Abstract
RP4 TrbB, an essential component of the conjugative transfer apparatus of the broad-host-range plasmid RP4, is a member of the PulE protein superfamily involved in multicomponent machineries transporting macromolecules across the bacterial envelope. PulE-like proteins share several well conserved motifs, most notable a nucleoside triphosphate binding motif (P-loop). Helicobacter pylori HP0525 also belongs to the PulE superfamily and is encoded by the pathogenicity island cag, involved in the inflammatory response of infected gastric epithelial cells in mammals. The native molecular masses of TrbB and HP0525 as determined by gel filtration and glycerol gradient centrifugation suggested a homohexameric structure in the presence of ATP and Mg2+. In the absence of nucleotides and bivalent cations, TrbB behaved as a tetramer whereas the hexameric state of HP0525 remained unaffected. Electron microscopy and image processing demonstrated that TrbB and HP0525 form ring-shaped complexes (diameter: 12 nm) with a central region (diameter: 3 nm) of low electron density when incubated in the presence of ATP and Mg2+. However, the TrbB average image appeared to be more elliptical with strong twofold rotational symmetry whereas HP0525 complexes are regular hexagons. Six well defined triangle-shaped areas of high electron density were distinguishable in both cases. Covalent crosslinking of TrbB suggests that the hexameric ring is composed from a trimer of dimers, because only dimeric, tetrameric, and hexameric species were detectable. The toroidal structure of TrbB and HP0525 suggests that both proteins catalyze a repetitive process, most probably translocating a cognate substrate across the inner membrane.
Conjugation is a widespread plasmid- or transposon-encoded process by which DNA is transferred from a bacterial donor into a recipient cell. Conjugative DNA transfer requires intimate cell–cell contact established by the mating pair formation (Mpf) system. Essential transfer (Tra) proteins of the Mpf system are responsible for assembly of conjugative pili thought to initiate the donor–recipient interaction. Mpf complexes have been proposed to consist of a membrane-spanning multicomponent complex. In the case of IncP plasmids, 12 plasmid-encoded proteins (TrbB–L of Tra2 and TraF of Tra1) are components of this complex (1, 2). TrbB, on which we are reporting here, is one of them. Conjugative plasmids belonging to several incompatibility (Inc) groups (e.g., IncP, IncN, IncW, Tra Ti, and IncX) display extensive similarities in gene organization and primary structure of Mpf components. Moreover, similarities to the T-DNA transfer systems of Ti/Ri plasmids have been explored (3–6) and recently extended to secretion systems that are not primarily dedicated to DNA transfer but to transport of multisubunit protein complexes from pathogenic bacteria to eukaryotic cells. Examples are the pertussis toxin export system Ptl of Bordetella pertussis (1), the cag pathogenicity island of Helicobacter pylori (7), the Dot/Icm system of Legionella pneumophila (8), and putative secretion systems from Rickettsia provazeckii (9), Actinobacillus actinomycetemcomitans, and Brucella spp. (10).
Thus, bacterial conjugation, the agrobacterial T-DNA transfer system, and the newly discovered pathogenicity-related secretion systems have been classified as secretion pathway type IV (11). It has been proposed that bacterial conjugation and the Agrobacterium-mediated T-DNA transfer are mechanistically related to type III secretion systems of plant and animal pathogens and to flagellum assembly (12, 13). In both type IV and type III systems, the secretion of specific macromolecules is mediated by multiprotein transmembrane complexes, and the genes encoding the proteins of the complexes are organized in a single cluster. Finally, a filamentous appendage is required for both secretion processes. It has been speculated that a pilus-like structure is also involved in the main terminal branch of the sec-dependent general secretion pathway type II. Similarity of the subunits of adhesive pili (type IV pili) of various bacterial pathogens and of proteins required for type IV piliation underlines this hypothesis (14, 15). A common feature of all complex secretion systems described so far is the presence of a cytoplasmic protein with NTPase motifs, reflecting the general requirement for an energy source to drive a transmembrane transport reaction. Most significantly, NTPases from type II and type IV systems are evolutionarily related, sharing four highly conserved motifs (16). These NTPases (the prototype for type IV systems is the agrobacterial VirB11 protein) are generally hydrophilic and lack obvious transmembrane domains and amino-terminal export signals. In the case of IncP and IncW plasmids, it is the only component of the Mpf complex that is soluble under native conditions. RP4 TrbB (17), Ti VirB11 (18, 19), and R388 TrwD (16) were localized in the cytoplasm. However, whereas RP4 TrbB is clearly a cytoplasmic protein, R388 TrwD and Ti VirB11 were also detected in the inner membrane fraction, and small amounts were found even in the outer membrane fraction.
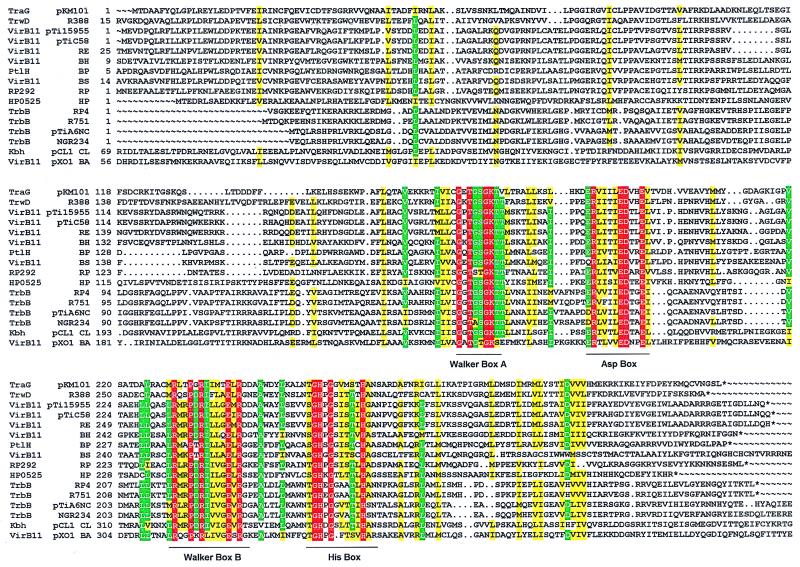
To obtain deeper insight into the structure of representatives of VirB11-like proteins (Fig. 1), we explored prototypes from two different type IV systems: TrbB, encoded by the conjugative IncPα plasmid RP4, and HP0525 from the cag pathogenicity island of H. pylori (20). Purified proteins were subjected to various physical and chemical techniques to probe their structure. The results demonstrate that both RP4 TrbB and H. pylori cag HP0525 form hexameric assemblies. Image processing of the electron micrographs illustrates that these hexamers are ring-shaped structures composed of six triangular monomers.
Figure 1.
Alignment of VirB11 family transport NTPases. Amino acid sequences were aligned by using the pileup program of the GCG program package version 9.1. boxshade 3.31 (http://www.ch.embnet.org/software/BOX_form.html) was used with the following parameters: red background and white letters, 100% conservation; green background and white letters, at least 75% conservation of identical residue; and yellow background, at least 75% conservation of similar residues. The extensions of the four common motifs are marked by horizontal lines. Accession numbers: TraG (pKM101), I79275; TrwD(R388), X81123; VirB11 (pTi15955), X06826; VirB11 (pTiC58), P07169; VirB11 (Rhizobium etli), AF176227; VirB11 (Bartonella henselae), AF182718; PtlH (B. pertussis), F47301; VirB11 (Brucella suis), AF141604; RP292 (Rickettsia prowazekii), AJ235271; HP0525 (Helicobacter pylori cag), AE00148; TrbB (RP4), M93696; TrbB (R751), U67194; TrbB (pTiA6NC), P54907; TrbB (Rhizobium sp. NGR234), AE000068; Kbh (pCl1 Chlorobium limicula), U77780; and VirB11 (pXO1 Bacillus anthracis), AF065404.
Materials and Methods
Enzymes and Proteins.
TrbB and HP0525 were purified as described (20). In brief, proteins were overexpressed after isopropyl β-d-thiogalactoside (IPTG) induction in soluble form in Escherichia coli SCS1 (Stratagene) carrying plasmids pMS54 or pWP4760, respectively (20). Brij-35/lysozyme extracts of induced cells were subjected to ammonium sulfate precipitation (60% saturation), and precipitated proteins were separated by chromatography on either heparin-Sepharose (TrbB) or HiTrap Q (HP0525) (Pharmacia). Further purification steps involved chromatography on hydroxyl-apatite (both proteins) and gel filtration on Superose-12 (HP0525 only). Electrophoretic purity of the final fractions (fraction IV) reached 99.8% and 98.3% for TrbB and HP0525, respectively (20). Molecular mass standards for gel filtration, glycerol gradient centrifugation, and SDS-polyacrylamide gel electrophoresis were from either Sigma or Boehringer Mannheim.
Reagents and Buffers.
Nucleoside triphosphates were from Boehringer Mannheim. Glutaraldehyde was obtained from Fluka. Buffer A, used for Superose-6 gel filtration, consisted of 20 mM Tris⋅HCl (pH 7.6), 0.1 M NaCl, 1 mM DTT, and 10% (wt/vol) glycerol. Buffer B contained additionally to buffer A 10 mM MgCl2 and 1 mM ATP. Buffers C and D, used for electron microscopy, were made of 20 mM Tris⋅HCl (pH 7.6), and 20 mM triethanolamine (pH 7.6), respectively, 0.1 M NaCl, 10 mM MgCl2, and 1 mM ATP. Buffer E, used for glycerol gradient centrifugation, consisted of 20 mM Tris⋅HCl (pH 7.6), 0.5 M NaCl, 2 mM DTT, 0.1% Brij-58, and 0.1 mM EDTA. Buffer F contained, additionally to buffer E, 0.5 mM dATP and 5 mM MgCl2. Buffer G was used for chemical crosslinking and contained 20 mM sodium phosphate buffer (pH 7.0), 1 mM DTT, 50 mM NaCl, 50% (vol/vol) glycerol, 10 mM MgCl2, and 0.5 mM dATP.
Superose-6 Gel Filtration.
TrbB (770 μg) and HP0525 (140 μg) were incubated in 200 μl of buffer A or B at 30°C for 10 min and applied to Superose-6 (10 × 300 mm) preequilibrated in buffer A or B. Proteins were eluted at a flow rate of 0.3 ml/min, and 0.5-ml fractions were collected. Fractions were electrophoresed in an SDS/15% polyacrylamide gel. Gels were stained with Serva Blue R.
Electron Microscopy and Image Processing.
Purified proteins (30–50 ng/μl final concentration) were incubated in buffer C (HP0525) or D (TrbB) for 10 min at 30°C. TrbB was fixed with 0.2% glutaraldehyde for 5 min. The material for electron microscopy was prepared by negative staining with 1% uranyl formate (TrbB) or 1% uranyl acetate (HP0525) (21) and observed in a Phillips CM100 electron microscope. Pictures were taken on Agfa SCIENTIA 23D56 sheets at ×73,000 magnification and 100 kV. The micrographs were digitized in an Eikonix charge-coupled device camera (model 1412) with a pixel size of 3.1 Å.
Image processing techniques available in the Xmipp software package (22) were used. Single-particle images were selected and extracted from the micrographs. Averaging of these noisy images leads to an increase in the signal-to-noise ratio that provides more detailed information on the structure of these macromolecules. To obtain a meaningful average image, the particles selected must be averaged in the same position and orientation. Thus, they were translationally and rotationally aligned by using cross-correlation methods and the reference-free pspc algorithm (23).
In all cases, the achieved resolution was evaluated by the spectra signal-to-noise ratio method (24) with the threshold set at 4. The average images were then low-pass filtered according to their respective resolution.
Results
Physical Properties of RP4 TrbB and cag HP0525.
Analysis of the purified TrbB and HP0525 protein by SDS/15% polyacrylamide gel electrophoresis indicated molecular masses of 32 kDa and 35 kDa, respectively (20). These values for the denatured and reduced proteins are in close agreement with the calculated value for TrbB of 34,995 Da and for HP0525 of 37,580 Da predicted from DNA sequence analysis. N-terminal sequence analysis of the purified proteins confirmed the amino acid sequence deduced from the nucleotide sequence.
The native molecular mass and friction coefficient of TrbB and HP0525 were calculated by using the Stokes radius determined from Superose-6 gel filtration and the sedimentation coefficient determined from glycerol gradient centrifugation (ref. 25; Table 1). Both proteins were incubated with or without dATP and Mg2+ to demonstrate their influence on oligomer formation. dATP was chosen because TrbB displayed its highest NTPase activity with dATP (20).
Table 1.
Molecular parameters of RP4 TrbB and Cag HP0525
| NTPase | Stokes radius, Å | Sedimentation coefficient, 10−13 s | Molecular mass, kDa | f/f0 |
|---|---|---|---|---|
| TrbB | ||||
| −dATP | 50 | 6.8 | 140 | 1.38 |
| +dATP | 55 | 9.7 | 220 | 1.46 |
| HP0525 | ||||
| −dATP | 52 | 9.7 | 208 | 1.52 |
| +dATP | 52 | 9.7 | 208 | 1.52 |
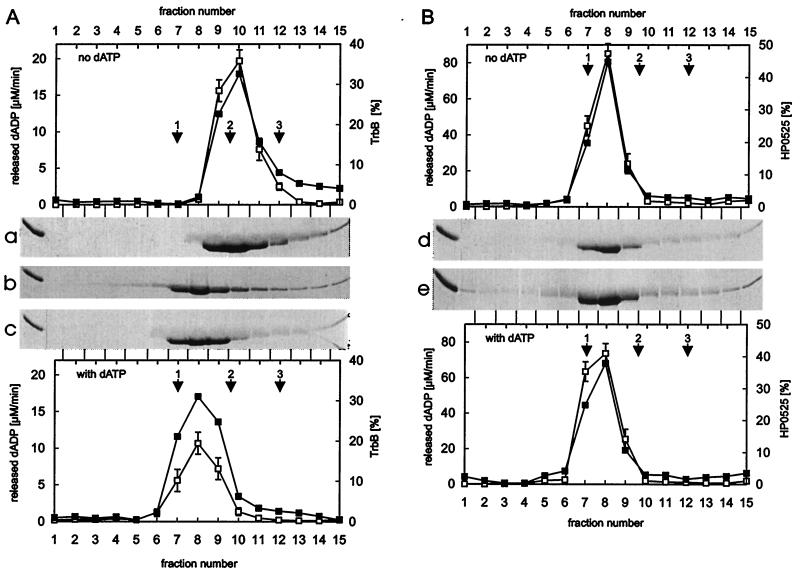
Combining data obtained from Superose-6 gel filtration and glycerol gradient centrifugation, native molecular masses of 220 kDa for TrbB and 208 kDa for HP0525 were calculated in the presence of dATP and Mg2+. Both results are consistent with TrbB and HP0525 behaving as hexamers of 35 kDa and 38 kDa subunits, respectively. In the absence of dATP and Mg2+, TrbB displayed the molecular mass of a tetramer, but smaller species were also evident (Fig. 2a) whereas HP0525 still behaved as a hexamer (Fig. 2d).
Figure 2.
Glycerol gradient centrifugation of TrbB and HP0525. Purified TrbB (A) [fraction IV, 150 μl, 435 μg; (20)] and purified HP0525 (B) [fraction IV, 150 μl, 435 μg; (20)] were laid on a 3.7-ml, 15–35% (wt/vol) linear glycerol gradient in buffer E or F. Centrifugation was at 270,000 × g for 15 h at 4°C. Before centrifugation, TrbB and HP0525 were incubated in buffer F for 10 min at 30°C. Fractions were tested for dATPase activity as published elsewhere (20, 30). Aliquots were electrophoresed in SDS/15% polyacrylamide gels. Gels were stained with Serva Blue R and scanned in the Personal Densitometer (Molecular Dynamics). The amount of protein was quantified by using the imagequant software. The stained gels of the glycerol gradient centrifugation in the absence of dATP and Mg2+ are shown in a (TrbB) and d (HP0525). Corresponding graphs are shown above the gels. Gels c (TrbB) and e (HP0525) are from glycerol gradient centrifugation in the presence of dATP and Mg2+. Corresponding graphs are shown below the gels. TrbB was also incubated with dADP and Mg2+, and glycerol gradient centrifugation was carried out in the presence of dADP and Mg2+ (b). In all graphs shown, open squares represent the ATPase activity of the fractions, filled squares, the amount of protein. Scales are given on the left and right hand sides of the graphs. Catalase (1: 240 kDa, s20,w = 11.3 S), aldolase (2: 185 kDa, s20,w = 7.8 S), and BSA (3: 68 kDa, s20,w = 4.4 S) were run in parallel as standards and marked by arrows.
Oligomers of TrbB and of HP0525, formed in the presence of dATP and Mg2+, sedimented with equal velocity in the glycerol gradient. The molecular mass of the TrbB monomer is smaller than that of HP0525 (35 kDa vs. 38 kDa). According to the results of gel filtration (TrbB)6, experiments, the Stokes radius of (HP0525)6 is smaller than that of (TrbB)6, indicating that (TrbB)6 and (HP0525)6 differ in their compactness. The observed compactness is also in agreement with the different frictional ratios and suggests that TrbB and HP0525 vary in their global shape.
RP4 TrbB and cag HP0525 Form Hexamers.
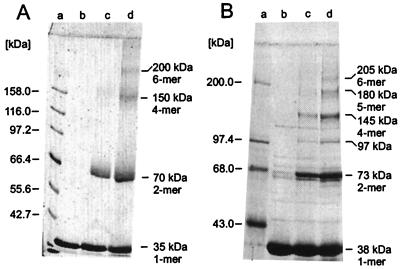
Four distinct polypeptide species of TrbB were obtained upon crosslinking the purified protein with glutaraldehyde, representing the monomer, dimer, tetramer, and hexamer of the protomer mass. This finding suggests that the TrbB hexamer might be composed as a trimer of dimers (Fig. 3A). For HP0525, six distinct polypeptide species with a molecular mass equal to integer multiples of the protomer mass were observed when glutaraldehyde crosslinking was applied to purified HP0525 (Fig. 3B). However, the polypeptide species corresponding to the trimer was underrepresented and differed from the theoretical mass by 17 kDa. In both cases, dimers were the most abundant species.
Figure 3.
Covalent crosslinking of TrbB and HP0525 subunits. TrbB protein (5 μM) and 15 μM HP0525, respectively, were incubated with various concentrations of glutaraldehyde in buffer G at 25°C for 90 min. Reactions were stopped with 3.5 M urea (final concentration). Denatured samples were electrophoresed in an SDS/10% polyacrylamide gel. Gels were stained with Serva Blue R. (A) Crosslinking of TrbB. Lanes: a, molecular mass markers; b, no glutaraldehyde; c, 100 μM; and d, 300 μM glutaraldehyde, respectively. (B) Crosslinking of HP0525. Lanes: a, molecular mass markers; b, no glutaraldehyde; c, 650 μM; and d, 1,300 μM glutaraldehyde, respectively.
The Hexamers of RP4 TrbB and cag HP0525 Are Ring Shaped.
To further investigate the architecture of TrbB and HP0525, electron microscopy was employed. When the proteins were incubated in buffer containing ATP and Mg2+ before examination by electron microscopy, both produced circular species with a central region where the staining agent penetrates (Fig. 4 a and b). TrbB structures proved to be less stable than the HP0525 ones, because a considerable amount of disassembled protein was present along with the ring-shaped molecules when glutaraldehyde fixation was not used (data not shown). In the absence of ATP and Mg2+, relatively few hexameric rings were detected for TrbB, indicating that ATP stabilizes the hexameric assembly (data not shown). TrbB was incubated with other nucleotides and Mg2+ and examined by electron microscopy to determine which nucleotides stabilize the hexameric rings. ATP, dATP, AMP, ATPγS, UTP, ADP, AMP-PNP, AMP-PCP, and GTP could stabilize the ring structure whereas no ring stabilization was observed with CTP or PPi (data not shown). These results demonstrate also that hydrolysis of nucleotides is not essential for the stabilization of TrbB hexameric rings. This finding is underlined by a glycerol gradient centrifugation experiment where TrbB behaved as a hexamer in the presence of dADP and Mg2+ (Fig. 2b).
Figure 4.
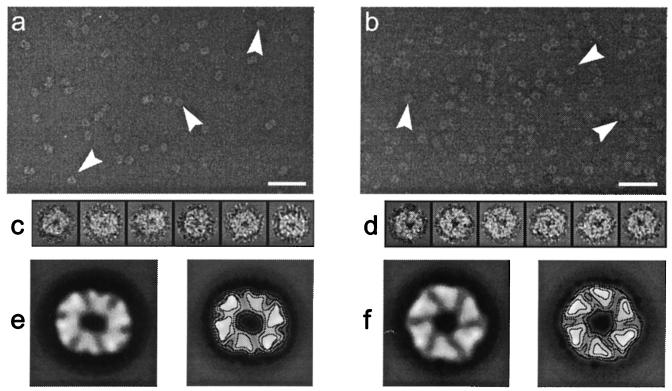
Typical electron micrographs of negatively stained TrbB (a) and HP0525 (b) samples in the presence of Mg2+ and ATP. The arrrowheads point to some characteristic particles selected from these fields. The bar represents 50 nm. Two galleries with the type of images that were used in the two-dimensional analysis are shown in c (TrbB) and d (HP0525). The global average images are shown in e (TrbB) and f (HP0525) and have been low-pass filtered to their final resolution (14 Å in both cases). The size of the frames of each image in the galleries and for the final average images is 20.0 × 20.0 nm.
As already indicated by glycerol gradient centrifugation and gel filtration, hexameric rings of HP0525 were detectable on electron micrographs in equal quality no matter whether NTP and Mg2+ were present or not (data not shown). Both physical methods suggested tetramer formation together with smaller species for TrbB in the absence of NTP and Mg2+. Under these conditions, heterogeneously sized particles with no obvious regularity were detectable on electron micrographs (data not shown).
RP4 TrbB and cag HP0525 Hexamers Consist of Triangular Monomers and Show a Defined Handedness.
Because rings of either TrbB or HP0525 were as stable with ATP as with dATP, both were incubated in buffer containing ATP and Mg2+ to take electron micrographs useful for image processing. A total amount of 6,773 images of TrbB and 4,855 images of HP0525 were extracted from the micrographs, constituting the initial sets (Fig. 4 c and d). Some general features are shared by TrbB and HP0525 hexamers, judging from the average images obtained (Fig. 4 e and f). In both cases, the average image is formed by six well defined pieces of mass, which is in full agreement with the predicted hexameric state of TrbB and HP0525 oligomers based on the previous physical studies and chemical crosslinking. Thus, the six pieces of mass could be directly related to monomers surrounding a central low-density region (around 3 nm in diameter), which is most likely a channel traversing the hexamer. The outer diameter for both average images is about 12 nm. The projection from the putative monomers renders triangular high-density areas, something that is more noticeable in the HP0525 average image (Fig. 4f). Another interesting characteristic of the two average images is that they possess a defined handedness.
Triangular pieces of HP0525 formed an almost perfect regular hexagon that seems to reflect a C6 macromolecular architecture (Fig. 4f). In contrast, the TrbB average image is elliptical, without a clear hexagonal profile but showing strong twofold rotational symmetry (Fig. 4e). Very similar average images where obtained with three independent preparations of TrbB and HP0525.
Discussion
Proteins related to RP4 TrbB and H. pylori cag HP0525 are found in a broad variety of organisms including Gram-negative and Gram-positive eubacteria and archaea (Fig. 1, ref. 16). Most of them are known to be part of complex machineries mediating the transport of macromolecules across the bacterial cell envelope, in some cases also across the cell wall of a bacterial or a eukaryotic recipient. Because of the homology of these proteins to nucleotide-binding proteins and the demonstrated influence of nucleotides on the proteins' properties, it seems likely that TrbB and HP0525 either directly energize the transport reaction or play a role in the assembly of the protein and/or DNA transport machinery.
Reported experimental evidence suggests that members of the VirB11 family form oligomers. Negative dominant effects of mutants of R388 TrwD (16) and Agrobacterium tumefaciens VirB11 (18, 26, 27) indicate that these proteins are functioning as multimers and/or that they are interacting with other components of the secretion machinery. A TrbB mutant in the Walker A box (K161A) also showed a transdominant effect (20). Here we report direct evidence that members of the VirB11 family indeed form oligomers because TrbB and HP0525 are hexameric rings.
Electron microscopy and two-dimensional image processing provide a direct insight into these hexameric structures, which share multiple features: they both exhibit six high-density triangular areas around a central channel of about 3 nm in diameter. The triangular pieces of mass are arranged in such a way that the averages have a defined handedness, implying that there is a preferred orientation for the particles on the carbon grid; otherwise, averaging of single images with opposite handedness would have led to an average image lacking handedness. Therefore, indirect evidence is provided that the two faces of the hexameric assembly differ significantly, accounting for the observed preference in orientation.
Despite their similar characteristics, the two average images are different in one aspect: whereas the HP0525 average shows a hexagonal macromolecule with a sixfold rotational symmetry and an apparent C6 architecture, TrbB average image exhibits strong twofold symmetry and elliptical shape. This symmetry is not expected for a homohexamer such as TrbB and contrasts with the structure of the closely related protein HP0525. Although an internal organization of the TrbB hexamer in two identical trimers could be proposed, the presence of only tetrameric and hexameric species in the crosslinking experiments makes this possibility very unlikely. Actually, TrbB could be a particle, similar to HP0525, whose top view is hexagonal but with a tendency to incline when it adsorbs to the carbon support. This inclination would change the appearance of the specimen in the micrographs, resulting in shortening of projection dimensions perpendicular to the inclination axis. This effect would create an ellipsoidal projection image from a nonellipsoidal specimen. The initial set of TrbB images would then be composed of a range of views, rather than a single top view of the TrbB hexamer. Averaging of such a heterogeneous set of views would yield an average image as the one obtained: elliptic and with a blurred hexagonal profile. Thus, the length of this axis of the hexagonal TrbB average (around 12 nm) would account for the diameter of the hexamer. Interestingly, this length coincides with the diameter of the HP0525 average image.
The orientational preferences of a particle are the result of a number of interrelated factors, including different features of the particle itself, such as the surface charge and topology (28). Therefore, there might be no obvious reason why TrbB has such orientational freedom whereas HP0525 shows a marked preference for a top-view orientation. However, gel filtration and glycerol gradient centrifugation suggested that TrbB is different from HP0525 in its global shape, which might confer to TrbB the tendency to incline on the carbon grid.
Another difference of TrbB and HP0525 is that the hexameric ring structure of TrbB is stabilized by nucleotides, whereas HP0525 hexamers are stable without nucleotides. A TrbB mutant protein in the Walker A Box (K161A) was still able to form hexamers in the presence of ATP and Mg2+ but lacked dATP, ATP, and GTP hydrolyzing activity, indicating that the binding and not the hydrolysis of NTPs stabilizes the ring structure (20).
Ring-like structured proteins are very prominent in nature, being able to accommodate a large range of activities and functions. Chaperones (e.g., GroEL), proteases (e.g., ClpP), replicative helicases (e.g., DnaB), the bacterial light-harvesting complex LH1–LH2, the F1-ATPase, and the regulatory protein of the cell cycle CksHs2 were shown to form ring-like structures (29). What are the features of such toroidal oligomers? A toroid separates a compartment or a channel with distinct chemical properties for catalytic activity. The oligomeric state of this structure provides a sequential arrangement of identical catalytic or binding sites, allowing successive binding and release of a substrate or a repeating catalytic activity against the substrate. According to the hexameric ring structure, TrbB and HP0525 might aid or catalyze a repetitive step/process. Successive rounds of NTP hydrolysis might support translocation of cognate substrate(s) across the inner membrane. The defined handedness of the TrbB and HP0525 images demonstrates that the hexameric rings have two sides with different properties. Thus, it may be assumed that these proteins form a channel that faces the hydrophilic cytoplasm on one and the hydrophobic membrane on the other side. Because TrbB is involved in the assembly of the membrane-spanning Mpf complex, components of this complex could be a substrate for TrbB. Alternatively, the transferred single DNA strand could also be considered as a substrate for TrbB. However, the latter possibility seems much less likely because PulE-like proteins are found in several secretion systems that are not involved in DNA transfer but solely in protein secretion.
Acknowledgments
We are grateful to Beth Traxler for critical reading of the manuscript and stimulating discussions. Hans Lehrach is acknowledged by S.K. and E.L. for generous support. The expert technical assistance of Marianne Schlicht was greatly appreciated. The work of S.K. was financially supported by Sonderforschungsbereich Grant 344/A8 of the Deutsche Forschungsgesellschaft. The research of M.B. and J.M.C. was supported by the Spanish Government Agency Comision Interministerial de Ciencia y Tecnologia through project BIO-98-0761. M.B. is recipient of a Formación de Personal Investigador postgraduate fellowship. The work of W.P. was financially supported by Biotech Grant ERB4001GT963065 of the European Union. W.P. thanks Paul Hooykaas for generous support and stimulating discussions.
Abbreviations
- Mpf
mating pair formation
- Inc
incompatibility
- Tra
transfer
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.050578697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.050578697
References
- 1.Pansegrau W, Lanka E. Prog Nucleic Acid Res Mol Biol. 1996;54:197–251. doi: 10.1016/s0079-6603(08)60364-5. [DOI] [PubMed] [Google Scholar]
- 2.Lessl M, Balzer D, Lurz R, Waters V L, Guiney D G, Lanka E. J Bacteriol. 1992;174:2493–2500. doi: 10.1128/jb.174.8.2493-2500.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lessl M, Balzer D, Pansegrau W, Lanka E. J Biol Chem. 1992;267:20471–20480. [PubMed] [Google Scholar]
- 4.Pansegrau W, Lanka E, Barth P T, Figurski D H, Guiney D G, Haas D, Helinski D R, Schwab H, Stanisich V A, Thomas C M. J Mol Biol. 1994;239:623–663. doi: 10.1006/jmbi.1994.1404. [DOI] [PubMed] [Google Scholar]
- 5.Pansegrau W, Schoumacher F, Hohn B, Lanka E. Proc Natl Acad Sci USA. 1993;90:11538–11542. doi: 10.1073/pnas.90.24.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lessl M, Lanka E. Cell. 1994;77:321–324. doi: 10.1016/0092-8674(94)90146-5. [DOI] [PubMed] [Google Scholar]
- 7.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, et al. Nature (London) 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 8.Vogel P J, Andrews H L, Wong S K, Isberg R R. Science. 1998;279:873–875. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- 9.Andersson S G E, Zomorodipour A, Andersson J O, Sicheritz-Pontén T, Alsmark U C M, Podowski R M, Kurland C G. Nature (London) 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 10.O'Callaghan D, Cazevielle C, Allardet-Servent A, Boschiroli M L, Bourg G, Foulongne P, Frutos P, Kulakov Y, Ramuz M. Mol Microbiol. 1999;33:1210–1220. doi: 10.1046/j.1365-2958.1999.01569.x. [DOI] [PubMed] [Google Scholar]
- 11.Salmond G P C. Annu Rev Phytopathol. 1994;32:181–200. [Google Scholar]
- 12.He S Y. Trends Microbiol. 1997;5:489–495. doi: 10.1016/S0966-842X(97)01163-3. [DOI] [PubMed] [Google Scholar]
- 13.Llosa M, Zambryski P. Trends Microbiol. 1998;6:98–99. doi: 10.1016/s0966-842x(98)01222-0. [DOI] [PubMed] [Google Scholar]
- 14.Pugsley A P. Mol Microbiol. 1993;9:295–308. doi: 10.1111/j.1365-2958.1993.tb01691.x. [DOI] [PubMed] [Google Scholar]
- 15.Hobbs M, Mattick J S. Mol Microbiol. 1993;10:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 16.Rivas S, Bolland S, Cabezón E, Goni F M, de la Cruz F. J Biol Chem. 1997;272:25583–25590. doi: 10.1074/jbc.272.41.25583. [DOI] [PubMed] [Google Scholar]
- 17.Grahn, A. M., Haase, J., Bamford, D. H. & Lanka, E. (2000), J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 18.Rashkova S, Spudich G M, Christie P J. J Bacteriol. 1998;179:583–591. doi: 10.1128/jb.179.3.583-591.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christie P J, Ward J E, Jr, Gordon M P, Nester E W. Proc Natl Acad Sci USA. 1989;86:9677–9681. doi: 10.1073/pnas.86.24.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krause, S., Pansegrau, W., Lurz, R., de la Cruz, F. & Lanka, E. (2000), J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 21.Steven A C, Trus B L, Maizel J V, Unser M, Parry D A D, Wall J S, Hainfeld J F, Studier F W. J Mol Biol. 1988;200:351–365. doi: 10.1016/0022-2836(88)90246-x. [DOI] [PubMed] [Google Scholar]
- 22.Marabini R, Masegosa I M, San Martín M C, Marco S, Fernández J J, de la Fraga L G, Vaquerizo C, Carazo J M. J Struct Biol. 1996;116:237–240. doi: 10.1006/jsbi.1996.0036. [DOI] [PubMed] [Google Scholar]
- 23.Marco S, Chagoyen M, de la Fraga L G, Carazo J M, Carrascosa J L. Ultramicroscopy. 1996;66:5–10. [Google Scholar]
- 24.Unser M, Trus B L, Steven A C. Ultramicroscopy. 1987;23:39–53. doi: 10.1016/0304-3991(87)90225-7. [DOI] [PubMed] [Google Scholar]
- 25.Siegel L M, Monty K J. Biochim Biophys Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- 26.Stephens K M, Roush C, Nester E. J Bacteriol. 1995;177:27–36. doi: 10.1128/jb.177.1.27-36.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou X-R, Christie P J. J Bacteriol. 1997;179:5835–5842. doi: 10.1128/jb.179.18.5835-5842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies. San Diego: Academic; 1996. pp. 54–67. [Google Scholar]
- 29.Hingorani M M, O'Donnell M. Curr Biol. 1998;8:R83–R86. doi: 10.1016/s0960-9822(98)70052-1. [DOI] [PubMed] [Google Scholar]
- 30.Scherzinger E, Ziegelin G, Bárcena M, Carazo J M, Lurz R, Lanka E. J Biol Chem. 1997;272:30228–30236. doi: 10.1074/jbc.272.48.30228. [DOI] [PubMed] [Google Scholar]