its biological effects. Among these effectors, the Raf-MEK-ERK pathway is the best characterized (31). Raf is the mitogen-activated protein (MAP) kinase kinase kinase. After its activation by Ras, it will phosphorylate and activate MAP kinase kinases (MEKs), which will in turn phosphorylate MAP kinase or extracellular signal-regulated kinases (ERKs). Once activated, ERKs can translocate into the nucleus, where they will regulate the activities of transcription factors such as AP-1 (31). The HBV X protein has been shown to activate this Ras-MEK-ERK signaling cascade (2). Ras becomes inactivated when its associated GTP is converted to GDP.
As Ras is an important signaling molecule activated by many external stimuli, we investigated its possible effects on the replication of HBV. Our results demonstrate that activated Ras can suppress the replication of HBV through the MAP kinase pathway. This suppression by Ras apparently occurs at the transcription level and involves multiple regions of the HBV genome. It is also independent of the HBV X protein. These results provide a mechanism to explain how external factors may regulate HBV replication.
MATERIALS AND METHODS
DNA plasmids.
The plasmids pCMV-RasV12 and pCMV-RasA15, which express constitutively active Ras and dominant negative Ras, respectively, with the immediate-early promoter of cytomegalovirus have been described previously (34). The HBV plasmid pWTD, which contains the head-to-tail dimer of the HBV genome joined at the unique EcoRI site, has also been described (22). pWTDX− is identical to pWTD except that a missense mutation has been introduced to abolish the initiation codon of the X protein and a premature termination codon has been introduced into the X protein coding sequence. These two mutations, which prevented the expression of the 16.5-kDa X protein (24, 35), do not affect the overlapping polymerase coding sequence. Plasmid pCpCAT contains the HBV sequence from nucleotides 953 to 1803. This sequence, which contains the ENI enhancer, the ENII enhancer, and the entire core promoter, was linked to the chloramphenicol acetyltransferase (CAT) reporter. pUCAT1, pUCAT2, pUCAT4, pUCAT7, pUCAT8, and pUCAT9 contain nucleotides 1403 to 1803, 1455 to 1803, 1476 to 1803, 1646 to 1803, 1688 to 1803, and 1703 to 1803, respectively, of the HBV sequence. Similarly, these HBV sequences were linked to the CAT reporter in these DNA constructs. The constructions of all of these CAT reporter constructs have been described previously (9, 13).
Transfection of cells.
Huh7 cells and HepG2 cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. These cells were transfected by the calcium phosphate precipitation method (26). In general, Huh7 cells were transfected for 16 h when they were 80% confluent, and HepG2 cells were transfected for 20 to 24 h when they were 40% confluent. pXGH5, a plasmid that expresses the human growth hormone (hGH) under the control of the metallothionine promoter, was often used for cotransfection to serve as an internal control and to monitor the transfection efficiency. The inclusion or absence of pXGH5 had no effect on the experimental results. Forty-eight hours after transfection, the incubation medium was harvested and analyzed for hGH activity with a commercial radioimmunoassay kit (Nichols Diagnostics), and the cells were lysed for either the CAT assay or DNA or RNA analysis. The CAT assay was conducted with our previous procedures (8), and CAT activities were normalized against the hGH transfection control. At least three independent transfection experiments were conducted for analysis of CAT activities.
Northern blot and Southern blot analyses.
Cells were rinsed with ice-cold phosphate-buffered saline and scraped off the plate into 1 ml of Trizole (Invitrogen). Total cellular RNA was extracted following the manufacturer's protocol. The RNA was then subjected to Northern blotting and analyzed with 32P-labeled HBV DNA and hGH cDNA probes. For Southern blot analysis, cells were lysed in 0.5 ml of TBS (10 mM Tris-HCl [pH 7.0], 150 mM NaCl) containing 0.5% Nonidet P40 (NP-40). After a brief centrifugation to remove the nuclei, CaCl2 and MgCl2 were added to the supernatant to a final concentration of 4 mM each. The supernatant was then treated with 5 μg of DNase I and 16 U of micrococcal nuclease (Amersham Biosciences) at 37°C for 1 h to remove the viral DNA that was not associated with the core particles. The reaction was stopped by the addition of 40 μl of 0.5 M EDTA, 25 μl of 10% sodium dodecyl sulfate, 25 μl of 5 M NaCl, 5 μl of 1 M Tris-HCl, pH 8.0, and 27 μg of proteinase K. After incubation at 65°C overnight, viral DNA was isolated by phenol extraction and ethanol precipitated. The DNA pellet was rinsed with 70% ethanol and resuspended in 20 μl of TE (10 mM Tris-HCl [pH 7.0], 1 mM EDTA) for Southern blot analysis with a 32P-labeled HBV DNA probe.
Western blot analysis of ERK1/2.
U0126 (Promega), the specific inhibitor for MEK1/2, was freshly dissolved in dimethyl sulfoxide (DMSO) before use. Huh7 cells were transfected with the HBV genome and pCMV-RasV12 or its parental vector pCDNA3 (Invitrogen). Thirty-two hours after transfection, cells were treated with U0126 or its control solvent DMSO for 16 h. Cells were then rinsed with phosphate-buffered saline, scraped off the plate into TBS containing 0.5% NP-40, and sonicated. Cell lysates were then subjected to Western blot analysis with anti-ERK1/2 or anti-activated ERK1/2 antibody (Promega).
RESULTS
Effects of Ras on HBV replication in the presence and absence of X protein.
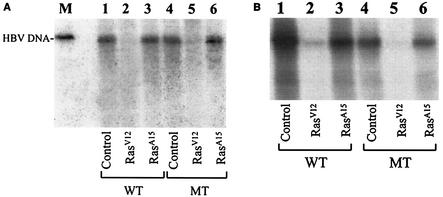
To investigate the possible effects of Ras on HBV replication, we cotransfected a head-to-tail dimer of the HBV genome with a plasmid that expressed either a constitutively active Ras (RasV12) or a dominant negative Ras (RasA15) into Huh7 cells, a well-differentiated human hepatoma cell line. RasV12 contains a glycine-to-valine substitution at amino acid 12. This substitution inhibits GTP hydrolysis and allows RasV12 to remain active. By contrast, RasA15 contains an alanine mutation at amino acid 15, binds to GDP, and suppresses Ras activity (7, 33). Two days after transfection, the replicated HBV DNA was isolated and analyzed by Southern blotting. As shown in Fig. 1A, an HBV DNA band of approximately 3.2 kb and a smear which represents the partially double-stranded HBV DNA could be detected in the gel in the absence of Ras. This result is similar to that in previous reports (1, 19, 20). Cotransfection with RasV12 significantly reduced the HBV DNA level, whereas cotransfection with RasA15 had no significant effect on the replication of HBV DNA. The reduction of HBV DNA replication by RasV12 is unlikely due to the nonspecific cytotoxic effect that might be generated by the expression of this protein, as the expression of human growth hormone (hGH) from pXGH5, a plasmid used for the cotransfection to serve as an internal control, was not affected (data not shown; also see below). These results indicated that the activated Ras could suppress HBV DNA replication.
FIG. 1.
Effects of Ras on HBV DNA replication. The wild-type (WT) HBV genome (lanes 1 to 3) and the mutant (MT) HBV genome, incapable of expressing only the 16.5-kDa X protein (lanes 4 to 6), was cotransfected with the control vector pCDNA3 (lanes 1 and 4), the RasV12-expressing plasmid pCMV-RasV12 (lanes 2 and 5), or the RasA15-expressing plasmid pCMV-RasA15 (lanes 3 and 6) into Huh7 cells (A) or HepG2 cells (B). The replicated HBV DNA was isolated from the core particles as described in Materials and Methods and analyzed by Southern blotting. In these transfection experiments, 5 μg of HBV DNA was cotransfected with 0.5 μg of the Ras expression plasmid into a 60-mm dish of cells. The 3.2-kb HBV DNA genome was run in lane M in A to serve as a size marker. Neither RasV12 nor RasA15 at the amount used affected the expression of hGH from pXGH5 when it was also used for cotransfection (data not shown).
The HBV X protein has previously been shown to activate Ras in hepatoma cells. Thus, it is conceivable that the X protein may interact with Ras to regulate HBV gene expression and replication. To investigate this possibility, we used an HBV DNA genome incapable of producing only the X protein for the cotransfection studies (24). As shown in Fig. 1A, abolishing the expression of the 16.5-kDa X protein did not affect HBV replication, nor did it affect the suppressive effect of RasV12 and the unapparent effect of RasA15 on HBV DNA replication. This result indicates that the X protein does not play a role in HBV replication in Huh7 cells. This is consistent with previous reports (4, 18, 23, 32).
To ensure that the effect of RasV12 on HBV DNA replication is not specific only to Huh7 cells, we also repeated the experiments with HepG2 cells, a well-differentiated human hepatoblastoma cell line. As shown in Fig. 1B, replication of the HBV DNA was reduced by RasV12 to an almost undetectable level, whereas the effect of RasA15 on HBV DNA replication was unapparent when the HBV DNA levels were normalized against the transfection efficiency (data not shown). These results indicated that the suppressive effect of RasV12 on HBV DNA replication was not specific to Huh7 cells. We also analyzed the effect of Ras on replication of the X-negative HBV genome in HepG2 cells. As shown in Fig. 1B, abolishing the expression of the 16.5-kDa X protein reduced the replication efficiency of the HBV genome in HepG2 cells. This is consistent with previous reports (16, 32). However, as in Huh7 cells, the X protein did not affect the suppressive activity of RasV12 on HBV DNA replication in HepG2 cells.
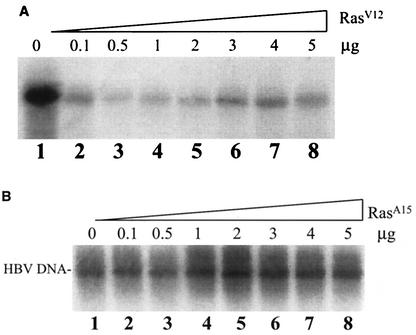
Dose-dependent effect of Ras on HBV DNA replication.
As the activities of Ras may be affected by its expression levels inside the cell, we examined the dose effects of RasV12 and RasA15 on HBV DNA replication. As shown in Fig. 2A, the cotransfection of 0.1 μg of RasV12 with the HBV genome into Huh7 cells resulted in a fivefold reduction in the amount of replicated HBV DNA. This reduction was even more substantial if the amount of RasV12 DNA was increased to 0.5 μg. Further increases in the RasV12 DNA amount led to a reduction of the suppressive effect. However, a sixfold reduction in HBV DNA replication efficiency was still observed when 5 μg of RasV12 was used in the cotransfection experiment. By contrast, as shown in Fig. 2B, the cotransfection of 0.1 μg or 0.5 μg of RasA15 DNA with the HBV genome into Huh7 cells had no apparent effect on HBV DNA replication. Further increases in RasA15 DNA resulted in a slight increase in the amount of replicated HBV DNA. Since RasA15 interfered with the activity of activated Ras, this slight enhancement of HBV DNA replication by RasA15 is consistent with a negative role of activated Ras in HBV DNA replication.
FIG. 2.
Dose effects of Ras on HBV DNA replication. The HBV DNA genome (5 μg) was cotransfected into a 60-mm dish of cells with either 5 μg of the control vector pCDNA3 (lane 1) or 0.1 to 5 μg (lanes 2 to 8) of a plasmid that expresses RasV12 (A) or RasA15 (B). When less than 5 μg of the Ras-expressing plasmid was used for cotransfection, the control vector was added to bring the amount of cotransfected DNA up to 5 μg. The experiments in A and B were conducted separately, and the film in A was exposed for a longer period of time than the film in B.
Suppression of HBV RNA transcription by activated Ras.
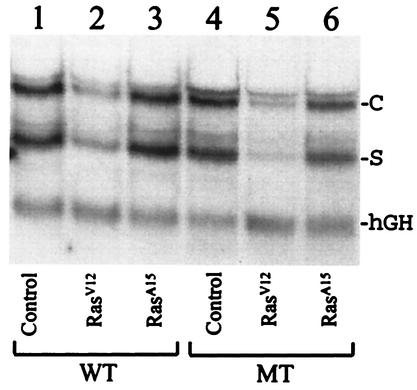
To understand how activated Ras suppressed HBV DNA replication, we performed Northern blotting experiments to analyze the transcription of HBV RNA. As shown in Fig. 3A, coexpression of RasV12 in Huh7 cells reduced the HBV RNA level sevenfold, whereas coexpression of RasA15 had no apparent effect on the HBV RNA level. The effect of RasV12 on HBV RNA was specific, as RasV12 had little effect on the hGH RNA level expressed from the control plasmid pXGH5. These results indicated that the suppression of HBV replication by activated Ras most likely occurred at the RNA level. Similar results were obtained with the X-negative HBV genome, suggesting that the X protein has no role in HBV RNA transcription in Huh7 cells.
FIG. 3.
Northern blot analysis of HBV RNA. The wild-type (WT) HBV DNA genome (8 μg) or the mutant (MT) HBV genome, incapable of expressing the X protein, was cotransfected with 8 μg of the control vector (lanes 1 and 4) or 7.2 μg of the control vector plus 0.8 μg of a plasmid that expresses either RasV12 (lanes 2 and 5) or RasA15 (lanes 3 and 6) into Huh7 cells. In addition, 4 μg of pXGH5 was also included in each transfection to monitor the transfection efficiency. Forty-eight hours after transfection, cells were lysed, and total cellular RNA was extracted for Northern blot analysis with both HBV DNA and hGH cDNA as probes. C and S mark the locations of HBV C gene and S gene transcripts, respectively, and hGH marks the location of the hGH RNA transcript. The RNA band that migrated slightly slower than the C gene transcripts was the 3.9-kb X gene transcript (13, 23) This RNA species was more apparent for the X-negative HBV genome.
Multiple Ras-responsive elements in HBV genome.
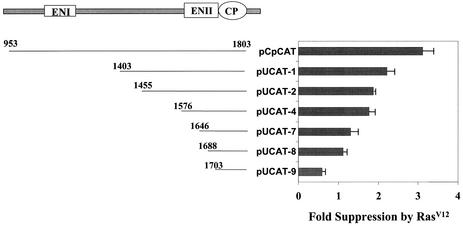
The results shown in Fig. 3 suggested a possible transcriptional regulation by Ras on the HBV genome. To investigate this possibility and to identify the possible Ras-responsive elements in the HBV genome, we used chloramphenicol acetyltransferase (CAT) as the reporter for the analysis. As the HBV C gene and S gene transcripts were similarly affected by Ras, we chose to focus our analysis on the core promoter and its upstream ENI and ENII enhancer elements. As shown in Fig. 4, there was a threefold suppressive effect by RasV12 when the core promoter as well as its upstream ENI and ENII enhancers was analyzed. This suppressive effect was reduced to slightly more than twofold when the ENI enhancer and its overlapping X promoter were deleted (pUCAT-1). Further deletion of the sequences upstream of the core promoter resulted in a successive decrease in the suppressive effect by RasV12. This suppressive effect was almost completely abolished when the sequence upstream of the core promoter was deleted to nucleotide 1688 in the ENII enhancer (pUCAT-8). Interestingly, further deletion to nucleotide 1703 (pUCAT-9) resulted in a nearly twofold increase in the core promoter activity by RasV12. These results suggested the presence of multiple RasV12-responsive elements in the HBV genome.
FIG. 4.
Deletion-mapping analysis of Ras-responsive elements in the HBV genome. The HBV core promoter (CP) as well as its upstream ENI and ENII enhancers is illustrated on the top. Sequences starting from various locations and ending at nucleotide 1803 were linked to the CAT reporter. The resulting reporter constructs were then cotransfected with an equal mass ratio of the control vector pCDNA3 or the RasV12 expression plasmid pCMV-RasV12 into Huh7 cells. pXGH5 was used for cotransfection to monitor the transfection efficiency. Fold suppression was determined by dividing the CAT activity obtained in the absence of RasV12 by that obtained in the presence of RasV12. The results represent the average of at least three independent transfection experiments.
Regulation of HBV gene expression by MAP kinase pathway.
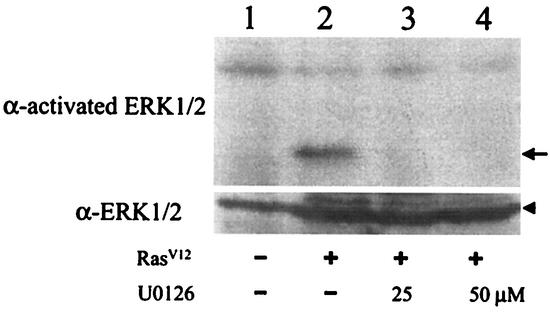
Activated Ras can activate Raf, which then activates MEK, which in turn activates ERK. To investigate whether this signaling cascade is responsible for the suppression of HBV DNA replication, we used U0126, a specific inhibitor of MEK1/2, in our studies. The HBV genome was cotransfected with the RasV12 expression plasmid into Huh7 cells, which was then treated with the control solvent DMSO, 25 μM U0126, or 50 μM U0126. To ensure that U0126 could indeed properly regulate MEK1/2 activities in these experimental conditions, we analyzed its effect on ERK1/2, which are the downstream effectors of MEK1/2. As shown in Fig. 5, RasV12 could activate ERK1/2, which were largely inactive in the absence of RasV12. The activation of ERK1/2 was blocked by U0126 at either 25 μM or 50 μM. These results indicated that RasV12 could activate ERK1/2 and that this activation was mediated by MEK1/2.
FIG. 5.
Suppression of ERK1/2 activation by U0126. Huh7 cells in a 60-mm dish were transfected with 5 μg of the control vector (lane 1) or the RasV12-expressing plasmid (lanes 2 to 4) and then treated with the control solvent DMSO (lanes 1 and 2) or 25 μM (lane 3) or 50 μM (lane 4) U0126 for 16 h. Cells were then lysed, and ERK1/2 were analyzed with either anti-activated ERK1/2 antibody (upper panel) or anti-ERK1/2 antibody (lower panel). The arrow marks the location of activated ERK1/2. The arrowhead marks the location of ERK1/2.
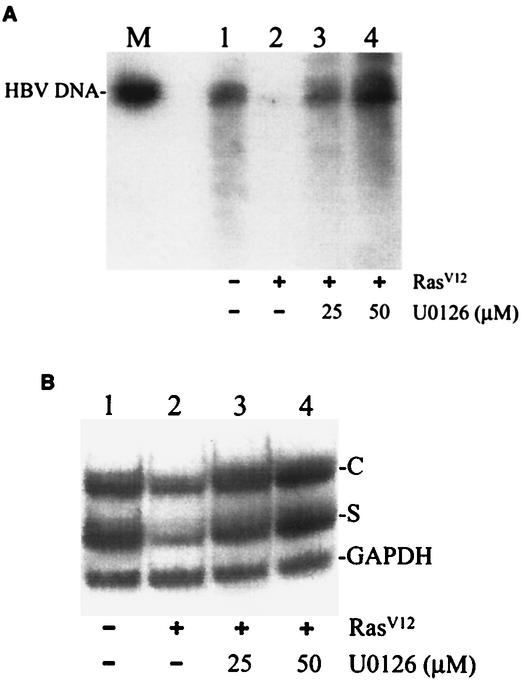
We then examined the effect of U0126 on the replication of HBV. As shown in Fig. 6A, in agreement with the results shown in Fig. 1, coexpression of the HBV DNA genome with RasV12 significantly suppressed the replication of HBV. However, in the presence of 25 μM U0126, this suppressive effect of RasV12 was almost entirely abolished. Increasing the U0126 concentration to 50 μM resulted in a further increase in the HBV DNA replication level. Similar results were also obtained when HBV RNA was analyzed. As shown in Fig. 6B, the reduction in the HBV RNA levels by RasV12 could be abolished by 25 μM or 50 μM U0126. Taken together, these results indicated that the suppressive effect of RasV12 on HBV replication was mediated by the MAP kinase pathway.
FIG. 6.
Effects of U0126 on HBV replication. Huh7 cells transfected with the control vector (lane 1) or the RasV12-expressing plasmid (lanes 2 to 4) were treated with DMSO (lanes 1 and 2) or 25 μM (lane 3) or 50 μM (lane 4) U0126 as described in Materials and Methods. Cells were then lysed, and the HBV DNA was isolated from core particles and analyzed by Southern blotting (A). Total cellular RNA was isolated and analyzed for HBV RNA by Northern blotting (B). In A, the 3.2-kb HBV genomic DNA fragment was run in lane M to serve as a size marker. In panel B, the C, S, and GADDH mark the locations of HBV C gene transcripts, HBV S gene transcripts, and GADDH RNA, respectively. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
DISCUSSION
Previous studies have demonstrated that the replication of HBV can be regulated by a variety of external stimuli, including hormones, growth factors, cytokines, and the cell cycle (5, 12, 14, 29, 30, 36). However, the molecular mechanisms of these regulations are largely unknown. Ras is a GTP-binding protein that can be activated by a number of these external factors. In this study, we demonstrated that activated Ras suppresses HBV DNA replication in a dose-dependent manner (Fig. 1 and 2A). The suppression of HBV DNA replication by activated Ras was most likely the result of suppression of HBV RNA transcription, because the HBV RNA levels were substantially reduced by activated Ras in cells (Fig. 3). The ability of activated Ras to regulate HBV gene transcription was confirmed by the reporter assays, in which RasV12 was found to suppress the core promoter activity (Fig. 4). In this reporter assay, RasV12 suppressed the core promoter activity at most threefold, whereas in the Northern blot analysis it reduced the C RNA level approximately sevenfold. This difference in suppression may be caused by the differences in the two experimental systems. For example, in the reporter studies, subgenomic DNA fragments were used. It remains possible, however, that RasV12 may also posttranscriptionally regulate HBV RNA, perhaps by enhancing its degradation.
In an attempt to identify the Ras-responsive elements in the HBV genome, we performed deletion-mapping experiments in the reporter studies. Our results revealed multiple responsive elements in the sequence upstream of the core promoter in the HBV genome (Fig. 4). It is likely that activation of the MAP kinase pathway by Ras leads to modification of the transcription factors that bind to different regions of the HBV genome. Alternatively, it is also likely that a specific cofactor that binds to the basal core promoter is affected by this signaling pathway. In this case, the overall suppressive effect of this cofactor on the core promoter will likely be the sum of its interaction with the transcription factors binding to the upstream regions of the core promoter. This explains why sequential deletion of these upstream sequences led to successive reductions in core promoter activity (Fig. 4). If this is indeed correct, then it is possible that the suppression of other HBV promoters is also mediated by a similar mechanism. An interesting finding is that the deletion of a sequence located between nucleotides 1688 and 1703 resulted in an approximately twofold increase in core promoter activity (Fig. 4). This sequence corresponds to the B1 region of the ENII enhancer and is recognized by a liver-specific orphan nuclear receptor named hBIF (25). It will be interesting to determine whether the activity of hBIF is affected by the activation of Ras.
The activation of Ras can lead to activation of the MAP kinase signaling pathway. This pathway is apparently responsible for the suppression of HBV replication, as blocking the activation of ERK1/2 by MEK1/2 with U0126 abolished the suppressive effects of RasV12 on HBV replication (Fig. 6). Although the HBV X protein has no apparent effect on HBV replication in Huh7 cells (Fig. 1A) (4, 18, 23, 32), it enhances HBV replication in HepG2 cells (Fig. 1B) (16, 28, 32). Previous studies indicate that the X protein can activate the Ras-MAP kinase signaling pathway (2, 3, 21). This activation was subsequently demonstrated to be mediated by the activation of Src (17), which was further found to be activated by the release of Ca2+ triggered by the X protein (6). Interestingly, the activation of Src but not the Ras-MAP kinase pathway was found to be responsible for the enhancement of HBV replication in HepG2 cells. Our results extend these previous findings and indicate that activation of the Ras-MAP kinase pathway can in fact have a suppressive effect on HBV replication. It remains to be determined why the simultaneous activation of Src and Ras by the X protein leads to the enhancement of HBV replication (Fig. 1B) (16, 32). It is likely that the plural effects of Src and Ca2+ diminish the suppressive effect of activated Ras on HBV replication, and therefore only the positive effect of Src and Ca2+ was observed. Alternatively, it is also possible that the modest activation of the Ras-MAP kinase pathway by the X protein renders the suppressive effect of Ras unapparent (16, 17). This second possibility is supported by our observation that dominant negative RasA15, which interferes with the activities of activated Ras, had only a marginal enhancing effect on HBV DNA replication (Fig. 2B). It is conceivable that, in the presence of a strong external stimulus for Ras such as insulin, the suppressive effect of Ras will override the enhancing effect of Src and Ca2+ and lead to the reduction of HBV replication (10, 12) (Fig. 1B). Further research in this area will certainly generate interesting results for understanding the interplay of different signaling pathways in HBV replication.
Acknowledgments
We thank Ben Yen and Jinah Choi for critical reading of the manuscript.
This work was supported by a research grant from the National Institutes of Health.
REFERENCES
- 1.Bchini, R., F. Capel, C. Dauguet, S. Dubanchet, and M. A. Petit. 1990. In vitro infection of human hepatoma (HepG2) cells with hepatitis B virus. J Virol. 64:3025-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benn, J., and R. J. Schneider. 1994. Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signaling cascade. Proc. Natl. Acad. Sci. USA 91:10350-10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benn, J., F. Su, M. Doria, and R. J. Schneider. 1996. Hepatitis B virus HBx protein induces transcription factor AP-1 by activation of extracellular signal-regulated and c-Jun N-terminal mitogen-activated protein kinases. J. Virol. 70:4978-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blum, H. E., Z. S. Zhang, E. Galun, F. von Weizsacker, B. Garner, T. J. Liang, and J. R. Wands. 1992. Hepatitis B virus X protein is not central to the viral life cycle in vitro. J. Virol. 66:1223-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borel, C., O. Schorr, I. Durand, F. Zoulim, A. Kay, C. Trepo, and O. Hantz. 2001. Initial amplification of duck hepatitis B virus covalently closed circular DNA after in vitro infection of embryonic duck hepatocytes is increased by cell cycle progression. Hepatology 34:168-179. [DOI] [PubMed] [Google Scholar]
- 6.Bouchard, M. J., L. H. Wang, and R. J. Schneider. 2001. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 294:2376-2378. [DOI] [PubMed] [Google Scholar]
- 7.Bourne, H. R., D. A. Sanders, and F. McCormick. 1991. The GTPase superfamily: conserved structure and molecular mechanism. Nature 349:117-127. [DOI] [PubMed] [Google Scholar]
- 8.Chen, M., S. Hieng, X. Qian, R. Costa, and J. H. Ou. 1994. Regulation of hepatitis B virus ENI enhancer activity by hepatocyte-enriched transcription factor HNF3. Virology 205:127-132. [DOI] [PubMed] [Google Scholar]
- 9.Chen, M., and J. H. Ou. 1995. Cell type-dependent regulation of the activity of the negative regulatory element of the hepatitis B virus core promoter. Virology 214:198-206. [DOI] [PubMed] [Google Scholar]
- 10.Chou, C. K., T. S. Su, C. M. Chang, C. P. Hu, M. Y. Huang, C. S. Suen, N. W. Chou, and L. P. Ting. 1989. Insulin suppresses hepatitis B surface antigen expression in human hepatoma cells. J. Biol. Chem. 264:15304-15308. [PubMed] [Google Scholar]
- 11.Ganem, D., and R. J. Schneider. 2001. Hepadnaviridae: the viruses and their replication, p. 3087. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 12.Gripon, P., C. Diot, A. Corlu, and C. Guguen-Guillouzo. 1989. Regulation by dimethyl sulfoxide, insulin, and corticosteroids of hepatitis B virus replication in a transfected human hepatoma cell line. J. Med. Virol. 28:193-199. [DOI] [PubMed] [Google Scholar]
- 13.Guo, W., M. Chen, T. S. Yen, and J. H. Ou. 1993. Hepatocyte-specific expression of the hepatitis B virus core promoter depends on both positive and negative regulation. Mol. Cell. Biol. 13:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hild, M., O. Weber, and H. Schaller. 1998. Glucagon treatment interferes with an early step of duck hepatitis B virus infection. J. Virol. 72:2600-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamata, T., and J. R. Feramisco. 1984. Epidermal growth factor stimulates guanine nucleotide binding activity and phosphorylation of ras oncogene proteins. Nature 310:147-150. [DOI] [PubMed] [Google Scholar]
- 16.Klein, N. P., M. J. Bouchard, L. H. Wang, C. Kobarg, and R. J. Schneider. 1999. Src kinases involved in hepatitis B virus replication. EMBO J. 18:5019-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein, N. P., and R. J. Schneider. 1997. Activation of Src family kinases by hepatitis B virus HBx protein and coupled signaling to Ras. Mol. Cell. Biol. 17:6427-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koike, K., and S. Takada. 1995. Biochemistry and functions of hepatitis B virus X protein. Intervirology 38:89-99. [DOI] [PubMed] [Google Scholar]
- 19.Lamberts, C., M. Nassal, I. Velhagen, H. Zentgraf, and C. H. Schroder. 1993. Precore-mediated inhibition of hepatitis B virus progeny DNA synthesis. J. Virol. 67:3756-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lan, Y. T., J. Li, W. Liao, and J. Ou. 1999. Roles of the three major phosphorylation sites of hepatitis B virus core protein in viral replication. Virology 259:342-348. [DOI] [PubMed] [Google Scholar]
- 21.Lee, S., C. Tarn, W. H. Wang, S. Chen, R. L. Hullinger, and O. M. Andrisani. 2002. Hepatitis B virus X protein differentially regulates cell cycle progression in X-transforming versus nontransforming hepatocyte (AML12) cell lines. J. Biol. Chem. 277:8730-8740. [DOI] [PubMed] [Google Scholar]
- 22.Li, J., V. E. Buckwold, M. W. Hon, and J. H. Ou. 1999. Mechanism of suppression of hepatitis B virus precore RNA transcription by a frequent double mutation. J. Virol. 73:1239-1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, J., and J. H. Ou. 2001. Differential regulation of hepatitis B virus gene expression by the Sp1 transcription factor. J. Virol. 75:8400-8406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, J., Z. Xu, Y. Zheng, D. L. Johnson, and J. H. Ou. 2002. Regulation of hepatocyte nuclear factor 1 activity by wild-type and mutant hepatitis B virus X proteins. J. Virol. 76:5875-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, M., Y. H. Xie, Y. Y. Kong, X. Wu, L. Zhu, and Y. Wang. 1998. Cloning and characterization of a novel human hepatocyte transcription factor, hB1F, which binds and activates enhancer II of hepatitis B virus. J. Biol. Chem. 273:29022-29031. [DOI] [PubMed] [Google Scholar]
- 26.Lu, W., S. Y. Lo, M. Chen, K. Wu, Y. K. Fung, and J. H. Ou. 1999. Activation of p53 tumor suppressor by hepatitis C virus core protein. Virology 264:134-141. [DOI] [PubMed] [Google Scholar]
- 27.Maassen, J. A., B. M. Burgering, R. H. Medema, A. P. Osterop, G. C. van der Zon, W. Moller, and J. L. Bos. 1992. The role of ras proteins in insulin signal transduction. Horm. Metab. Res. 24:214-218. [DOI] [PubMed] [Google Scholar]
- 28.Melegari, M., P. P. Scaglioni, and J. R. Wands. 1998. Cloning and characterization of a novel hepatitis B virus X binding protein that inhibits viral replication. J. Virol. 72:1737-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ozer, A., V. I. Khaoustov, M. Mearns, D. E. Lewis, R. M. Genta, G. J. Darlington, and B. Yoffe. 1996. Effect of hepatocyte proliferation and cellular DNA synthesis on hepatitis B virus replication. Gastroenterology 110:1519-1528. [DOI] [PubMed] [Google Scholar]
- 30.Pasquetto, V., S. F. Wieland, S. L. Uprichard, M. Tripodi, and F. V. Chisari. 2002. Cytokine-sensitive replication of hepatitis B virus in immortalized mouse hepatocyte cultures. J. Virol. 76:5646-5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peyssonnaux, C., and A. Eychene. 2001. The Raf/MEK/ERK pathway: new concepts of activation. Biol. Cell 93:53-62. [DOI] [PubMed] [Google Scholar]
- 32.Seeger, C. 1997. The hepatitis B virus X protein: the quest for a role in viral replication and pathogenesis. Hepatology 25:496-498. [DOI] [PubMed] [Google Scholar]
- 33.Stewart, S., and K. L. Guan. 2000. The dominant negative Ras mutant, N17Ras, can inhibit signaling independently of blocking Ras activation. J. Biol. Chem. 275:8854-8862. [DOI] [PubMed] [Google Scholar]
- 34.Wang, H. D., A. Trivedi, and D. L. Johnson. 1997. Hepatitis B virus X protein induces RNA polymerase III-dependent gene transcription and increases cellular TATA-binding protein by activating the Ras signaling pathway. Mol. Cell. Biol. 17:6838-6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu, Z., T. S. Yen, L. Wu, C. R. Madden, W. Tan, B. L. Slagle, and J. H. Ou. 2002. Enhancement of hepatitis B virus replication by its X protein in transgenic mice. J. Virol. 76:2579-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeh, C. T., H. T. Chiu, C. M. Chu, and Y. F. Liaw. 1998. G1 phase dependent nuclear localization of relaxed-circular hepatitis B virus DNA and aphidicolin-induced accumulation of covalently closed circular DNA. J. Med. Virol. 55:42-50. [DOI] [PubMed] [Google Scholar]
- 37.Yen, T. S. 1996. Hepadnaviral X protein: review of recent progress. J. Biomed. Sci. 3:20-30. [DOI] [PubMed] [Google Scholar]
- 38.Yuh, C. H., and L. P. Ting. 1990. The genome of hepatitis B virus contains a second enhancer: cooperation of two elements within this enhancer is required for its function. J. Virol. 64:4281-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]