Abstract
AU-rich elements (AREs) located in the 3′ untranslated region target the mRNAs encoding many protooncoproteins, cytokines, and lymphokines for rapid degradation. HuR, a ubiquitously expressed member of the embryonic lethal abnormal vision (ELAV) family of RNA-binding proteins, binds ARE sequences and selectively stabilizes ARE-containing reporter mRNAs when overexpressed in transiently transfected cells. HuR appears predominantly nucleoplasmic but has been shown to shuttle between the nucleus and cytoplasm via a novel shuttling sequence HNS. We report generation of a mouse monoclonal antibody 3A2 that both immunoblots and immunoprecipitates HuR protein; it recognizes an epitope located in the first of HuR's three RNA recognition motifs. This antibody was used to probe HuR interactions with mRNA before and after heat shock, a condition that has been reported to stabilize ARE-containing mRNAs. At 37°C, approximately one-third of the cytoplasmic HuR appears polysome associated, and in vivo UV crosslinking reveals that HuR interactions with poly(A)+ RNA are predominantly cytoplasmic rather than nuclear. This comprises evidence that HuR directly interacts with mRNA in vivo. After heat shock, 12–15% of HuR accumulates in discrete foci in the cytoplasm, but surprisingly the majority of HuR crosslinks instead to nuclear poly(A)+ RNA, whose levels are dramatically increased in the stressed cells. This behavior of HuR differs from that of another ARE-binding protein, hnRNP D, which has been implicated as an effector of mRNA decay rather than mRNA stabilization and of the general pre-RNA-binding protein hnRNP A1. We interpret these differences to mean that the temporal association of HuR with ARE-containing mRNAs is different from that of these other two proteins.
The regulation of mRNA stability is an important aspect of eukaryotic gene expression. AU-rich elements (AREs) found in the 3′ untranslated region are the best characterized signals that target a variety of short-lived mRNAs, including those of protooncogenes, cytokines, and lymphokines, for rapid decay in mammalian cells (1). It has long been known that the rate of ARE-mediated decay is itself subject to regulation. Conditions of stress [e.g., heat shock (2), exposure to short-wavelength UV light (3), or hypoxia (4)], cell stimulation [e.g., T cell activation (5) or stimulation of mast cells by ionomycin (6)], and oncogenic transformation (7) have all been shown to stabilize ARE-containing mRNAs.
Although many proteins that selectively bind AREs have been identified and characterized, so far only two, HuR and heterogeneous nuclear ribonucleoprotein (hnRNP) D, have been demonstrated to affect mRNA stability in vivo. HuR (8) is a ubiquitously expressed member of the embryonic lethal abnormal vision (ELAV) family of RNA-binding proteins (9). Three other closely related neural proteins, called HelN1 [or HuB (10, 11)], HuC (11) and HuD (12), are target antigens in paraneoplastic neuropathy [Hu syndrome (13)]. HuR is a 36-kDa polypeptide that contains three classical RNA-binding domains [RNA recognition motifs (RRMs)] (8): the first two have been implicated in ARE recognition, whereas the third has been suggested to bind the poly(A) tail of target mRNAs (14). HuR is relatively low abundance, about 5 × 105 molecules per HeLa cell (15). Overexpression of HuR in transiently transfected mammalian cells leads to stabilization of reporter mRNAs carrying AREs in their 3′ untranslated regions (16, 17). Although immunofluorescence studies show HuR to be predominantly nuclear in interphase cells, it has been demonstrated to shuttle between the nucleus and cytoplasm (16). Its shuttling signal, HNS, located in the hinge region between its second and third RRMs (18), appears similar but not identical to the M9 shuttling sequence first identified in hnRNP A1 (19). It has been suggested that HuR may initially bind ARE-containing mRNAs in the nucleus and accompany them to the cytoplasm, providing ongoing protection from the decay machinery (18).
HnRNP D (also called AUF1) exists in four isoforms ranging from 37 to 45 kDa as a result of alternative splicing (20, 21) and is approximately 10-fold more abundant than HuR. It has been reported to be a DNA- (22) as well as an RNA-binding protein (23). HnRNP D contains two RRMs (which are not affected by the alternative splicing) and is present in both the nucleus and the cytoplasm (24). Ectopic expression of hnRNP D (particularly the p37 and p42 isoforms) in hemin-induced human erythroleukemic cells enhanced rapid mRNA decay directed by the ARE (25). Moreover, induction of hsp70 after heat shock, down-regulation of the ubiquitin-proteasome pathway, or inactivation of the E1 ubiquitinating enzyme all resulted in hnRNP D movement to the nucleus of human HeLa cells accompanied by a block in ARE-mediated mRNA decay (26).
Because HuR and hnRNP D apparently play opposite roles in the stabilization/decay of ARE-containing mRNAs, we set out to examine the behavior and associations of HuR before and after heat shock of HeLa cells. We find that HuR and hnRNP D differ from each other not only in their cellular localization after heat shock but also in their polysome association and RNA binding; hnRNP A1, a general pre-mRNA-binding protein, which is about 100-fold more abundant than HuR (27), exhibits behavior different from both HuR and hnRNP D. Our results highlight the distinctions between these three important RNA-binding proteins, all of which are believed to be nucleocytoplasmic shuttling proteins (16, 25, 28), in their contributions to the trafficking and fate of ARE-containing mRNAs.
Materials and Methods
Antibodies.
The mAb 3A2 was obtained by immunizing BALB/c mice over a 7-mo period with His-tagged recombinant HuR in monophosphoryl lipid A-synthetic trehalose dicorynom (MPL/TDM) adjuvant (RIBI Immunochemical Research, Hamilton, MT). The injection schedule consisted of four subcutaneous injections of 50 μg native protein followed by three injections of 50 μg of denatured protein and a final intraperitoneal injection of 50 μg of native protein. Denatured antigen was prepared by adding an equal volume of 126 mM Tris⋅HCl (pH 6.8)/20% glycerol/4% SDS/0.005% bromophenol blue to recombinant HuR, heating the sample to 100°C for 3 min, cooling to room temperature, and adding an equal volume of MPL/TDM adjuvant. Monoclonal antibodies were isolated as described previously (29), except that rapid screening of initial hybridoma supernatants was performed by ELISA, and those scoring positive by ELISA were subsequently examined by immunoblot and cell immunofluorescence analyses. The heavy chain subclass of mAb 3A2 is IgG1.
Anti-hnRNP D antibodies were raised against peptide CMSEEQFGGDG, which corresponds to the N terminus of both human and murine hnRNP D (21), with a cysteine added to facilitate coupling. The peptide was synthesized at the W. H. Keck facility (Yale University School of Medicine) and coupled to maleimide-activated keyhole limpet hemocyanin (KLH) carrier protein (Pierce) according to the manufacturer's instructions; coupling efficiency was greater than 85% as estimated by measuring the free sulfhydryl concentration with Ellman's reagent (Pierce). The KLH conjugate was dialyzed against PBS containing 0.9 M NaCl. Rabbits were immunized and boosted once 3 wk later and at 2-wk intervals thereafter with 100 μg per injection. Antibodies were affinity purified on protein A Sepharose (Pharmacia).
Construction and Expression of HuR Plasmids.
HuR plasmids were synthesized by using oligonucleotides that contain EcoRI (upstream) and NotI (downstream) restriction sites adjacent to the coding regions and pcDNA3-HuR (16) as the template. After PCR amplification, the products were digested and cloned into pGEX-5X-2 (Pharmacia), forming in-frame fusions with GST. Oligonucleotides used to clone pGEX-5X-2/HuR were as follows: 5′-GTCTCGGGAATTCCCTCTAATGGTTATGAAGA CCACATG-3′ and 5′-GTCTCGACGCCGGCGTTATTATTT GTGGGACTTGTTGGTTTT-3′. The same upstream oligonucleotide and the following downstream oligonucleotides were used to create constructs lacking RRM 3: 5′-GTCTCGACGCCGGCGTTATTAGGAGGAGGCGTTTCCTGGCACGTT-3′; lacking RRM 3 and the hinge region: 5′-GTCTCGACGCCG GCGTTATTACTGGTTGGGGTTGGCTGCAAACTT-3′; and lacking RRM 3, the hinge region, and RRM 2: 5′-GTCTCG ACGCCGGCGTTATTATGAGCTCGGGCGAGCATACG ACAC-3′. The same downstream oligonucleotide used in the construction of pGEX-5X-2/HuR and the following upstream oligonucleotides were used to create the mutant lacking the N terminus: 5′-GTCTCGGGAATTCCCAGAACGAATTTGATCGTCAACTAC-3′, and the mutant lacking the N terminus and RRM 1: 5′-GTCTCGGGAATTCCCGAGGTGATCAAAGACGCCAACTTG-3′. Expression plasmids were transformed into BL21 cells and grown, induced, and lysed as described (30). The proteins were purified according to the GST fusion purification protocol supplied by Pharmacia.
Cell Culture, Immunofluorescence, and in Situ Hybridization.
Adherent HeLa cells were grown in DMEM supplemented with 10% FBS, penicillin, and streptomycin. Before heat shock and to maintain the pH, Hepes (pH 7.9) was added to 10 mM. Afterward the cells (on plates) were floated in a 45°C water bath for 1 h and either fixed for immunofluorescence or used to prepare extracts as described below.
For immunofluorescence, HeLa cells were grown on coverslips overnight as described above. They were treated as reported (16) by using the following antibody dilutions: 3A2 monoclonal anti-HuR, 1:300; Y12 monoclonal [anti-Sm (31)], 1:1,000; Y10B monoclonal [anti-rRNA (31)], 1:1,000; 4B10 monoclonal [anti-hnRNP A1 (28)], 1:1,000; anti-La monoclonal (32), 1:500; anti-hnRNP D polyclonal, 1:40. Quantitations used the NIH image 1.62 program.
The 35-mer oligo(dT) probe was 3′-end labeled with digoxigenin according to the protocol supplied by Boehringer Mannheim. The cells were prepared as described (33) and the probe hybridized and detected with rhodamine conjugated antidigoxigenin antibody (Boehringer Mannheim) (34).
Analysis of HuR–RNA Interactions and Western Blots.
In vivo crosslinking and analysis of crosslinked proteins proceeded exactly as reported (35).
Cell lysates were prepared and immunoprecipitated exactly as described (36).
Polysome gradients were performed as reported (37) on cytoplasmic extracts from 4 × 108 HeLa cells.
For Western blots, samples were fractionated on 12% denaturing gels, transferred to nitrocellulose, and probed with antibodies (38) at the following dilutions: 3A2, 1:30,000; 4B10 (anti-hnRNP A1), 1:1,000; anti-hnRNP D, 1:200; 4F10 (hnRNP C), 1:500. The secondary antibody was either horseradish peroxidase (HRP)-conjugated donkey anti-rabbit or HRP-conjugated donkey anti-mouse (Pierce). Blots were developed by using the ECL system (Amersham) according to the manufacturer's directions and quantitated as for immunofluorescence.
Results
A Monoclonal Antibody Specific for HuR.
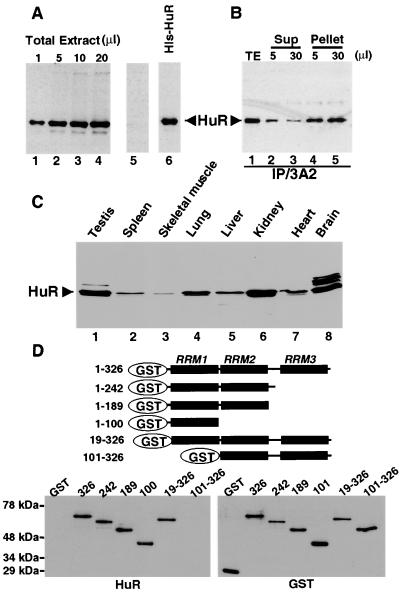
To study HuR function and behavior during heat shock, monoclonal antibodies were generated against recombinant His-tagged human HuR (kindly prepared by C. Fan). Screening of the clones was based on two criteria: (i) that the antibody react with recombinant His-tagged or GST-tagged HuR in immunoblotting, and (ii) that it produce the same staining pattern as polyclonal anti-HuR antibodies (16) in immunofluorescence microscopy. One resulting monoclonal antibody, designated 3A2, detected recombinant HuR in immunoblots (Fig. 1A, lane 6) and reacted selectively with a 36-kDa band in whole cell extract from HeLa cells (lanes 1 to 4) that comigrates with the band identified by anti-HuR polyclonal antibodies (Fig. 1B, lane 1). This band sometimes appears as a doublet, perhaps because of uncharacterized posttranslational modifications. When the 3A2 antibody was preincubated with an excess of recombinant HuR for 1 h at room temperature before use in Western blot analysis, the 36-kDa band was no longer detectable (Fig. 1A, lane 5). Addition of increasing amounts of the 3A2 antibody to HeLa cell extract revealed that it can immunoprecipitate at least 90% of the HuR (Fig. 1B). This result also indicates that the same protein is recognized by 3A2 and by our polyclonal anti-HuR antibody.
Figure 1.
The anti-HuR monoclonal antibody, 3A2, recognizes RRM 1 of HuR (A). The indicated amounts of HeLa total extract at 5 × 104 cells per microliter (lanes 1–4, 5 μl in lane 5) and 5 ng of recombinant human HuR were electrophoresed on 12% polyacrylamide gels, transferred to nitrocellulose, and probed with the 3A2 antibody. In lane 5, the monoclonal antibody was preincubated with 1 μg of recombinant HuR before use. The band migrating faster than HuR is a degradation product (16). (B) Immunoprecipitation was performed by using the indicated amounts of the 3A2 antibody (5 μl, lanes 2 and 4, and 30 μl, lanes 3 and 5) and the same amounts of total extract from 2 × 106 HeLa cells (lane 1). The Western blot was probed with polyclonal anti-HuR (16). The band above HuR migrates at the position of the immunoglobulin heavy chain. (C) Extracts from different human tissues (CLONTECH) were fractionated, blotted, and probed with the 3A2 antibody. (D) Mapping the HuR epitope recognized by 3A2. (Upper) Schematic representation of HuR truncations, which were expressed in Escherichia coli as GST fusion proteins. (Left) HuR amino acid positions included (full length, 326 amino acids). (Lower) Immunoblots of full-length and truncated GST-HuR fusion proteins were probed by using the 3A2 antibody (HuR blot) or rabbit anti-GST polyclonal antibody (GST blot).
HuR is ubiquitously expressed (9) but at different levels in different mammalian cell lines (16). We used the 3A2 monoclonal antibody to survey various human tissues by immunoblotting (Fig. 1C). Whereas a single predominant HuR band is detected in most tissues, a second more slowly migrating species is seen in testis (Fig. 1C, lane 1) and three in brain (lane 8). These additional bands probably correspond to other ELAV family members. We have observed that 3A2 reacts with recombinant proteins corresponding to human HuD and HelN1 (HuB), but not with HuC (kindly provided by I. Laird-Offringa, University of Southern California) (data not shown).
We also tested the 3A2 antibody against extracts from various species (data not shown). A 36-kDa band comigrating with that in HeLa cell extract (human) is detected in NIH 3T3 cell extract (mouse) and in Xenopus egg nuclear extract [elrA (39)]. A 40-kDa band is seen in Drosophila extract [ELAV (39)]. No crossreacting proteins were detected in yeast or Caenorhabditis elegans by using 3A2, although the C. elegans database does predict a homolog of about 55 kDa, which is detected by our polyclonal anti-HuR antibody (C.M.B. and C. Weiss, unpublished data). The 3A2 epitope therefore appears to be conserved in vertebrates and insects but not in lower eukaryotic organisms.
To locate the epitope recognized by the 3A2 monoclonal antibody, we prepared a series of truncated forms of recombinant HuR fused to GST (Fig. 1D). Analysis by immunoblotting revealed that 3A2 recognizes all HuR mutants except the one lacking the first RRM (Fig. 1D, HuR blot, lane 101–326). Use of anti-GST polyclonal antibodies showed that all the HuR mutant proteins were equally loaded and appropriately transferred (GST blot). We conclude that the 3A2 epitope is located in RRM 1 of HuR, which is highly conserved among all ELAV proteins [82% identity between HuR, HuD, and Hel-N1/N2, and 72% identity between HuR and HuC (39)].
Heat Shock Partially Relocalizes HuR to the Cytoplasm and poly(A)+ RNA to the Nucleus.
Previous studies showed that HuR shuttles between the nucleus and the cytoplasm (16). Because ARE-containing mRNAs have been reported to be stabilized after heat shock (2, 26), we asked whether this stress condition affects HuR subcellular localization. Immunofluorescence experiments were performed by using the 3A2 antibody on HeLa cells before and after 1 h at 45°C (Fig. 2). Although a variety of times and temperatures have been reported to induce heat shock in mammalian cells in culture, these were chosen because they yielded 95% cell viability after return to 37°C. No change in the total amount of HuR was detected after heat shock of HeLa cells (data not shown).
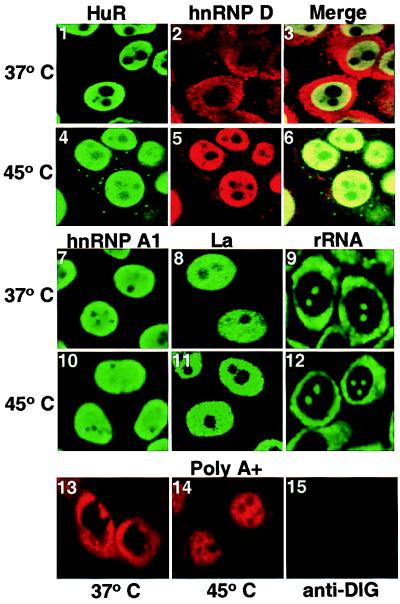
Figure 2.
Heat shock induces cytoplasmic relocalization of HuR and poly(A)+ RNA sequestration in the nucleus. (1–12) Localization of HuR, hnRNP D, hnRNP A1, La protein, and ribosomal RNA before and after heat shock. HeLa cells were grown on coverslips, heat shocked, fixed, permeabilized, and immunofluorescently stained as described in Materials and Methods. In 1–6, the cells were singly or doubly stained with 3A2 and/or polyclonal anti-hnRNP D and detected by using fluorescein isothiocyanate- or rhodamine-labeled secondary antibody, respectively. (13–15) The distribution of poly(A)+ RNA was examined in HeLa cells before and after heat shock by using 5 ng/μl digoxigenin-labeled oligo(dT35) probe (13, 14) and a 1/200 dilution of sheep antidigoxigenin Fab-rhodamine antibody (Boehringer). Cells in 15 were treated the same way as those in 13 and 14, except that during the hybridization step they were incubated without the digoxigenin-labeled oligo(dT35) probe. All images were captured by using the Bio-Rad MRC-1024 confocal microscope.
In agreement with previous reports (16), at 37°C HuR appears predominantly nucleoplasmic (excluded from the nucleoli) (Fig. 2 1). However, after heat shock about 12–15% (as determined by immunofluorescence and cell fractionation) of HuR is detected in discrete foci in the cytoplasm (Fig. 2 4); this pattern is different from the more uniform distribution of ribosomes throughout the cytoplasm (9 and 12). Neither hnRNP A1 (28) nor the La protein (32) exhibits this peculiar behavior but remains entirely nucleoplasmic (Fig. 2 10 and 11), suggesting that general nuclear leakage is not occurring. Furthermore, pretreatment of the cells with cycloheximide for 2 h before heat shock does not alter HuR's appearance in cytoplasmic foci (data not shown), demonstrating that the foci are not caused by de novo synthesis of HuR. Rather, these results suggest that the reimport of HuR into the nucleus may be impaired after heat shock. The foci indeed disperse, and HuR returns to the nucleoplasm when the heat-shocked cells are returned to 37°C for several hours even in the presence of cycloheximide (data not shown).
For comparison, we examined the behavior of the other ARE-binding protein strongly implicated in controlling mRNA decay, hnRNP D. The results in Fig. 2 confirm previous observations that heat shock causes hnRNP D, which normally resides in both the nucleus and cytoplasm, to concentrate in the nucleus (26). The anti-hnRNP D antibody was raised in rabbits against a synthetic decapeptide bearing the N-terminal sequence of hnRNP D; this antibody recognizes all hnRNP D isoforms. The superimposed images of Fig. 2 6 further show that the HuR foci that appear in the cytoplasm after heat shock do not colocalize with the anti-hnRNP D signal. We conclude that HuR and hnRNP D, although they are both ARE-binding proteins, exhibit quite different subcellular relocalization after heat shock.
In yeast, poly(A)+ RNA has been shown to accumulate in the nucleus after heat shock, suggesting a block in the nuclear export of mRNA (40). We localized poly(A)+ sequences in HeLa cells before and after heat shock by hybridization of digoxigenin-labeled oligo dT (Fig. 2 13 and 14). As in yeast, the poly(A)+ RNA signal shifts from predominantly cytoplasmic and perinuclear to predominantly nuclear, suggesting the retention of mRNA in the nucleus after heat shock.
HuR Associates with Polysomes.
To investigate whether the altered localization of HuR might be correlated with the stabilization of ARE-containing mRNAs after heat shock, we examined the distribution of HuR in polysome gradients (Fig. 3). Although only a tiny fraction of the total cellular HuR resides in the HeLa cytoplasm at 37°C (about 2% by immunofluorescence), approximately one-third of these molecules cosediment with polysomes (Fig. 3, HuR blot, 37°C, MgCl2, fractions 16–19). As anticipated, EDTA treatment dissociates the polysomes and causes the HuR peak to shift upwards in the gradient (Fig. 3, HuR blot, 37°C, EDTA). After 1 h at 45°C (Fig. 3, HuR blot, 45°C, MgCl2), the polysome peak was significantly diminished because of the stress-associated inhibition of translation (41). Little HuR appeared in the polysomes, but a much greater fraction of the HuR sedimented to the bottom of the tube (Fig. 3, HuR blot, 45°C, MgCl2, fraction B), which may reflect precipitation of the HuR-containing cytoplasmic foci visualized by immunofluorescence after heat shock (Fig. 2).
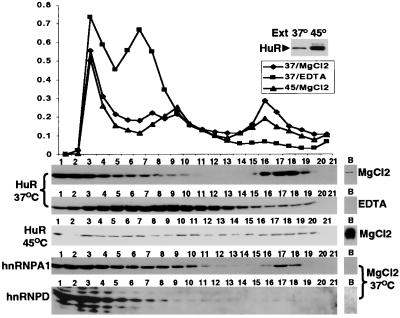
Figure 3.
Localization of HuR, hnRNP A1, and hnRNP D in polysome gradients. Upper shows absorbance profiles of 15% to 45% sucrose gradients fractionating cytoplasmic extract from 4 × 108 HeLa cells before (37°C) and after heat shock (45°C) in the absence (MgCl2) or presence of 10 mM EDTA. Inset shows a Western blot of a portion of the extracts probed with the 3A2 antibody. The fraction numbers correspond to the lanes in the Western blots below the polysome profiles. The same 37°C blot was sequentially probed with the antibodies to different proteins. The lanes labeled B show material recovered from the bottom of the centrifuge tube after collection of the fractions. Higher resolution absorbance traces revealed that 40S, 60S, and 80S ribosomes are in fractions 5–6, 7–8, and 9–10, respectively.
One concern was that HuR might appear to peak with the polysomes simply because it leaks from the nucleus and then binds mRNA during preparation of the cell extract. To investigate this possible artifact, we examined the profiles of several other RRM-containing RNA-binding proteins in the same gradients: hnRNP A1 and hnRP D (Fig. 3). Neither of these other proteins associates to nearly the same extent as HuR with the polysome region of the gradient. Instead, only about 8% of the hnRNP A1 in the cytoplasmic extract is detected in polysomes (Fig. 3, hnRNP blot, A1 37°C, fractions 15–17). Surprisingly, despite its cytoplasmic abundance and its ability to crosslink to ARE sequences in vitro (42), an even tinier fraction of hnRNP D (about 0.05% above background) cosediments with polysomes (Fig. 3, hnRNP D blot, 37°C, fractions 15–16). After heat shock, both hnRNP A1 and hnRNP D were completely absent from the polysomes (data not shown). Although these divergent observations do not rule out the possible artifactual binding of HuR to polysomal RNA, they strongly suggest that the appearance of HuR in polysomes is related to its specific binding to ARE-containing mRNAs undergoing translation.
HuR Crosslinks to Cytoplasmic poly(A)+ RNA Before, but to Nuclear poly(A)+ RNA After Heat Shock.
Finally, to ask whether the heat shock-induced cytoplasmic foci contain HuR bound to mRNA or in an RNA-free state, we examined the in vivo interaction of HuR with poly(A)+ RNA by UV crosslinking. HeLa cells were grown at 37°C or heat shocked at 45°C for 1 h and each sample divided into two portions, one of which was exposed to UV light for 3.5 min. Poly(A)+ RNA was then isolated from the nuclear and cytoplasmic fractions of each of the four samples by oligo(dT)-cellulose chromatography, by using conditions that strip even the most avidly binding noncrosslinked proteins from the RNA (35). The eluted poly(A)+ RNAs were subsequently treated with RNase and crosslinked RNA-binding proteins detected by Western blotting.
Fig. 4 (hnRNP A1 blot, lanes 7 and 9) shows that the control protein hnRNP A1 is detected crosslinked to RNA (at about the 1% level) almost exclusively in the nuclear fraction, both before and after heat shock. This is consistent with its predominantly nuclear localization by immunofluorescence under both conditions (Fig. 2 7 and 10) (43) and with previous reports by using this methodology (35). HnRNP D cannot be detected crosslinking to either nuclear or cytoplasmic poly(A)+ RNA in vivo (less than about 0.03%); no difference is seen after heat shock (Fig. 4, hnRNP D blot, lanes 6–9).
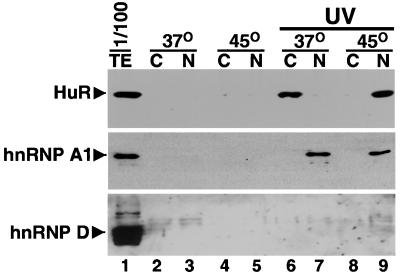
Figure 4.
HuR interaction with poly(A)+ RNA shifts from the cytoplasm to the nucleus after heat shock. HeLa (5 × 106) cells, grown at 37°C (lanes 2, 3, 6, and 7) or incubated for 1 h at 45°C (lanes 4, 5, 8, and 9) were exposed to Stratalinker UV for 15 min to induce covalent RNA–protein crosslinks in vivo. Poly(A)+ RNA-protein crosslinked complexes were then isolated from the cytoplasmic (C) and nuclear (N) fractions by chromatography on oligo(dT) in the presence of 1% SDS and β-mercaptoethanol (28) to strip away noncovalently bound proteins; the absence of signal in lanes 2–5 shows this strategy to be successful. Crosslinked proteins were released by digestion with RNase, resolved by SDS/PAGE; and HuR, hnRNP A1 and hnRNP D were detected by sequential probing of the same blot with the respective antibodies. In lanes 2–5, the cells were treated exactly the same, but the UV treatment was omitted. Lane 1 shows the proteins present in 1/100 of the 37°C UV-treated extract before separation into the nuclear and cytoplasmic fractions. These experiments were reproduced three times.
In contrast to hnRNP A1 and hnRNP D, HuR crosslinks quite efficiently (about 1%) to poly(A)+ RNA in the cytoplasm (Fig. 4, HuR blot, lane 6) and virtually not at all in the nucleus (lane 7) at 37°C. After heat shock, the situation is reversed: although there is more cytoplasmic HuR, HuR crosslinks predominantly to poly(A)+ RNA (at about the same level) in the nuclear fraction (lane 9). Western blots revealed that the expected 15% of HuR was indeed recovered in the cytoplasmic fraction (data not shown), arguing that the heat shock-induced cytoplasmic foci were not cofractionating with nuclei, thereby producing a misleading conclusion. These unanticipated results confirm HuR's interaction with polysomal mRNA before heat shock and suggest that it is released from mRNA binding and coalesces into the cytoplasmic foci seen in Fig. 2 4 in an RNA-free form in the stressed cells. Moreover, the crosslinking of HuR to the poly(A)+ RNA, which accumulates in the nucleus on heat shock (Fig. 2 14), argues that HuR does indeed bind ARE-containing mRNAs initially in the nucleus but becomes trapped with them there when mRNA export is compromised in cells subjected to stress.
Discussion
Using a monoclonal antibody, we have compared the behavior of HuR before and after heat shock to that of two more abundant RNA-binding proteins, hnRNP A1 and hnRNP D. Although only a few percent of HuR is in the cytoplasm of HeLa cells cultured at 37°C, in vivo binding of HuR to cytoplasmic mRNA can be detected both in polysome gradients and by UV crosslinking. After heat shock, 10–15% of HuR concentrates in cytoplasmic foci, but HuR surprisingly crosslinks to nuclear poly(A)+ RNA. In contrast, hnRNP A1 appears nuclear and crosslinks predominantly to nuclear RNA both before and after heat shock, whereas hnRNP D does not significantly associate with poly(A)+ RNA in either cellular compartment under any condition.
Finding a substantial fraction of HuR on polysomes (Fig. 3) is consistent with the idea that HuR binds ARE-containing mRNAs and protects them from the decay machinery while undergoing translation. Previously, other members of the Hu family of proteins, which are localized in both the nucleus and the cytoplasm, have been reported to bind mRNA (39, 44). Our inability to detect hnRNP D in polysomes or crosslinked to poly(A)+ RNA (Figs. 3 and 4) is consistent with the possibility that once bound, it targets an mRNA for such rapid decay that it is immediately released (26). Although there have been many reports of hnRNP D being UV crosslinked to ARE sequences in vitro (42), our negative in vivo results suggest that caution should be applied to the interpretation of these observations.
Comparison of the polysome and crosslinking data (Figs. 3 and 4) for HuR vs. hnRNP A1 argues that HuR may bind ARE-containing mRNAs only immediately before their export from the nucleus. Both proteins have been definitively shown to shuttle between the nucleus and cytoplasm (16, 28), but the crosslinking of hnRNP A1 to nuclear RNA only and its comparatively low abundance on polysomes indicate that it may be quickly released from the mRNA after exit. HuR, in contrast, cannot normally be detected in association with the major nuclear hnRNP complex by coimmunoprecipitation (M.S.S., unpublished observations), but its binding to nuclear RNA is revealed when the export of mRNA from the nucleus is inhibited by heat shock (Figs. 2 and 4).
We have observed that heat shock causes a buildup of poly(A)+ RNA in the nucleus of HeLa cells (Fig. 2), just as in yeast (40). This accumulation could be caused by a loss of binding to the protein(s) responsible for mRNA transport through the nuclear pore, because of interference with that protein's ability to exit the nucleus, or simply because of competition from heat shock mRNAs (45). Because the immunofluorescent staining pattern of hnRNP A1, as well as its crosslinking to nuclear RNA, does not change after heat shock, it could be that hnRNP A1 no longer shuttles and is largely responsible for mRNA export. This remains to be tested. The behavior of hnRNP A1 after heat shock contrasts with that of a yeast hnRNP-like protein, which no longer binds mRNA after heat shock (46). The finding that the crosslinking of hnRNP A1 does not increase even when more RNA accumulates in the nucleus after heat shock suggests that the amount of hnRNP A1 relative to RNA may be limiting.
Clearly, the cellular trafficking of HuR and of hnRNP D are both perturbed after heat shock. Whereas hnRNP D is largely cytoplasmic under normal conditions, it accumulates in the nuclei of stressed cells (26), which has been suggested to sequester it away from the mRNA and thereby stabilize ARE-containing messages. It is not known whether hnRNP D shuttles. HuR, on the other hand, has a shuttling sequence HNS (18) that bears some similarity to the M9 sequence of hnRNP A1 (19, 47). The observation that the nuclear import of HuR but not of hnRNP A1 is inhibited after heat shock (Fig. 2) argues that HNS and M9 could be distinct shuttling sequences recognized by different import and export receptors. Alternatively, heat shock may generate novel interactions of HuR or of hnRNP A1 with other cellular components that differentially alter the ability of each protein to shuttle. Again, testing the activity of HNS (both in its native context in HuR and attached to a reporter protein) to direct nucleocytoplasmic shuttling after heat shock should provide insights that distinguish between these possibilities.
Although we had hoped that investigating HuR interactions with RNA after heat shock might reveal greater binding correlated with stabilization of ARE-containing messages, we could not detect associations between HuR and cytoplasmic mRNA in heat-shocked cells (Figs. 3 and 4). It may be that the reported stabilization of certain short-lived mRNAs (2) is simply because of their sequestration in the nucleus after heat shock. In the future, it will be necessary to analyze HuR interactions with specific ARE-containing mRNAs, in particular those whose nuclear export and translation continue after heat shock, as has been done for hnRNP D (26).
Acknowledgments
We thank Linda Green of the Hybridoma Core Facility at the University of Florida for producing the 3A2 monoclonal antibody. We are grateful to Dr. S. Pinol Roma for many helpful discussions concerning the in vivo crosslinking experiments and for providing the anti-hnRNP A1 antibody, 4B10. We thank Wojciech Przylecki for assistance in preliminary experiments, Drs. P. Silver, K. Tycowski, and Y. Huang for comments on the manuscript, Dr. G. Dreyfuss for providing the 4F4 antibody, and Dr. I. Laird-Offringa for providing HuB, HuC, and HuD proteins. This work was supported by grant P01CA16038 from the National Institutes of Health. C.M.B. and A.E. were supported by National Institutes of Health Predoctoral Training Program GM07223. J. Steitz is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- ARE
AU-rich element
- RRM
RNA recognition motif
- hnRNP
heterogeneous nuclear ribonucleoprotein
Note Added in Proof
After submission of this manuscript, Kedersha et al. (48) reported that TIA-1 and TIAR, two other RNA-binding proteins, colocalize with poly(A)+ RNA in cytoplasmic stress granules (SGs) after heat shock. Despite the fact that we are not able to detect any interaction between HuR and cytoplasmic poly(A)+ RNA after heat shock, we cannot exclude the possibility that the foci seen in Fig. 2 4 and Fig. 2 6 are SGs; this is under investigation.
References
- 1.Chen C Y, Shyu A B. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 2.Andrews G K, Harding M A, Calvet J P, Adamson E D. Mol Cell Biol. 1987;7:3452–3458. doi: 10.1128/mcb.7.10.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorospe M, Wang X, Holbrook N J. Mol Cell Biol. 1998;18:1400–1407. doi: 10.1128/mcb.18.3.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levy N S, Chung S, Furneaux H, Levy A P. J Biol Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- 5.Lindsten T, June C H, Ledbetter J A, Stella G, Thompson C B. Science. 1989;244:339–343. doi: 10.1126/science.2540528. [DOI] [PubMed] [Google Scholar]
- 6.Ming X F, Kaiser M, Moroni C. EMBO J. 1998;17:6039–6048. doi: 10.1093/emboj/17.20.6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirsch H H, Nair A P, Backenstoss V, Moroni C. J Biol Chem. 1995;270:20629–20635. doi: 10.1074/jbc.270.35.20629. [DOI] [PubMed] [Google Scholar]
- 8.Ma W J, Cheng S, Campbell C, Wright A, Furneaux H. J Biol Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. [DOI] [PubMed] [Google Scholar]
- 9.Good P J. Proc Natl Acad Sci USA. 1995;92:4557–4561. doi: 10.1073/pnas.92.10.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jain R G, Andrews L G, McGowan K M, Pekala P H, Keene J D. Mol Cell Biol. 1997;17:954–962. doi: 10.1128/mcb.17.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akamatsu W, Okano H J, Osumi N, Inoue T, Nakamura S, Sakakibara S, Miura M, Matsuo N, Darnell R B, Okano H. Proc Natl Acad Sci USA. 1999;96:9885–9890. doi: 10.1073/pnas.96.17.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung S, Eckrich M, Perrone-Bizzozero N, Kohn D T, Furneaux H. J Biol Chem. 1997;272:6593–6598. doi: 10.1074/jbc.272.10.6593. [DOI] [PubMed] [Google Scholar]
- 13.Darnell R B. Proc Natl Acad Sci USA. 1996;93:4529–4536. doi: 10.1073/pnas.93.10.4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma W J, Chung S, Furneaux H. Nucleic Acids Res. 1997;25:3564–3569. doi: 10.1093/nar/25.18.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myer V E, Fan X C, Steitz J A. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan X C, Steitz J A. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng S S, Chen C Y, Xu N, Shyu A B. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan X C, Steitz J A. Proc Natl Acad Sci USA. 1998;95:15293–15298. doi: 10.1073/pnas.95.26.15293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michael W M, Choi M, Dreyfuss G. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 20.Pinol-Roma S, Choi Y D, Matunis M J, Dreyfuss G. Genes Dev. 1988;2:215–227. doi: 10.1101/gad.2.2.215. [DOI] [PubMed] [Google Scholar]
- 21.Dempsey L A, Li M J, DePace A, Bray-Ward P, Maizels N. Genomics. 1998;49:378–384. doi: 10.1006/geno.1998.5237. [DOI] [PubMed] [Google Scholar]
- 22.Dempsey L A, Hanakahi L A, Maizels N. J Biol Chem. 1998;273:29224–29229. doi: 10.1074/jbc.273.44.29224. [DOI] [PubMed] [Google Scholar]
- 23.Swanson M S, Dreyfuss G. EMBO J. 1988;7:3519–3529. doi: 10.1002/j.1460-2075.1988.tb03228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W, Wagner B J, Ehrenman K, Schaefer A W, DeMaria C T, Crater D, DeHaven K, Long L, Brewer G. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loflin P, Chen C Y, Shyu A B. Genes Dev. 1999;13:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laroia G, Cuesta R, Brewer G, Schneider R J. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 27.Kiledjian M, Burd C, Portman D, Dreyfuss G. Structure and Function of hnRNP Proteins. Oxford: IRL; 1994. p. 127. [Google Scholar]
- 28.Pinol-Roma S, Dreyfuss G. Nature (London) 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 29.Anderson J T, Paddy M R, Swanson M S. Mol Cell Biol. 1993;13:6102–6113. doi: 10.1128/mcb.13.10.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frangioni J V, Neel B G. Anal Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- 31.Lerner E A, Lerner M R, Janeway C A, Jr, Steitz J A. Proc Natl Acad Sci USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith P R, Williams D G, Venables P J, Maini R N. J Immunol Methods. 1985;77:63–76. doi: 10.1016/0022-1759(85)90184-x. [DOI] [PubMed] [Google Scholar]
- 33.Watkins J L, Murphy R, Emtage J L, Wente S R. Proc Natl Acad Sci USA. 1998;95:6779–6784. doi: 10.1073/pnas.95.12.6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forrester W, Stutz F, Rosbash M, Wickens M. Genes Dev. 1992;6:1914–1926. doi: 10.1101/gad.6.10.1914. [DOI] [PubMed] [Google Scholar]
- 35.Pinol-Roma S, Adam S A, Choi Y D, Dreyfuss G. Methods Enzymol. 1989;180:410–418. doi: 10.1016/0076-6879(89)80114-4. [DOI] [PubMed] [Google Scholar]
- 36.Gallouzi I E, Parker F, Chebli K, Maurier F, Labourier E, Barlat I, Capony J P, Tocque B, Tazi J. Mol Cell Biol. 1998;18:3956–3965. doi: 10.1128/mcb.18.7.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao F B, Keene J D. J Cell Sci. 1996;109:579–589. doi: 10.1242/jcs.109.3.579. [DOI] [PubMed] [Google Scholar]
- 38.Ausubel F M, Brent R, Kingson R E, Moore D D, Seidan J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. Boston: Wiley; 1994. [Google Scholar]
- 39.Antic D, Keene J D. Am J Hum Genet. 1997;61:273–278. doi: 10.1086/514866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, Liang S, Tartakoff A M. EMBO J. 1996;15:6750–6757. [PMC free article] [PubMed] [Google Scholar]
- 41.Duncan R, Hershey J W. J Biol Chem. 1984;259:11882–11889. [PubMed] [Google Scholar]
- 42.DeMaria C T, Brewer G. J Biol Chem. 1996;271:12179–12184. doi: 10.1074/jbc.271.21.12179. [DOI] [PubMed] [Google Scholar]
- 43.Weighardt F, Cobianchi F, Cartegni L, Chiodi I, Villa A, Riva S, Biamonti G. J Cell Sci. 1999;112:1465–1476. doi: 10.1242/jcs.112.10.1465. [DOI] [PubMed] [Google Scholar]
- 44.Keene J D. Proc Natl Acad Sci USA. 1999;96:5–7. doi: 10.1073/pnas.96.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stutz F, Rosbash M. Genes Dev. 1998;12:3303–3319. doi: 10.1101/gad.12.21.3303. [DOI] [PubMed] [Google Scholar]
- 46.Krebber H, Taura T, Lee M S, Silver P A. Genes Dev. 1999;13:1994–2004. doi: 10.1101/gad.13.15.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fridell R A, Truant R, Thorne L, Benson R E, Cullen B R. J Cell Sci. 1997;110:1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- 48.Kedersha N L, Gupta M, Li W, Miller I, Anderson P. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]