Abstract
Hepatitis C virus (HCV) infection is thought to mostly become chronic and rarely resolves. HCV infection was serologically screened in 4,984 samples from Ghanaian blood donors, and 1.3% prevalence was found. At least 53% of confirmed anti-HCV carriers had no detectable viral RNA and were considered to have cleared the virus and recovered from the infection. Confirmation was authenticated by the presence of antibodies specific to at least two viral antigens, mostly NS3 and E2. Reactivity to HCV core antigens was lower in Ghanaian than United Kingdom blood donors. The minority of chronically infected donors carried a viral load significantly lower than an unselected comparative group of United Kingdom blood donors (2.5 × 105 versus 2.9 × 106 IU/ml; P = 0.004). HCV genotype 2 was largely predominant (87%). Sequence clustering was similarly broad in the E1/E2 and NS5 regions. The phylogenetic diversity and the incapacity to distinguish subtypes within genotype 2 in our and others' West African strains suggested that West Africa may be the origin of HCV genotype 2. The genetic diversity extended to the identification of strains clearly separated from known subtypes of genotype 2 and genotype 1. One strain appears to be part of a new HCV genotype. HCV infection in Ghana is characterized by a high rate of recovery and the predominance of broadly divergent genotype 2 strains.
Hepatitis C virus (HCV) is the major etiological agent of posttransfusion non-A, non-B hepatitis. According to World Health Organization (WHO) estimates, approximately 3% of the world population may be infected with HCV (20). The prevalence of HCV infection varies widely according to the location and the population studied (28). In sub-Saharan Africa, HCV prevalence has been reported to be less than 1% in southern African countries (43, 45) and to range between 1.7 and 27.5% in central Africa (5, 25, 29) and between 1.4 and 7% in West and East Africa (1, 10, 36, 39). The variations observed between studies appear related not only to the heterogeneity of the populations investigated but also to the methods used to detect HCV infection (36). More population-based studies using highly sensitive and specific assays are necessary to evaluate the exact magnitude of HCV infection in sub-Saharan Africa.
After an initial exposure to HCV, infection may resolve or evolve to chronic infection, resulting in a variety of outcomes ranging from no symptoms to end-stage liver disease (15, 41). Studies performed in Western and Far Eastern countries showed that about 80% of the HCV infections evolve to chronic infection (15, 41). However, considering that primary infection is predominantly asymptomatic and that antibodies become undetectable over months or years in a proportion of those who spontaneously clear the virus (37), the infection recovery rate may be underestimated. A few recent studies from East Asia and sub-Saharan Africa involving a limited number of patients reported recovery rate ranging between 30 and 89% (17, 36, 38, 43, 45). The nature and the relative importance of the host and viral factors determining the outcome of HCV infection are not well understood. Host factors that may play a role include cellular immunity (40, 49) and host genetic determinants (7, 12). Viral factors include genetic heterogeneity (14), viral load (46), and possibly genotype (3, 17), although this last factor remains controversial (50).
Genetic variants of HCV have been classified into six phylogenetically distinct genotypes, each containing multiple subtypes (33). There is a marked difference in the distribution of the genotypes and subtypes worldwide. The geographic distribution and diversity of HCV genotypes may provide important indications about the origin of HCV (35). In addition, the identification of HCV genotypes and subtypes may have implications in the efficacy of diagnostic assays. In West Africa, preliminary results suggest a predominance of genotype 2.
This study was designed to determine the ratio between HCV chronic infection and recovery in samples from blood donors in Kumasi, Ghana. In studying viral strains from these individuals, new aspects of the molecular distribution of HCV in West Africa emerged.
MATERIALS AND METHODS
Samples.
Serum or plasma samples from 4,984 blood donors were collected and screened for anti-HCV by enzyme immunoassay (EIA) at the Komfo Anokye Teaching Hospital blood bank in Kumasi, Ghana. Reactive samples were stored at −20°C and shipped in dry ice to the Laboratory of Molecular Virology, Division of Transfusion Medicine, Cambridge, United Kingdom, to confirm the presence of anti-HCV and to screen for HCV RNA (36). Serological and molecular investigations were often limited by the volume of plasma sample available (1 to 1.5 ml). This study was approved by the University of Science and Technology School of Medical Sciences committee on human research publication and ethics, Kumasi, Ghana. For comparison, samples from a study of HCV and human immunodeficiency virus (HIV) infection in 50,000 first-time blood donors conducted in the United Kingdom and previously published (8) were used.
Serological screening.
Samples reactive with Murex anti-HCV version 4.0 EIA (Murex Biotech SA Ltd, Kyalami, South Africa) were retested, and repeatable reactive samples were tested with a second anti-HCV EIA from SANOFI (SANOFI, Marnes la Coquette, France). Both EIAs were performed according to the manufacturers' instructions.
Reactivity with two independent locally performed EIAs defined confirmed positivity, but samples were subsequently retested with a third-generation recombinant immunoblot assay (RIBA HCV 3.0 SIA; Chiron, Emeryville, Calif.) at the Laboratory of Molecular Virology (Division of Transfusion Medicine, University of Cambridge, Cambridge, United Kingdom) according to the manufacturer's instructions. This assay is based on two recombinant proteins (NS5 Ag and c33c Ag: NS3) and three synthetic peptides (c22p from core and c100p/5-1-1p from NS4) immobilized as four individual bands onto test strips. Reactivity to at least two of the four HCV antigens was considered a positive result; no reactivity was considered a negative result, and reactivity to only one antigen was considered an indeterminate result.
A second confirmatory test was performed on anti-HCV-positive samples using a Murex prototype HCV confirmatory assay (Murex Biotech, Dartford, United Kingdom). The assay is a line immunoassay in which purified recombinant antigens or peptides representing the core, envelope, NS3, NS4, and NS5 regions are immobilized onto a supported nitrocellulose membrane in discrete bands. The immobilized antigens are from HCV serotypes 1, 2, and 3. In addition, three control lines are present: an anti-human band to act as a sample addition monitor and two human immunoglobulin G (IgG) lines set at 1+ positive and 3+ positive to aid in the scoring of the strips by eye. Criteria for positive, indeterminate, or negative result are as for the RIBA 3.0 assays.
Additional EIA testing.
Plasmas were screened for anti-core and anti-NS3 reactivity by EIA using three core peptides and two NS3 recombinant proteins mixtures, respectively. The core peptides were designated C1 (MSTNPKPQRKTKRNTNRR-C), C2 (KTKRNTNRRPQDVKF-C), and C3 (GQIVGGVYLLPRRGP-C) and synthesized from HCVJK1G genotype 1b (18). The NS3 recombinant proteins 00117-V (amino acids 1450 to 1643) and 00118-V (amino acids 1209 to 1643) were obtained from ViroGen (ViroGen, Watertown, Mass.). 00117-V and 00118-V NS3 antigens are derived from a HCV genotype 1b strain and from a Russian HCV strain not genotyped, respectively. Briefly, 100 μl of the core peptide mixture (10 μg of each/ml) or the two recombinant NS3 protein mixture (1 μg of each/ml) in sodium carbonate (pH 9.6) was added to a Nunc-Immuno microtiter plate (Maxisorp, Nalge nunc International) and incubated overnight at 4°C. The EIA procedure was then carried out as previously described (18). Levels of antibody reactivity were expressed as a sample/cutoff ratio (S/CO). The cutoff was calculated for each individual plasma as the mean of the absorbance values of three non-HCV peptides plus six standard deviations.
In order to limit background, IgGs were purified from the African plasmas by affinity chromatography with protein G Sepharose using the MabTrap kit (Amersham Pharmacia Biotech AB, Uppsala, Sweden) and then tested for anti-HCV core and NS3 reactivity. Protein concentration of the column eluate was determined in each fraction using the BCA Protein assay (Pierce, Rockford, Ill.) according to the manufacturer's instructions.
HCV RNA testing.
HCV RNA was screened using the transcription-mediated amplification (TMA)-based HIV/HCV RNA multiplex assay (Procleix HIV-1/HCV assay, Gen-Probe Inc., San Diego, Calif., and Chiron Blood Testing, Emeryville, Calif.) according to the manufacturer's instructions, as previously described (8).
Reverse transcription (RT) nested PCR for HCV RNA.
Viral RNA was extracted from 140 μl of plasma with a QIAamp Viral RNA Mini Kit (Qiagen Ltd, Crawley, United Kingdom) according to the manufacturer's instructions.
RT was carried out in a final volume of 20 μl containing 50 units of murine leukemia virus reverse transcriptase (Applied Biosystems, Warrington, United Kingdom), 20 units of RNasin (Promega, Southampton, United Kingdom), 0.08 units of random hexamers, and 1 mM each deoxynucleotide triphosphate (dNTP). The mixture was incubated at 42°C for 60 min and 95°C for 5 min.
The E1/E2 region of the HCV genome was amplified using nested PCR as previously described (26). Part of the NS5B region was amplified using two different assays. First, primer pairs HCV243-HCV242 and HCV554-HCV555 were used as described (9). When no amplified product was obtained, a second nested PCR was performed using the outer primers JT1 (5′-GGGTTTGGGGCNAAGGAGGT-3′, 5′ position 7970) and JT2 (5′-CTCTCAGGTTYCGCTCGTCC-3′, 5′ position 8678) and the inner primers JT3 (5′-CCAAAAATGAGGTGTTYTG-3′, 5′ position 8085) and JT4 (5′-TGGCTYTCTGAGATGACRACC-3′, 5′ position 8646). Primer positions are relative to the sequence of the HCV strain J6 (GenBank accession no. D00944). Briefly, first-round PCR was performed in 50 μl containing 2.25 mM MgCl2, 0.2 mM dNTPs, 1 μM each primer, 2.5 units of Amplitaq (Applied Biosystems), and 5 μl of cDNA. The same conditions were used in the second-round PCR, but the reaction was performed in 20 μl containing 4 μl of first round-amplified product. The cycling profile was identical for the two rounds and consisted of a predenaturation at 95°C for 5 min followed by 30 cycles at 95°C for 30s, 58°C for 1 min, and 72°C for 1 min. A final extension was performed at 72°C for 10 min.
HCV RNA quantification.
Viral RNA was quantified using the Mx4000 Multiplex Quantitative PCR System (Stratagene, La Jolla, Calif.). The sequences of the primers MAD-1 (5′-TGCTAGCCGAGTAGYGTTGG-3′) and MAD-2 (5′-ACTCGCAAGCACCCTATCAG-3′) and the probe MAD-3 (5′-ACCACAAGGCCTTTCGCGAC-3′) were designed from the conserved 5′ untranslated region (UTR) of the HCV genome. The fluorogenic probe was 5′ labeled with FAM (6-carboxyfluorescein) and 3′ labeled with TAMRA (6-carboxy-N-tetramethylrhodamine). Amplification was performed with the Brilliant two-step quantitative RT-PCR core reagent kit (Stratagene, La Jolla, Calif.). An RT reaction mixture contained 1× core RT buffer, 2 mM each dNTP, 2 μM MAD-2 primer, 20 units of StrataScript reverse transcriptase, 20 units of RNasin, and 10 μl of template RNA preparation per 20-μl reaction. The reaction was carried out at 25°C for 5 min, 45°C for 60 min, and 95°C for 3 min. The quantitative PCR mixture contained 1× core PCR buffer, 5 mM MgCl2, 0.8 mM each dNTPs, 1.8 μM each primers, 0.2 μM fluorogenic probe, 3.0 × 10−4 mM reference dye (carboxy-X-rhodamine; ROX), 2.5 U of SureStart Taq polymerase, and 10 μl of template cDNA preparation per 50-μl reaction. After an initial incubation at 95°C for 10 min, fifty two-step cycles of 1 min at 60°C and 30 s at 95°C were carried out. For each run, duplicates of a tenfold serial dilution of WHO International Standard for HCV RNA for NAT assays 96 and 790 (NIBSC, Potters Bar, United Kingdom) containing 5 to 5,000 IU of HCV genome per RT reaction were analyzed. The sensitivity of the method was estimated using the WHO standard 96/790 and Probit software at 150 IU/ml.
HCV genotyping.
E1/E2 and NS5B amplicons were purified in a 1.5% agarose gel using the QIAquick gel extraction kit (Qiagen Ltd), and 10 ng of purified DNA was used in a sequencing reaction. In addition, E1/E2 amplicons were cloned into the TA vector pCR 4-TOPO (Invitrogen Ltd, Paisley, United Kingdom). Following transformation into One Shot TOP10 chemically competent Escherichia coli (Invitrogen Ltd.), clones were isolated using a QIAprep Spin Miniprep kit (Qiagen Ltd.). Recovered plasmid DNA was tested for insert by restriction digest with EcoRI. All assays were performed according to the manufacturers' instructions. Purified amplicons and plasmid DNA were sequenced using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction kit, version 2.0, and the ABI PRISM 3100 genetic analyzer (Applied Biosystems) according to the manufacturer's instructions.
Phylogenetic analyses were performed using the PHYLIP software package and previously genotyped HCV sequences from the GenBank data base as references (genotype 1: GenBank nos. AF009606, AJ132996, D10749, D14853, D90208, M62321, M67463, U1214, and X61596; genotype 2: AB030907, AB031663, AF238484, AF238486, D00944, D10988 and D50409; genotype 3: AF046866, D17763, D28917, D49374, D63821 and X76918; genotype 4: Y11604; genotype 5: AF064490 and Y13184; and genotype 6: D63822, D84262, D84263, D84264, D84265 and Y12083). Briefly, alignments were created using the CLUSTAL X program and edited using the Se Al program, version 2.0. Distance matrices were produced by DNADIST using the Kimura two-parameter setting and further analyzed in NEIGHBOR, using the neighbor-joining setting. The significance of clustering was evaluated by bootstrapping (1,000 replicates) using the programs SEQBOOT and CONSENSE. Bootstrap values of ≥80% were considered significant.
Nucleotide sequence accession numbers.
The GenBank accession numbers of nucleotide sequences analyzed in this study are AY236365 to AY236408.
RESULTS
HCV infection in Ghanaian blood donors.
Samples from 4,984 Ghanaian blood donors were initially tested for HCV antibody, and 150 were reactive (3%). Reactive samples were retested with an alternative anti-HCV EIA, and 63 (1.3%) were reactive with both independent assays. These 63 blood donors were mainly males (82%) and ranged in age between 16 and 53 years (median age, 29 years).
By TMA, 111 of 140 donations (79.3%) were HCV RNA negative and 29 samples (20.7%) contained HCV RNA. Ten samples were not tested due to volume limitation. The 5′ UTR region of HCV was successfully amplified in all the TMA-positive samples, with an independent nested PCR assay used for confirmation (data not shown).
To further assess the presence of antibody to HCV in samples that were seropositive but HCV RNA negative, two anti-HCV confirmatory assays were used, namely, RIBA HCV 3.0 and the Murex prototype HCV confirmatory assay. Forty-four donations with S/CO values of ≥2 in both EIAs were tested (Table 1). For comparison, 15 RNA-positive samples were also tested. Sixty-seven samples were not tested due to volume limitation (12 samples) or an S/CO value of <2 (55 samples). All 15 RNA-positive samples were confirmed with both assays. Using RIBA, 9 HCV RNA-negative donations (20.5%) were confirmed to be anti-HCV positive, whereas 24 donations (54.5%) were confirmed with the Murex assay. Among the Murex-confirmed donations, 13 were RIBA indeterminate and two were RIBA negative. Only one donation positive with two independent screening EIAs was negative with both confirmatory assays.
TABLE 1.
Comparison of two anti-HCV confirmatory assays
| Chiron RIBA HCV 3.0 SIA result | Murex prototype HCV confirmatory assay result
|
Total | ||
|---|---|---|---|---|
| Positive | Indeterminate | Negative | ||
| Positive | 24 | 0 | 0 | 24 (41%) |
| Indeterminate | 13 | 10 | 3 | 26 (44%) |
| Negative | 2 | 6 | 1 | 9 (15%) |
| Total | 39 (66%) | 16 (27%) | 4 (7%) | 59 |
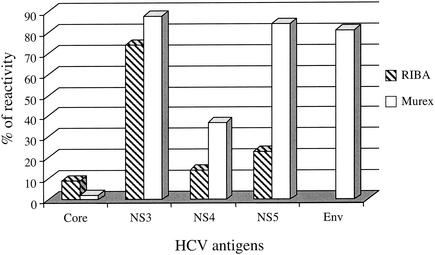
In order to analyze the discrepancies between the RIBA and Murex assays, the individual antigen reactivity was compared in 43 HCV RNA-negative donations (the donation found negative with the two confirmatory assays was excluded from the analysis) (Fig. 1). In both RIBA and Murex assays, a weak reactivity was observed with the core antigen. Four samples (9%) reacted with the core antigen in the RIBA assay, and one reacted in the Murex assay. In contrast, the NS3 antigen showed the highest rate of reactivity in both assays (74% reactivity in RIBA and 88% reactivity in Murex). The Murex assay also showed a higher frequency of reactivity with the NS5 antigen (84% reactive) than with RIBA (23% reactive). In addition, the E2 envelope protein present in the Murex assay, but not in RIBA, was reactive in 35 (81%) of the samples tested. Since a confirmed result with either RIBA or Murex confirmatory assay requires at least two reactive bands, most samples tested were indeterminate with RIBA. In contrast, the presence of two highly reactive antigens in the Murex assay (NS3 and E2) identified most of the samples as confirmed.
FIG. 1.
Antigen reactivity of 43 HCV RNA-negative Ghanaian blood donations in two anti-HCV confirmatory assays.
Among the 19 samples classified indeterminate in either RIBA and/or Murex assays, we hypothesized that some had been truly infected and had lost antibody reactivity over time and others were not infected but displayed false reactivity. To distinguish between the two possibilities, in-house assays developed with core peptides and commercial recombinant NS3 proteins were used. In particular, it was assumed that should reactivity with core peptides be observed, the likelihood of genuine contact with HCV would be considerably increased. Of the 19 samples indeterminate by reactivity to NS3 antigen in the confirmatory assays, 18 also reacted with the recombinant NS3 EIA. In addition, nine of these NS3-reactive samples reacted with the mixture of core peptides derived from the genotype 1a prototype HCV sequence.
If the confirmed antibody-positive rate was extrapolated to include all of the 29 RNA-positive samples, 38 samples (9 RNA-negative samples and 29 RNA-positive samples) were confirmed to be antibody positive by RIBA, giving a recovery rate of 9 of 38, or 23.7%. Using Murex assay in addition to RIBA, the number of confirmed antibody-positive samples increased to 53 (24 RNA-negative and 29 RNA-positive samples) and the recovery rate became 24 of 53, or 45%. According to the rule that confirmation is defined by reactivity with two distinct viral antigens, the nine samples indeterminate with RIBA and/or Murex assay but reactive with recombinant NS3 and core peptide EIAs were included, bringing the number of confirmed seropositive samples to 62 (33 RNA-negative and 29 RNA-positive samples). The percentage of recovery from HCV infection became 33 of 62, or 53%. No significant difference in age or sex was observed between blood donors that were both seropositive and RNA positive and blood donors that were RNA negative, nor was significant difference observed within the RNA-negative group between donors with confirmed or indeterminate serology.
The poor reactivity of Ghanaian samples with the genotype 1a-derived core antigen was further investigated in 38 HCV RNA-containing samples from Ghana (n = 26) or the United Kingdom (n = 12) with a peptide anti-core EIA. The anti-NS3 EIA was used as control. All samples were reactive with the mixture of NS3 recombinant proteins, and the S/CO values were not significantly different between the two groups. In contrast, the United Kingdom samples, relative to the Ghanaian samples, showed a significantly higher rate (92 and 50% reactive, respectively; P = 0.035) and level of reactivity with the core peptides (comparing S/CO values) (P < 0.0001).
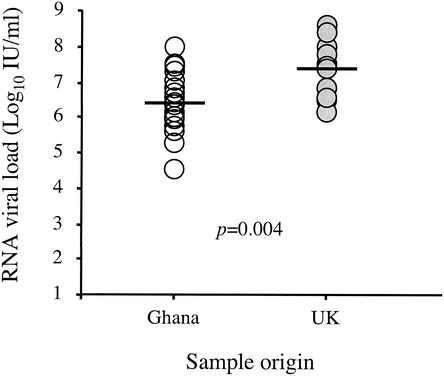
To assess whether such difference of reactivity was correlated to viral load, HCV RNA was quantified in the United Kingdom and Ghanaian donations using real-time quantitative RT-PCR. Six Ghanaian samples were not tested due to plasma volume limitation. As shown in Fig. 2, the HCV RNA load detected in the 20 Ghanaian donations was significantly lower than that in the 12 United Kingdom donations. The median RNA viral load was 2.5 × 105 IU/ml of plasma in the Ghanaian donations (range, 3.3 × 103 to 9.0 × 106 IU/ml) compared to 2.9 × 106 IU/ml in the United Kingdom donations (range, 1.5 ×105 to 4.1 × 107 IU/ml) (P = 0.004). However, no correlation was found between plasma viral load and reactivity with core peptides.
FIG. 2.
RNA viral load in HCV-infected blood donors from Ghana and the United Kingdom. The horizontal bars denote median values.
An alternative explanation for such difference in HCV genotype 1-derived peptides reactivity was a different distribution of HCV genotypes between Ghana and the United Kingdom. Of the 11 United Kingdom samples reactive with the screening mixture of core peptides, six were infected with HCV genotype 1a, two with genotype 1b, one with genotype 3a, and two with genotype 4a (GenBank accession numbers AF488352, AF488360, AF488355, AF488356, AF488364, AF488370, AF488366, AF488375, AF488369, AF488371, and AF488372). The United Kingdom nonreacting donation was infected with HCV genotype 1a. Two United Kingdom donations infected with HCV genotype 1a and 3a showed the lowest anti-core reactivity (S/CO values of 3.19 and 2.64, respectively). Molecular analysis was then performed to characterize the HCV strains infecting Ghanaian blood donors. This explanation, however, is unlikely, since genotypes 1, 2, and 4 share the same consensus sequence for the core peptides used in our assay.
Molecular characterization of Ghanaian HCV strains.
The E1/E2 and NS5B regions from 23 and 21 Ghanaian donations, respectively, were sequenced (GenBank accession numbers, AY236365 to AY236408). Five samples were not tested due to plasma volume limitation. Nucleotide sequences of the E1/E2 (513 bp) and NS5B (271 bp) regions were obtained by direct sequencing, and phylogenetic analysis was performed using 31 reference sequences representative of HCV genotypes 1 to 6.
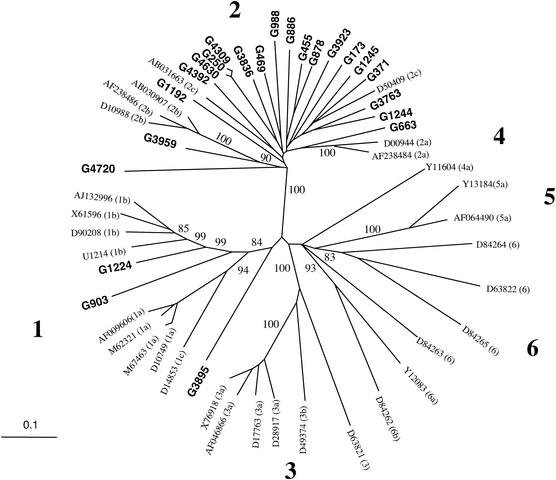
Twenty (87%) of the Ghanaian strains clustered with genotype 2 reference sequences, with bootstrap values of 100% over 1,000 replicates (Fig. 3). However, these strains did not clearly cluster with any of subtypes 2a, 2b, or 2c, nor did they form a distinct cluster within clade 2. Only strain G3959 was clearly related to subtype 2b (bootstrap score, 90%), with 74 to 75% sequence homology with the three 2b reference strains.
FIG. 3.
Phylogenetic relationship between HCV E1/E2 sequences. The 23 Ghanaian sequences were aligned with 31 HCV reference strains from the GenBank data base. The six major lineages of HCV are indicated by the boldface numbers along the branches; reference strains are identified by their GenBank accession numbers, and subtypes of the viruses are also reported in parenthesis (classification according to Salemi and Vandamme [35]). The Ghanaian sequences determined in this study are indicated by boldface characters; only significant bootstrap values (≥80%) are shown.
Strain G4720 clustered as a divergent genotype 2 subtype, but the bootstrap value was too low to draw any firm conclusion. G4720 had 64 to 68% sequence homology with the other Ghanaian strains and the type 2 reference sequences. Sequence homology between all the other Ghanaian and genotype 2 reference sequences ranged between 67 and 84%.
Three strains, G250, G4309, and G4630, showed remarkably similar sequences (sequence homology, 97, 98 and 99%). The possibility of cross-contamination during sample processing was checked and eliminated. Investigations conducted in Ghana established that these three samples were collected from the same donor at different times over a period during which the information regarding HCV status was not available at the time of donation and could not be delivered to the individual.
Two other strains, G1224 and G903, clustered with genotype 1, with bootstrap values of 84% over 1,000 replicates (Fig. 3). G1224 was clearly a divergent 1b subtype, exhibiting 69 to 71% sequence homology with the four 1b subtype references used in this analysis. Sequence homology between the 1b subtype references ranged between 80 and 82%. Strain G903 formed a distinct branch within genotype 1, with the highest sequence homology being with 1b strains but being low (≤67%). Bootstrap values of 99% over 1,000 replicates assigned to this node supported G903 possibly constituting a new subtype of genotype 1. Finally, the Ghanaian strain G3895 was the most divergent, sharing less than 70% E1/E2 sequence homology with all other Ghanaian and reference sequences studied. Strain G3895 did not cluster with any known HCV genotypes with any significant bootstrap values (Fig. 3).
Sequences of 170 amino acids, including the 27-amino-acid-long HVR1 region, were deduced from the PCR consensus sequence for each Ghanaian isolate (data not shown). Within HVR1, 44.7 ± 9% amino acid identity was observed between the Ghanaian strains. Positions 2, 23, and 26 were conserved as threonine, glycine, and glutamine, respectively. Strain G371 had a deletion of one residue at position 4, and strain G663 had an additional tyrosine residue at the end of the E1 protein. The comparison of HVR1 sequences using the Kyte and Doolittle method (http://us.expasy.org/cgi-bin/protscale.pl) indicated very similar hydrophobicity profiles among the HCV strains, regardless of the genotype. Twenty sequences were globally basic, and one (G3895), containing the same number of acidic and basic residues, was globally neutral. The secondary structures of HVR1 sequences were predicted by using the Chou-Fasman, Robson-Garnier, and double prediction methods available at the NPSA website (http://npsa-pbil.ibcp.fr). Common characteristics in the secondary structure were consistently predicted for the HVR1 sequences despite the high amino acid variability displayed (data not shown). In all sequences except G988, β sheet conformations were predicted around positions 15 to 19. A turn was also predicted in the region between positions 20 and 24 in almost all sequences except G4392.
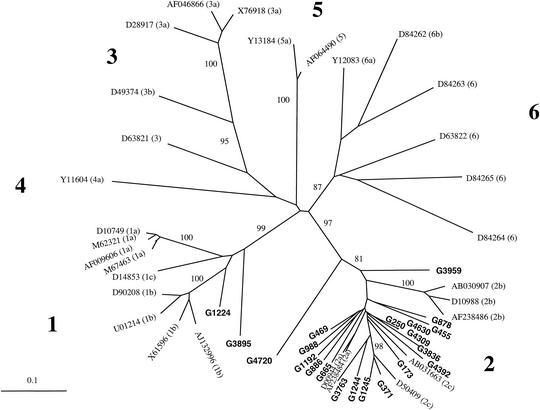
The approach used for the analysis of the E1/E2 sequences was also applied to the NS5B sequences. The phylogenetic tree of the NS5B region confirmed the topology observed in the E1/E2 region analysis, namely, that 18 of 21 (86%) of the Ghanaian sequences clustered with genotype 2, with bootstrap values of 97% on 1,000 replicates (Fig. 4). As observed in the E1/E2 region, the Ghanaian sequences did not form a specific cluster within genotype 2. The only significant cluster consisted of a subtype 2c reference (D50409) and the Ghanaian strains G371 and G1245, supported by bootstrap values of 98%. However, the other type 2c reference strain, AB031663, was not included in that cluster. Again, G4720 was the most divergent isolate within the type 2 group, sharing 72 to 74% sequence homology with the other genotype 2 sequences. All the other type 2 sequences shared 75 to 93% homology. Bootstrap values of 81% over 1,000 replicates supported the individuality of G4720 within genotype 2. The NS5B sequence of G3959 was also distant from the other genotype 2 sequences. Strain G1224 clustered with subtype 1b references (83 to 91% sequence homology) (Fig. 4). However, the relatedness of G1224 with subtype 1b references was not supported by significant bootstrap values. In contrast, the bootstrap value of strain G3895 with genotype 1 was 99% over 1,000 replicates. However, G3895 remained highly divergent from the genotype 1 references. Sequence homology between G3895 and the references ranged from 73 to 80% but ranged from 79 to 97% among the genotype 1 references. No amplified product was obtained for G903 despite several attempts, suggesting the lack of annealing of NS5B-specific PCR primers with G903 sequence.
FIG. 4.
Phylogenetic relationship between HCV NS5B sequences. The 21 Ghanaian sequences were aligned with the corresponding sequences from the 31 HCV reference strains shown in Fig. 3. The results are displayed as described in the legend for Fig. 3.
The deduced NS5B amino acid sequences were aligned with the corresponding sequence of the reference HCV genotype 1a (GenBank accession number, M62321) and used to develop the RIBA HCV 3.0 SIA assay (data not shown). The partial 101-amino-acid-long sequences obtained for the African strains accounted for 17% of the whole NS5B protein but contained three crucial motifs for the RNA-dependent RNA polymerase (RdRp) located at positions 2640 to 2645, 2702 to 2711, and 2737 to 2739 (positions numbered according to M62321 polyprotein). Seventy residues (69%) were conserved among all the Ghanaian sequences irrespective of their genotype. Concerning the RdRp activity-associated motifs, DTRCFD (positions 2640 to 2645) and GDD (positions 2737 to 2739) motifs were conserved in both Ghanaian sequences. However, among the type 2 Ghanaian strains, some variability was observed within the motif SVGLTTSCGN (positions 2702 to 2711). At position 2705, phenylalanine instead of leucine was found in five sequences (25%), and at position 2709, cysteine was replaced by methionine in 18 sequences (90%) and by alanine and leucine in the two remaining sequences. Finally, proline instead of threonine was observed at position 2706 in the G3959 sequence. Residues of glycine (position 2703), threonine (position 2707), and asparagine (position 2711) were conserved in all sequences as previously reported for plus-strand RNA virus polymerases.
Characterization of highly divergent Ghanaian HCV viral strains.
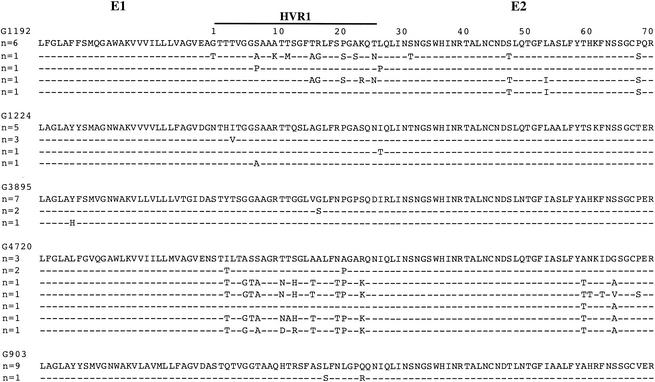
In order to determine whether or not the high sequence divergence observed for strains G4720, G1224, G903, and G3895 corresponded to a single quasispecies and was not the consequence of a complex mixture of multiple infections, the E1/E2 amplified product was cloned. Ten clones from each sample were sequenced and phylogenetically analyzed (Fig. 5). Genotype 2 strain G1192 was also sequenced as control. For each sample tested, sequences were closely related, with sequence homology ranging from 98 to 99% for G1224, from 99 to 100% for G903, from 98 to 100% for G3895, from 95 to 99% for G4720, and from 94 to 100% for G1192. Phylogenetic analysis showed that clone sequences of each isolate clustered together and with the corresponding PCR-derived sequence (data not shown). When the sequences of the clones were included in the E1/E2 phylogenetic analysis, the topology and bootstrap values were similar to those obtained with the PCR-derived consensus sequences shown in Fig. 2. These data confirmed the genomic divergence of the whole quasispecies from these four strains.
FIG. 5.
Alignment of E1/E2 amino acid sequences of five HCV Ghanaian variants. The sequences of ten clones were aligned for each variant, and the number of identical sequences is indicated on the left; for each variant, the major sequence was taken as reference. Residues identical to the major sequence are indicated by dashes. To facilitate the viewing, the HVR1 region is indicated; numbering of the residues starts at the NH2 terminus of E2.
To further support the uniqueness of two of these four strains, a 1.8-kb-long region of HCV genome encoding the 5′ UTR and core, E1, and part of E2 regions of strains G903 and G4720 was amplified and sequenced (GenBank accession numbers, AY236365 and AY236366). Phylogenetic analyses confirmed the results obtained in the E1/E2 region, namely, a branching of G903 and G4720 distinct from all identified subtypes within genotypes 1 and 2, respectively. These results were confirmed by bootstrap values of 99 and 93% on 1,000 replicates for G903 and G4720, respectively (data not shown). Comparing G903 and G4720 with genotype 1 and 2 references, respectively, the highest sequence homology was observed in the 5′ UTR (96 ± 0.5% and 96 ± 0.7%, respectively), the next highest (89 ± 3% and 93 ± 3%) was observed in the core region, and the lowest was observed in the E1 (75 ± 8% and 80 ± 9%) and E2 (70 ± 7.5% and 72 ± 9.8%) regions. Similar diversity was observed in the deduced amino acid sequences, but the general hydrophilic patterns and the predicted secondary structure profiles of core, E1, and E2 proteins were conserved among the viruses studied. G4720 and G903 exhibited four and three substitutions, respectively, among the 44 first residues of the core that are used as antigen in the serology confirmatory assay.
DISCUSSION
Using two separate EIA screening assays, an anti-HCV prevalence of 1.3% was found in blood donors from Kumasi, Ghana (this study and reference 36). This result is consistent with HCV seroprevalence ranging between 0.9 and 6.7% reported from previous studies in Ghana and neighboring West African countries (1, 4, 10, 19, 48). The differences observed are probably related to the populations studied (blood donors, children, pregnant women, general population) and the testing algorithm used (one or more EIA screening assays followed or not by a confirmatory assay).
Hepatitis C virus infection may resolve or evolve to chronic infection, resulting in a variety of outcomes ranging from no symptoms to end-stage liver disease (15, 41). From studies performed in Western and Far Eastern countries, it is generally assumed that 15 to 20% of HCV infections resolve spontaneously (15, 41). Using RIBA 3.0, a similar recovery rate of 23.7% was observed in confirmed seropositive Ghanaian blood donors. The higher recovery rates (45 and 56%) reported in studies of large groups of women from Ireland (21) and Germany (37), respectively, who were infected with anti-D immunoglobulins cannot really be compared with other and our data sets, since all individuals in each cohort were infected with the same HCV strains instead of a variety of strains. Two recent studies from Japan and Taiwan involving a limited number of patients with acute hepatitis C reported recovery rates of 30 to 36% (17, 38). When the sensitivity of the confirmation procedure was improved by using the Murex assay and in-house anti-NS3 and anti-core EIAs, the recovery rate increased to 53%. This recovery rate in Ghana might be underestimated, since 67 samples that tested anti-HCV positive with two independent EIAs did not undergo further confirmation and consequently were not included in the final analysis. However, with the S/CO of these samples being borderline (<2.0), the likelihood of false positivity was high. This result is consistent with previous observations (36) and with a recent study reporting spontaneous HCV clearance in 8 of 14 patients after acute hepatitis C (24). In sub-Saharan Africa, a limited number of studies also reported an absence of detectable viral RNA in 41 to 89% of seropositive individuals from Togo (2), South Africa (43), and Namibia (45). However, the absence of confirmatory assay and the heterogeneity of the EIAs used make the comparison between studies difficult. The estimation of recovery rate relies on the performance of two types of assays: genomic detection of viral RNA by amplification and serological confirmatory assays. While there are international standards and quality assurance schemes enabling establishment of the sensitivity of genomic amplification assays, no such standard exists for serological confirmation. With RIBA 3.0 being the first, and virtually only, confirmatory assay, serological confirmation depends on the performance of this test. A number of studies have pointed out the shortcomings of this assay (11). Both the choice of antigen and the genotype 1a of the proteins used in RIBA appear to influence the confirmation rate.
It was argued that viral clearance was overestimated in African samples due to false-positive serological results or to false-negative HCV RNA results. It was also suggested that false-positive results were more frequent in African populations than in blood donors from Western countries (39). In this study, this possibility was eliminated, as only samples reactive with either three separate EIAs or two EIAs and at least one confirmatory assay were considered to be carrying antibodies to HCV (Table 1). In addition, the high performance of the TMA assay to detect HCV RNA (100% specificity and sensitivity of 15 to 20 IU/ml for genotypes 1 to 6) found in a previous study reduced the possibility of false-negative results (8). The assays and algorithms used in this study provided a diagnostic accuracy at least equal to, if not better than, that of most studies conducted previously.
Indirect evidence supporting a higher frequency of recovery from HCV infection is provided by the significantly lower viral load observed in this population compared to a population of United Kingdom blood donors (Fig. 2). This difference might be related to host or viral factors. Host factors include humoral and cellular immune response. In this study, only humoral factors were examined, in particular the reactivity to individual viral proteins (Fig. 1). All RNA-containing samples strongly reacted with each viral protein, structural and nonstructural. Finding dissociated reactivity in samples from recovered patients is not surprising, since in HCV infection, as in other viral infections, such as hepatitis B virus, some antibodies persist (anti-HBc) and others are short-lived (anti-HBs). From the data collected here, anti-NS3 seems to persist, while anti-core appears short-lived (Fig. 1). Single reactivity to NS3 may represent either nonspecific reactivity or resolved infection with antibodies to the other HCV proteins no longer detectable. It has been reported that the HCV-specific antibody response gradually decreased and eventually disappeared after recovery from HCV infection (37). NS3 and core antibodies were the last remaining specificities in cases of seroreversion (23). It is possible that the rate of recovery indicated in this study might be underestimated, as NS3-only reactive samples could indicate ancient HCV infection. An alternative explanation to this poor reactivity might be related to sequence and epitope differences between recombinant antigens derived from the genotype 1a prototype sequence and sera from patients infected with genotype 2 prevalent in West Africa.
Viral clearance and clinical outcome during HCV infection has been strongly associated with a vigorous and multispecific CD4+ and CD8+ T-cell response to the virus (40, 49). In addition, several reports indicate that in West and South Africa, HCV infection is less frequently associated with chronic liver disease than in North America, Europe, and Japan (1, 13, 32). Therefore, we hypothesize that HCV-infected West African people may mount an early and efficient immune response resulting in a high frequency of viral clearance. Further studies are in progress in our laboratory to investigate the HCV-specific cellular immune response in Ghanaians with or without viremia.
Host genetic factors may also influence viral infection outcome. For example, HLA-DR alleles have been associated with spontaneous hepatitis B virus clearance and progression of HIV-1 infection (27, 42). To date, little is known about the potential role of such host factors in spontaneous HCV clearance. However, few studies suggested that host HLA class II genotype (particularly alleles DQB1*, DRB1*, and DQA1*) might be an important factor in determining the outcome of HCV infection (12). Since the distribution and the associations of HLA alleles may differ between populations, this aspect is also currently investigated.
Viral factors that influence hepatitis C infection outcome remain poorly understood. HCV genotype, particularly genotype 1b, has been already implicated in determining the clinical outcome of patients with chronic hepatitis C and the response to antiviral treatments (3, 50). Among the Ghanaians who showed evidence of active infection, the most common HCV genotype was genotype 2 (87%). It may suggest that virus clearance is more efficient in people infected with genotype 2 than with other genotypes due to a lower level of replication of genotype 2 strains. However, no significant difference in viral load between genotypes was previously reported with sensitive quantification assays (16, 30). In addition, several studies showed no significant relationship between HCV genotypes, replication properties, HCV clearance, and disease outcome (6, 31, 38, 50). A higher rate of chronicity among patients infected with genotype 2c than with genotype 1b was recently reported (22). The high recovery rate and low viral load in HCV chronic infection observed in Ghanaian blood donors (Fig. 2) do not appear related to viral factors but rather to host factors.
The distribution of HCV genotypes in Africa is still largely unknown. Studies in a few African countries lead to the general picture that genotypes 4 and 5 predominate in North and South Africa, respectively. The epidemiological situation in central Africa is poorly defined. Recent studies reported that genotypes 1 and 2, rather than genotype 4 (as previously thought), are predominant (29). In Ghana, we found that genotype 2 is associated with 87% of chronic HCV infections, while genotype 1 accounts for the remaining 13% (Fig. 3 and 4). This predominance of genotype 2 is consistent with data obtained in Ghana and neighboring countries (Benin, Burkina Faso, and Guinea) on the basis of short NS5B sequences (19, 34, 48). We conducted analyses of longer NS5B as well as E1/E2 sequences that indicated nearly identical clustering irrespective of the region sequenced. The data clearly indicated that the epidemiology of HCV infection in Ghana was dominated by genotype 2.
In contrast to the clear HCV genotype distribution in Ghana, the subtype distribution appears more complex. Ghanaian HCV strains, whether of genotype 1 or 2, exhibited considerable genetic diversity when compared to each other or to reference strains representative of subtypes 1a, 1b, 1c, 2a, 2b, or 2c. Genetic diversity seemed to be widespread along the whole HCV genome, since it was not limited to the E1/E2 region already described as hypervariable (47) but was also observed in the more conserved NS5B region (Fig. 4). Due to this genetic variability, all Ghanaian strains, except one identified as subtype 1b, could not be definitively assigned to an identified subtype, nor did they form a specific distinct cluster (Fig. 3 and 4). A similar subtype wide distribution was obtained from short sequences of HCV NS5B from the neighboring countries Benin, Burkina Faso, and Guinea (19, 34). Moreover, when the Ghanaian NS5B sequences and the corresponding aligned sequences from Benin, Burkina Faso, and Guinea available in the data banks were phylogenetically analyzed together, a similar genetic distribution of the West African strains type 2 was observed. None of the African sequences clustered with subtypes 2a to c, irrespective of their geographical origin. The overall data suggest that the broad genetic diversity of genotype 2 is a characteristic of HCV infection in West Africa.
The phylogenetic analyses also showed that the distribution of the HCV Ghanaian strains described here was almost starlike within genotype 2 (Fig. 3 and 4). There are two possible biological interpretations for this tree shape. The first is a recent and rapidly expanding epidemic of HCV type 2 in Ghana. Unequal evolutionary rates among the different genotypes of HCV have been reported, and HCV genotype 2 appears to evolve faster than the average of the other genotypes (35). However, no epidemiological data are supporting the hypothesis of an epidemic spread of HCV in Ghana. The second hypothesis is that an ancient endemicity of HCV type 2 in Ghana and probably in West Africa led to the emergence of an undifferentiated multiplicity of subtypes over a long-term evolution process. A similar endemic spread seems to have occurred in the Indian subcontinent with genotype 3 and in Southeast Asia with genotype 6. HCV types 3 and 6 include a great number of subtypes characterized by extensive sequence variability (33). From extensive phylogenetic analysis of published sequences in both E1/E2 and NS5B regions, classified subtypes of genotype 2 tended to be geographically restricted. Subtype 2a was common in East Asia, subtype 2b was common in Northern America, and subtype 2c was common in Europe. None of these subtypes have been clearly identified in West Africa in this or other studies (19, 34, 48). These observations, with the predominance and the wide genetic diversity of the West African genotype 2 strains, suggest that HCV genotype 2 is possibly indigenous to this part of Africa and, consequently, that subtypes 2a to c may derive from West African ancestors. A similar epidemiological situation has been previously documented for the human T-cell lymphotropic virus type I (44). The wide genetic diversity showed by all West African type 2 strains is compatible with ancient export of specific strains from which other strains derived in restricted subtypes away from West Africa. This could be related to a lack of contact between host groups or to the high rate of recovery from HCV infection found in West Africa. In contrast, the relatively low proportion of HCV genotype 1 strains compared to genotype 2, and their divergence from previously classified subtypes 1a to c, may indicate that these strains were introduced in Ghana from other areas, possibly central Africa or Europe.
Sequence similarity or evolutionary distance analyses have yielded the existence of six major genotypes of HCV. However, the nature and the minimal length of the subgenomic fragments sufficiently informative to correctly classify new HCV subtypes within these genotypes remained controversial and resulted in considerable confusion in the past. In a recent attempt at standardization, it was proposed that new genotypes or subtypes should be based on nucleotide sequences of at least two coding regions, preferably of complete genome sequences, and that they should be based on phylogenetic analysis rather than percent identity (33). The analysis of core, E1, and NS5B genomic regions using the neighbor-joining method has been confirmed as a reliable method for the (re)assignment of most new HCV strains (35). According to these guidelines, the present analysis was limited to the two subgenomic regions encoding part of the E1 and the E2 proteins and NS5B. In this study, it appears that a precise subtype classification of the Ghanaian strains will require longer fragments due to the genetic variability exhibited. In the E1/E2 and NS5B phylogenetic trees, the relative position of most Ghanaian strains varied from one tree to the other, further suggesting a lack of subclustering (Fig. 3 and 4). Interestingly, the two subtype 2c reference sequences did not cluster together in both phylogenetic analyses.
Some Ghanaian strains clearly departed from this broad genotype distribution and were not related to mixtures of variants from multiple infections (Fig. 5). G903 and G4720 strains were significantly divergent compared to the other Ghanaian and reference strains. This divergence from genotypes 1 and 2 was confirmed by the phylogenetic analysis of a 1.8-kb sequence encoding the 5′UTR region and the core, E1, and (partially) E2 proteins. A third Ghanaian strain (G3895) could not be definitively classified within the six HCV genotypes following the guidelines mentioned above. The high genetic diversity of G3895 was also indirectly evidenced by the fact that a longer 5′ UTR-E2 sequence could not be amplified using oligonucleotide primers sufficiently conserved to detect strains of all genotypes. From the data available, and being fully aware that a definitive (sub)type assignment of new HCV sequences requires full genome analysis, we suggest that G903 and G4720 strains belong to new subtypes within genotypes 1 and 2, respectively, and that G3895 strain may be part of a new HCV genotype. Additional phylogenetic analyses based on full genome sequences are in progress to definitively characterize these variant strains.
In conclusion, sensitive anti-HCV confirmatory assays have reveal a higher rate of spontaneous HCV clearance than previously reported, and HCV infection in Ghana is characterized by the predominance of HCV genotype 2, with broad genetic diversity that might represent the origin of genotype 2 strains disseminated worldwide.
Acknowledgments
We thank J. Koziarz from Abbott Laboratories, North Chicago, Ill., for providing a grant to screen antibodies to HCV; J. W. Acheampong from the Department of Medicine at Komfo Anokye Teaching Hospital, Kumasi, Ghana, for his support throughout the study; and M. Salemi for helpful discussion on the phylogenetic analysis.
REFERENCES
- 1.Acquaye, J. K., and D. Tettey-Donkor. 2000. Frequency of hepatitis C virus antibodies and elevated serum alanine transaminase levels in Ghanaian blood donors. West Afr. J. Med. 19:239-241. [PubMed] [Google Scholar]
- 2.Agbodjan, E., M. Prince-David, T. Nicot, C. Dagnra, and F. Denis. 1995. Serologic and genomic research by PCR of hepatitis C virus in different populations in Lome (Togo). Bull. Soc. Pathol. Exot. 88:219-224. [PubMed] [Google Scholar]
- 3.Amoroso, P., M. Rapicetta, M. E. Tosti, A. Mele, E. Spada, S. Buonocore, G. Lettieri, P. Pierri, P. Chionne, A. R. Ciccaglione, and L. Sagliocca. 1998. Correlation between virus genotype and chronicity rate in acute hepatitis C. J. Hepatol. 28:939-944. [DOI] [PubMed] [Google Scholar]
- 4.Ampofo, W., N. Nii-Trebi, J. Ansah, K. Abe, H. Naito, S. Aidoo, V. Nuvor, J. Brandful, N. Yamamoto, D. Ofori-Adjei, and K. Ishikawa. 2002. Prevalence of blood-borne infectious diseases in blood donors in Ghana. J. Clin. Microbiol. 40:3523-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basaras, M., A. Santamaria, M. Sarsa, E. Gutierrez, Y. de Olano, and R. Cisterna. 1999. Seroprevalence of hepatitis B and C, and human immunodeficiency type 1 viruses in a rural population from the Republic of Equatorial Guinea. Trans. R. Soc. Trop. Med. Hyg. 93:250-252. [DOI] [PubMed] [Google Scholar]
- 6.Benvegnù, L., P. Pontisso, D. Cavalletto, F. Noventa, L. Chemello, and A. Alberti. 1997. Lack of correlation between hepatitis C virus genotypes and clinical course of hepatitis C virus-related cirrhosis. Hepatology 25:211-215. [DOI] [PubMed] [Google Scholar]
- 7.Bigger, C. B., K. M. Brasky, and R. E. Lanford. 2001. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J. Virol. 75:7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Candotti, D., A. Richetin, B. Cant, J. Temple, C. Sims, I. Reeves, J. A. Barbara, and J.-P. Allain. 2003. Evaluation of a transcription-mediated amplification-based HCV and HIV-1 RNA duplex assay for screening individual blood donations: a comparison with a minipool testing system. Transfusion 43:215-225. [DOI] [PubMed] [Google Scholar]
- 9.Chan, S. W., F. McOmish, E. C. Holmes, B. Dow, J. F. Peutherer, E. Follett, P. L. Yap, and P. Simmonds. 1992. Analysis of a new hepatitis C virus type and its phylogenetic relationship to existing variants. J. Gen. Virol. 73:1131-1141. [DOI] [PubMed] [Google Scholar]
- 10.Combe, P., G. La Ruche, D. Bonard, T. Ouassa, H. Faye-Kette, F. Sylla-Koko, and F. Dabis; DYSCER-CI study group. 2001. Hepatitis B and C infections, human immunodeficiency virus and other sexually transmitted infections among women of childbearing age in Cote d'Ivoire, West Africa. Trans. R. Soc. Trop. Med. Hyg. 95:493-496. [DOI] [PubMed] [Google Scholar]
- 11.Courouce, A. M., L. Noel, F. Barin, M. H. Elghouzzi, F. Lunel, M. L. North, and W. Smilovici. 1998. A comparative evaluation of the sensitivity of five anti-hepatitis C virus immunoblot assays. Vox Sang. 74:217-224. [PubMed] [Google Scholar]
- 12.Cramp, M. E., P. Carucci, J. Underhill, N. V. Naoumov, R. Williams, and P. T. Donaldson. 1998. Association between HLA class II genotype and spontaneous clearance of hepatitis C viraemia. J. Hepatol. 29:207-213. [DOI] [PubMed] [Google Scholar]
- 13.Dusheiko, G. M. 1992. Hepatocellular carcinoma in Africans. Ital. J. Gastroenterol. 24:146-147. [PubMed] [Google Scholar]
- 14.Farci, P., A. Shimoda, A. Coiana, G. Diaz, G. Peddis, J. C. Melpoder, A. Strazzera, D. Y. Chien, S. J. Munoz, A. Balestrieri, R. H. Purcell, and H. J. Alter. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339-344. [DOI] [PubMed] [Google Scholar]
- 15.Freeman, A. J., G. Marinos, R. A. Ffrench, and A. R. LLoyd. 2001. Immunopathogenesis of hepatitis C virus infection. Immunol. Cell Biol. 79:515-536. [DOI] [PubMed] [Google Scholar]
- 16.Hawkins, A., F. Davidson, and P. Simmonds. 1997. Comparison of plasma virus loads among individuals infected with hepatitis C virus (HCV) genotypes 1, 2, and 3 by Quantiplex HCV RNA assay version 1 and 2, Roche Monitor assay, and an in-house limiting dilution method. J. Clin. Microbiol. 35:187-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang S.-J., S.-D. Lee, R.-H. Lu, C.-W. Chu, J.-C. Wu, S.-T. Lai, and F.-Y. Chang. 2001. Hepatitis C viral genotype influences the clinical outcome of patients with acute posttransfusion hepatitis C. J. Med. Virol. 65:505-509. [PubMed] [Google Scholar]
- 18.Jackson, P., J. Petrik, G. J. M. Alexander, G. Pearson, and J.-P. Allain. 1997. Reactivity of synthetic peptides representing selected sections of hepatitis C virus core and envelope proteins with a panel of hepatitis C virus-seropositive human plasma. J. Med. Virol. 51:67-79. [DOI] [PubMed] [Google Scholar]
- 19.Jeannel, D., C. Fretz, Y. Traore, N. Kohdjo, A. Bigot, E. Pê Gamy, G. Jourdan, K. Kourouma, G. Maertens, F. Fumoux, J.-J. Fournel, and L. Stuyver. 1998. Evidence for high genetic diversity and long-term endemicity of hepatitis C virus genotypes 1 and 2 in West Africa. J. Med. Virol. 55:92-97. [PubMed] [Google Scholar]
- 20.Journal of Viral Hepatology.1999. Global surveillance and control of hepatitis C. Report of a W. H. O. Consultation organized in collaboration with the Viral Hepatitis Prevention Board, Antwerp, Belgium. J. Viral Hepat. 6:35-47. [PubMed] [Google Scholar]
- 21.Kenny-Walsh, E., and the Irish Hepatology Research Group. 1999. Clinical outcomes after hepatitis C infection from contaminated anti-D immune globulin. N. Engl. J. Med. 340:1228-1233. [DOI] [PubMed] [Google Scholar]
- 22.Kondili, L. A., P. Chionne, A. Constantino, U. Villano, C. Lo Noce, F. Pannozzo, A. Mele, S. Giampaoli, and M. Rapicetta. 2002. Infection rate and spontaneous seroreversion of anti-hepatitis C virus during the natural course of hepatitis C virus infection in the general population. Gut 50:693-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanotte, P., F. Dubois, S. Le Pogam, C. Guerois, B. Fimbel, Y. Bacq, Y. Gruel, A. Goudeau, and F. Barin. 1998. The kinetics of antibodies against hepatitis C virus may predict viral clearance in exposed hemophiliacs. J. Infect. Dis. 178:556-559. [DOI] [PubMed] [Google Scholar]
- 24.Larghi, A., M. Zuin, A. Crosignani, M. L. Ribero, C. Pipia, P. M. Battezzati, G. Binelli, F. Donato, A. R. Zanetti, M. Podda, and A. Tagger. 2002. Outcome of an outbreak of acute hepatitis C among healthy volunteers participating in pharmacokinetics studies. Hepatology 36:993-1000. [DOI] [PubMed] [Google Scholar]
- 25.Laurent, C., D. Henzel, C. Mulanga-Kabeya, G. Maertens, B. Larouzé, and E. Delaporte. 2001. Seroepidemiological survey of hepatitis C virus among commercial sex workers and pregnant women in Kinshasa, Democratic Republic of Congo. Int. J. Epidemiol. 30:872-877. [DOI] [PubMed] [Google Scholar]
- 26.Lawal, Z., J. Petrik, V. S. Wong, G. J. Alexander, and J.-P. Allain. 1997. Hepatitis C virus genomic variability in untreated and immunosuppressed patients. Virology 228:107-111. [DOI] [PubMed] [Google Scholar]
- 27.Magierowska, M., I. Theodorou, P. Debre, F. Sanson, B. Autran, Y. Riviere, D. Charron, and D. Costagliola. 1999. Combined genotypes of CCR5, CCR2, SDF1, and HLA genes can predict the long-term nonprogressor status in human immunodeficiency virus-1-infected individuals. Blood 93:936-941. [PubMed] [Google Scholar]
- 28.Memon, M. I., and M. A. Memon. 2002. Hepatitis C: an epidemiological review. J. Viral Hepat. 9:84-100. [DOI] [PubMed] [Google Scholar]
- 29.Ndjomou, J., B. Kupfer, B. Kochan, L. Zekeng, L. Kaptue, and B. Matz. 2002. Hepatitis C virus infection and genotypes among human immunodeficiency virus high-risk groups in Cameroon. J. Med. Virol. 66:179-186. [DOI] [PubMed] [Google Scholar]
- 30.Persico, M., E. Persico, R. Suozzo, S. Conte, M. De Seta, L. Coppola, B. Palmentieri, F. C. Sasso, and R. Torella. 2000. Natural history of hepatitis C virus carriers with persistently normal aminotransferase levels. Gastroenterology 118:760-764. [DOI] [PubMed] [Google Scholar]
- 31.Prati, D., C. Capelli, A. Zanella, F. Mozzi, P. Bosoni, M. Pappalettera, F. Zanuso, L. Vianello, E. Locatelli, C. de Fazio, G. Ronchi, E. del Ninno, M. Colombo, and G. Sirchia. 1996. Influence of different hepatitis C virus genotypes on the course of asymptomatic hepatitis C virus infection. Gastroenterology 110:178-183. [DOI] [PubMed] [Google Scholar]
- 32.Reid, A. E., and T. J. Liang. 1995. Association of hepatitis C virus and hepatocellular carcinoma in the United States. Princess Takamatsu Symp. 25:41-49. [PubMed] [Google Scholar]
- 33.Robertson, B., G. Myers, C. Howard, T. Brettin, J. Bukh, B. Gaschen, T. Gojobori, G. Maertens, M. Mizokami, O. Nainan, S. Netesov, K. Nishioka, T. Shin-i, P. Simmonds, D. Smith, L. Stuyver, and A. Weiner. 1998. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. Arch. Virol. 143:2493-2503. [DOI] [PubMed] [Google Scholar]
- 34.Ruggieri, A., C. Argentini, F. Kouruma, P. Chionne, E. D'Ugo, E. Spada, S. Dettori, S. Sabbatani, and M. Rapicetta. 1996. Heterogeneity of hepatitis C virus genotype 2 variants in West Central Africa (Guinea Conakry). J. Gen. Virol. 77:2073-2076. [DOI] [PubMed] [Google Scholar]
- 35.Salemi, M., and A.-M. Vandamme. 2002. Hepatitis C virus evolutionary patterns studied through analysis of full-genome sequences. J. Mol. Evol. 54:62-70. [DOI] [PubMed] [Google Scholar]
- 36.Sarkodie, F., M. Adarkwa, Y. Adu-Sarkodie, D. Candotti, J. W. Acheampong, and J.-P. Allain. 2001. Screening for viral markers in volunteer and replacement blood donors in West Africa. Vox Sang. 80:142-147. [DOI] [PubMed] [Google Scholar]
- 37.Takaki, A., M. Wiese, G. Maertens, E. Depla, U. Seifert, A. Liebetrau, J. L. Miller, M. P. Manns, and B. Rehermann. 2000. Cellular immune responses persist and humoral responses decrease two decades after recovery from a single-source outbreak of hepatitis C. Nature 6:578-582. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka, E., and K. Kiyosawa. 2000. Natural history of acute hepatitis C. J. Gastroenterol. Hepatol. 15(Suppl.):E97-E104. [DOI] [PubMed] [Google Scholar]
- 39.Tess, B. H., A. Levin, G. Brubaker, J. Shao, J. E. Drummond, H. J. Alter, and T. R. O'Brien. 2000. Seroprevalence of hepatitis C virus in the general population of northwest Tanzania. Am. J. Trop. Med. Hyg. 62:138-141. [DOI] [PubMed] [Google Scholar]
- 40.Thimme, R., J. Bukh, H. C. Spangenberg, S. Wieland, J. Pemberton, C. Steiger, S. Govindarajan, R. H. Purcell, and F. V. Chisari. 2002. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc. Natl. Acad. Sci. USA 99:15661-15668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas, D. L., J. Astemborski, R. M. Rai, F. A. Anania, M. Schaeffer, N. Galai, K. Nolt, K. E. Nelson, S. A. Strathdee, L. Johnson, O. Laeyendecker, J. Boitnott, L. E. Wilson, and D. Vlahov. 2000. The natural history of hepatitis C virus infection: host, viral and environmental factors. JAMA 284:450-456. [DOI] [PubMed] [Google Scholar]
- 42.Thursz, M. R., D. Kwiatkowski, C. E. Allsopp, B. M. Greenwood, H. C. Thomas, and A. V. Hill. 1995. Association between an MHC class II allele and clearance of hepatitis B virus in the Gambia. N. Engl. J. Med. 332:1092-1093. [DOI] [PubMed] [Google Scholar]
- 43.Tucker, T. J., M. Voight, A. Bird, S. Robson, B. Gibbs, J. Kannemeyer, M. Galloway, R. E. Kirsch, and H. Smuts. 1997. Hepatitis C virus infection rate in volunteer blood donors from the Western Cape—comparison of screening tests and PCR. S. Afr. Med. J. 87:603-605. [PubMed] [Google Scholar]
- 44.Van Dooren, S., E. Gotuzzo, M. Salemi, D. Watts, E. Audenaert, S. Duwe, H. Ellerbrok, R. Grasmann, E. Hagelberg, J. Desmyter, and A.-M. Vandamme. 1998. Evidence for a post-Columbian introduction of human T-cell lymphotropic virus in Latin America. J. Gen. Virol. 79:2695-2708. [DOI] [PubMed] [Google Scholar]
- 45.Vardas, E., F. Sitas, K. Seidel, A. Casteling, and J. Sim. 1999. Prevalence of hepatitis C virus antibodies and genotypes in asymptomatic, first-time blood donors in Namibia. Bull. W. H. O. 77:965-972. [PMC free article] [PubMed] [Google Scholar]
- 46.Villano, S. A., D. Vlahov, K. E. Nelson, S. Cohn, and D. L. Thomas. 1999. Persistence of viraemia and the importance of long-term follow-up after acute hepatitis C infection. Hepatology 29:908-914. [DOI] [PubMed] [Google Scholar]
- 47.Vizmanos, J. L., C. J. Gonzalez-Navarro, F. J. Novo, M. P. Civeira, J. Pietro, A. Gullon, and M. Garcia-Delgado. 1998. Degree and distribution of variability in the 5′ untranslated, E1, E2/NS1 and NS5 regions of the hepatitis C virus (HCV). J. Viral Hepat. 5:227-240. [DOI] [PubMed] [Google Scholar]
- 48.Wansbrough-Jones, M. H., E. Frimpong, B. Cant, K. Harris, M. R. W. Evans, and C. G. Teo. 1998. Prevalence and genotype of hepatitis C virus infection in pregnant women and blood donors in Ghana. Trans. R. Soc. Trop. Med. Hyg. 92:496-499. [DOI] [PubMed] [Google Scholar]
- 49.Wedemeyer, H., X.-S. He, M. Nascimbeni, A. R. Davis, H. B. Greenberg, J. H. Hoofnagle, T. J. Liang, H. Alter, and B. Rehermann. 2002. Impaired effector function of hepatitis C virus-specific CD8+ T cells in chronic hepatitis C virus infection. J. Immunol. 169:3447-3458. [DOI] [PubMed] [Google Scholar]
- 50.Zein, N. N. 2000. Clinical significance of hepatitis C virus genotypes. Clin. Microbiol. Rev. 13:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]